Abstract
β-Defensins are cationic antimicrobial peptides expressed in epithelia. They exhibit antibacterial, antifungal, and antiviral properties. Defensins are a component of the innate immune response, and it has been proposed that they have a protective role in the oral cavity. Previous studies have shown that human β-defensin 1 (hBD-1) is constitutively expressed in oral epithelial cells but that expression varies between individuals. We tested the hypothesis that genetic variations in defensin peptide expression may be associated with opportunistic infections. This may be critical in the immunocompromised patient population, in which innate immune responses may have a relatively more important role. Oral Candida carriage status and the presence of six single-nucleotide polymorphisms (SNPs) in the DEFB1 gene encoding hBD-1 were evaluated in type I diabetic patients (n = 43) and nondiabetic controls (n = 50). Genomic DNA was obtained from buccal swabs. Portions of the DEFB1 gene were amplified, and each SNP was analyzed by a TaqMan assay, standardized with control DNA of known genotype. Candida carriage status was determined from unstimulated saliva on CHROMagar plating medium. A low level of Candida carriage was defined as ≤350 CFU/ml. A high level of Candida carriage was seen in 44% of the diabetic subjects but only in 28% of the nondiabetic controls (P < 0.05). C. albicans predominated; however, diabetic subjects, especially those with high levels of carriage, showed an increased proportion of Candida glabrata and C. tropicalis. There was a strong association between an SNP in the 5′ untranslated region (C→G at position −44) and Candida carriage in both groups. Among individuals in the diabetic population who had the SNP allele 2 (G), 58% had low CFU, while 6% had high CFU. The C→G SNP at position −44 is associated with low levels of Candida carriage. The resultant odd ratios are statistically significant for a protective effect (odd ratios, 25 for diabetic subjects and 8.5 for nondiabetic subjects). These results indicate that genetic variations in the DEFB1 gene encoding hBD-1 may have a major role in mediating and/or contributing to susceptibility to oral infection.
The body surface provides a protective barrier against mechanical and microbial insult. This microbial barrier may be especially significant in the mucosa of the oral cavity and other mucosae, where the potential for bacterial and fungal infections is great. Innate host defenses are a critical aspect of the maintenance of oral health. These defenses include multiple salivary factors, secreted antibodies, and epithelial products (40). One family of epithelial antimicrobial peptides is the β-defensins. The β-defensins are small cationic amphipathic peptides (30 to 48 amino acids) with a signature motif of six cysteines and a beta-sheet structure stabilized by three disulfide bonds. They exhibit a broad spectrum of activity against gram-positive and gram-negative bacteria, fungal species, and viruses (for reviews, see reference 3, 8, and 22). There are 28 human β-defensins (33); however, expression of only four β-defensins has been characterized. The genes encoding these four peptides and α-defensins are located on chromosome 8 (8p 22-23) (23). These include human β-defensin 1 (hBD-1), which is constitutively expressed; hBD-2 and -3, which are inducibly expressed by microbial cell components within the oral cavity, skin, and internal epithelia; and hBD-4, which is primarily expressed in the testes (9). The expression of hBD-1 varies between individuals (20), while the expression of hBD-2 is enhanced in response to infection and inflammation (14, 19, 25).
The finding that hBD-1 is variably expressed and that other β-defensins may be differentially induced led to the hypothesis that individuals may exhibit increased susceptibility to infection, with this increased susceptibility dependent upon on genetically determined expression of the DEFB genes encoding β-defensins. Candidiasis is the most common oral fungal infection in humans. Candida albicans and related species are opportunistic pathogens that produce infections ranging from non-life-threatening mucocutaneous disturbances to invasive diseases involving almost all organs. The frequency of serious mycological infections is increasing worldwide and is due to a “multiplicity of predisposing factors which facilitate the conversion of the commensal Candida to a parasitic existence” (30, 31). The increase in candidal infection is associated with and may be an indication of immune deficiency, as seen in the high proportion of cases of oral pharyngeal candidiasis among human immunodeficiency virus-positive patients and those with AIDS (11). The progression from colonization to mucosal candidal infection is dependent upon host defenses (4) and the ability of the Candida organism to circumvent the host surveillance mechanism. Oral candidiasis is the result of yeast overgrowth and penetration of the epithelial mucosal protective barrier and is evident in patients with various conditions that exhibit immunosuppression including diabetes. Diabetic patients have an increased susceptibility to Candida and other fungal infections (1, 37) and, specifically, increased rates of oral candidal infections (15, 21, 35, 42, 43). The etiology for this occurrence has only been hypothesized and has been related to metabolic changes, insulin levels, medications, neutrophil suppression, and behavioral variables.
Nevertheless, not all diabetic patients experience fungal infections or exhibit signs and symptoms associated with fungal infection (5). This phenomenon is also evident in the nondiabetic population, in which some individuals have a greater propensity for fungal infection than others. This variability in pathogenic response may be related to variations in host innate immune defenses such as histatins (36), lactoferrins and lysozyme (32), and defensins. The development of candidal infection is in part determined by the mechanical adhesion of the Candida cells to the epithelium and subsequent colonization. This adhesion abrogates the normal clearance mechanisms of the host. The β-defensins are expressed by epithelial cells and convey potent antimicrobial properties. The fungicidal activity of β-defensin is based upon direct contact and disruption of the fungal cell membrane. Therefore, alterations in peptide composition and expression and the resultant functions could be significant determinants in Candida-based pathogenesis. In this report we show an association of Candida carriage with a single-nucleotide polymorphism (SNP) in the DEFB1 gene encoding the constitutively expressed hBD-1 peptide.
MATERIALS AND METHODS
Subjects.
Forty-three subjects with type I insulin-dependent diabetes mellitus (IDDM) from the Diabetology Clinic, Harborview Medical Center, Seattle, Wash., were enrolled in the study. The inclusion and exclusion criteria for this study required the subjects to be at least 18 years old and not to have taken any antibiotic or antifungal medications for the last 10 weeks. Female subjects could not be pregnant at the time of the study. Subjects needed to produce 3 to 5 ml of unstimulated saliva in 15 min (i.e., the subjects could not be xerostomic). The subjects were all patients of record of the clinic, and all had a primary diagnosis of type I IDDM. The subjects were examined for oral Candida yeast infection, and a thorough head-and-neck examination was performed. Buccal cells were collected with a cytobrush swab, and 5 ml of unstimulated saliva was collected for determination of Candida carriage status and Candida species determination. In patients with dentures, the dentures were removed for saliva collection, but the surfaces of the dentures were not sampled. The subjects were questioned about prior fungal infections and a history of antibiotic and antifungal usage. Laboratory data, prescribed medications, and pertinent medical information were recorded.
Fifty nondiabetic subjects were recruited from the Basic Assessment Clinic, Department of Oral Medicine, School of Dentistry, University of Washington, Seattle. All subjects were prospective University of Washington School of Dentistry patients and were scheduled to undergo an initial dental examination. All subjects enrolled in the study were nondiabetic, were at least 18 years of age, and had not been on antibiotics or antifungal medications for at least 10 weeks prior to sampling. Recruitment and testing for all subjects were in compliance with the Human Subjects Institutional Review Board of the University of Washington, Seattle.
Procedures. (i) Clinical examination.
An oral examination and a head-and-neck examination were performed and included determination of status of dentition or denture-bearing surfaces, determination of soft tissue morphology and detection of lesions or other occult signs of pathology, determination of the morphology and surface characteristics of the tongue, palpation of masticatory muscle, and examination of facial and cervical lymph nodes. External signs and lesions (angular chelitis) were also noted and recorded. A medical history, medication history, and present laboratory data were also recorded. The subjects were questioned about recent dental treatment and oral complaints. All emergent care was referred to the appropriate dental specialty. One observer (R.J.J.) collected the oral examination and history data; therefore, intraobserver error was not considered.
(ii) Candida carriage determination.
Candida carriage status was determined from unstimulated whole saliva. Subjects were requested to rinse twice with 25 ml of sterile water and expectorate. They were then instructed to allow saliva to collect and then expectorate into a sterile 50-ml tube until 3 to 5 ml of saliva was collected. Non-iodet Nonidet P-40 (0.5% [by volume]; Sigma Inc., St. Louis, Mo.) was added to each sample as an antiviral agent. All samples were stored on ice until they could be plated later the same day. A total of 200 μl of saliva was plated on CHROMagar medium and incubated at 30°C for 48 h. The numbers of CFU were calculated by automated analysis with the Stratagene Eagle Eye system, and the numbers of CFU per milliliter of saliva were determined. Designation of the high-level and low-level cutoff at 350 CFU/ml was empirically determined. Species of Candida were identified colorimetrically and morphologically (29). The CHROMagar medium has a specific color for each Candida species (C. albicans is green, C. tropicalis is blue or black, and C. glabrata is smooth pink). This unique feature allows the rapid, specific, and sensitive identification of species (29). Residual saliva was stored in glycerol at −70°C for further analysis.
(iii) Genomic DNA.
Genomic DNA was extracted from buccal cells collected by sampling with a cytobrush by using a DNA extraction kit (Epicentre, Madison, Wis.). The DNA was purified with the Qiagen QIA Quick kit (Qiagen Inc., Valencia, Calif.). The genomic DNA was stored at −70°C until sequencing or SNP analysis was performed.
(iv) SNP analysis.
Polymorphic sites in genomic DNA were analyzed by high-throughput site-specific TaqMan assays. The primers, probes, and assay conditions were developed at Albany Molecular Research Inc., Bothell, Wash. Molecular probes were designed to specifically determine SNP sites that we had previously identified by conventional sequencing (18) and listed in Table 1. The six sites that were assayed included sites 322 (−379), 668 (−44), 692 (−20), 1654 (V37I), 1741(C66S), 1836 (polyadenylation site). Numbering is in accordance with GenBank accession nos. U50930 and U50931; the information in parentheses refers to the position relative to the beginning of the protein sequence (ATG) start site or to the altered amino acid. The primers and PCR conditions used for amplification by the TaqMan assays are listed in Table 1. The 7700 sequence detection system (Perkin-Elmer, Foster City, Calif.), a 96-well, laser and fiber optic reading instrument, was used for high-throughput analysis of genomic DNA samples. Fluorophores VIC (a proprietary fluorescent reporter dye) and 6-carboxyfluorescein (FAM) and a quencher (6-carboxy-N,N,N′,N′-tetramethylrhodamine [TAMRA]) were incorporated into the specific wild-type VIC and mutant FAM probes for SNP determination. The TaqMan 5′ nuclease assay is a homogeneous technique for determination of nucleotide mutations or polymorphisms (34). This assay is based upon the binding of a fluorescence-labeled probe to the specific single-stranded DNA segment that contains the SNP site. The fluorescent probe is nonfluorescing when it is in close proximately to the quencher (Förester resonance energy transfer). When the tag (probe) is freed from the primer, a characteristic fluorescent signal is detectable and the quenching effects are negated. The use of an exonuclease in the amplification reaction releases the fluorescent label if binding occurs, and the resultant signal is determined as a function of the allele sequence.
TABLE 1.
Primers, probes, and conditions for hBD-1 SNP TaqMan assaysa
| SNP and primers and probesb | Sequencec | Tm (°C) | PCR conditions; amplican size (Tm) |
|---|---|---|---|
| SNP 322 (−379) (T→A) | |||
| 322 F primer | 5′-CAAGGGAAGAGGGTGAAGTTTGAG-3′ | 60.2 | 50°C for 2 min. 95°C for 10 min. and 40 cycles of 95°C for 15 s and 64°C for 1 min; 174 bp (78.2°C) |
| 322 R primer | 5′-TGATGGGGTTTCTGGAACAGGC-3′ | 59.6 | |
| 322 WT probe | 5′-CAGAGCTTCCCTGTGGCTCTCC-3′ | 67.0 | |
| 322 MUT probe | 5′-CAGAGCTTCCCAGTGGCTCTCC-3′ | 67.0 | |
| SNP 668 (−44) (C→G) | |||
| 668 F primer | 5′-GACGAGGTTGTGCAATCCACCAG-3′ | 60.2 | 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 64°C for 1 min; 156 bp (77.9°C) |
| 668 R primer | 5′-GCAGAAGGTAGGAAGTTCTCATGGCG-3′ | 62.2 | |
| 668 WT probe | 5′-AGCCAGCCTCTCCCCAGTTCC-3′ | 68.3 | |
| 668 MUT probe | 5′-AGCCAGCGTCTCCCCAGTTCC-3′ | 66.3 | |
| SNP 692 (−20) (A→G) | |||
| 692 F primer | 5′-GACGAGGTTGTGCAATCCACCA-3′ | 60.2 | 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 62°C for 1 min, 156 bp (77.9°C) |
| 692 R primer | 5′-GCAGAAGGTAGGAAGTTCTCAT-3′ | 62.2 | |
| 692 WT probe | 5′-TGGCAGGCAACACTCAGGATTTC-3′ | 67.8 | |
| 692 MUT probe | 5′-TGGCAGGCAACACCCAGGAT-3′ | 68.3 | |
| SNP 1654 (G→A) V371 | |||
| 1654 F primer | 5′-CGTTGCAGCTACAAGCCATGAGTC-3′ | 59.8 | 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 62°C for 1 min; 157 bp (75.2°C) |
| 1654 R primer | 5′-TGAATTTTGGTAAAGATCGGGCAGG-3′ | 61.1 | |
| 1654 WT probe | 5′-CAATTGCGTCAGCAGTGGAGG-3′ | 66.3 | |
| 1654 MUT probe | 5′-CAATTGCATCAGCAGTGGAGGG-3′ | 67.2 | |
| SNP 1741 (T→A) C66S | |||
| 1741 F primer | 5′-TTCTGCCTGCCCGATCTTTACC-3′ | 59.1 | 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 62°C for 1 min; 107 bp (73.5°C) |
| 1741 R primer | 5′-TTTCACTTCTGCGTCATTTCTTCTGG-3′ | 59.7 | |
| 1741 WT probe | 5′-CCAAGTGCTGCAAGTGAGCTGAG-3′ | 66.8 | |
| 1741 MUT probe | 5′-CCAAGTGCAGCAAGTGAGCTGAG-3′ | 66.8 | |
| SNP 1836 (A→G) poly A site | |||
| 1836 F primer | 5′-GCTGAGAGTGACCAGAAGAAATGACG-3′ | 60.4 | 50°C for 2 min, 95°C for 10 min, 40 cycles of 95°C for 15 s and 62°C for 1 min; 164 bp (71.5°C) |
| 1836 R primer | 5′-AACCCATGCACCCTAACCCCC-3′ | 60.5 | |
| 1836 WT probe | 5′-CTTTTGAAGTATACCTCCTTTGGGCC-3′ | 65.0 | |
| 1836 MUT probe | 5′-CTTTTGAAGTGTACCTCCTTTGGGC-3′ | 65.2 |
Abbreviations: Tm, melting temperature; WT, wild type; MUT, mutant.
Probes for the wild type have a 5′ VIC label (proprietary fluorophore) and 3′ TAMRA quencher probes for the mutants have a 5′- FAM label and 3′ TAMRA quencher. The information in parentheses represents the position relative to the beginning of the protein sequence start site or to the altered amino acid.
SNP sites are indicated in boldface in the sequences.
Statistical analysis.
All single-nucleotide data were evaluated for Hardy-Weinberg equilibrium. Chi-square analysis for observed and expected occurrences of all SNPs was performed, with a P value of <0.05 indicating significance. The Fisher exact test with P values of <0.05 was used for determination of statistical significance of the demographic data. Relative risk analysis and logistic and linear regression analyses were calculated for determination of odds ratios and the influence of confounding variables on the outcome, respectively. Nonparametric chi-square analysis for the significance of Candida carriage and species differentiation was calculated, with a P value of <0.05 indicating significance.
RESULTS
Demographics.
The study enrolled 43 subjects with type I IDDM and 50 nondiabetic subjects. Demographic data are summarized in Table 2. The ratio of men to women for the diabetic population was 60:40, and that for the nondiabetic population was 40:60. The mean age for both groups was approximately the same. The diabetic group had a larger proportion of smokers (46%) compared to the proportion among the nondiabetic subjects (22%) and a larger proportion of subjects with dentures (30%) compared to the proportion among the nondiabetic subjects (2%). Each of these features was statistically significant (P < 0.05). The ethnicities of the subjects were not closely matched because selection of subjects was based upon the availability and the efficient utilization of clinical resources. The control group had a significantly larger proportion of Caucasians (66%) than the diabetic group (28%), while the diabetic group had a significantly larger proportion of African Americans (34%) than the nondiabetic group (4%).
TABLE 2.
Demographics and Candida carriage in patients with type I IDDM and nondiabeticsa
| Characteristic | Diabetic subjects with ≤350 CFU/ml | Diabetic subjects with >350 CFU/ml | Totals for diabetic subjects | Nondiabetic subjects with ≤350 CFU/ml | Nondiabetic subjects with >350 CFU/ml | Totals for nondiabetic subjects |
|---|---|---|---|---|---|---|
| Gender (no. of men, no. of women) | 14, 10 | 12, 7 | 26, 17 | 12, 24 | 4, 9 | 17, 33 |
| Age (yr) | ||||||
| Mean | 50.1 | 47.6 | 49 | 46.5 | 42.3 | 45.4 |
| Median | 51 | 47.0 | 50 | 49 | 43.5 | 45.5 |
| Range | 18-82 | 30-59 | 18-82 | 18-77 | 20-65 | 18-77 |
| No. of subjects who are smokers | 9 | 11 | 20 | 4 | 7 | 11 |
| No. of subjects with dentures | 6 | 7 | 13 | 0 | 1 | 1 |
| Abnormal oral examination | 6 | 7 | 13 | 1 | 3 | 4 |
| Previous antifungal therapy | 3 | 4 | 7 | 0 | 0 | 0 |
| History of fungal infection | 3 | 4 | 7 | 0 | 1 | 1 |
| Oral symptoms | 2 | 2 | 5 | 1 | 0 | 1 |
| No. of subjects by race: | ||||||
| Caucasian | 7 | 5 | 12 | 21 | 12 | 33 |
| African American | 7 | 8 | 15 | 2 | 0 | 2 |
| Hispanic | 2 | 1 | 3 | 6 | 1 | 7 |
| Japanese | 1 | 0 | 1 | 3 | 0 | 3 |
| Chinese | 1 | 0 | 1 | 3 | 0 | 3 |
| Filipino | 3 | 0 | 3 | 1 | 1 | 2 |
| Cambodian | 0 | 4 | 4 | 0 | 0 | 0 |
| Ethiopian | 3 | 0 | 3 | 0 | 0 | 0 |
| Indonesian | 0 | 1 | 1 | 0 | 0 | 0 |
| Other | 0 | 0 | 0 | 0 | 0 | 0 |
Data are for 43 subjects with type I IDDM and 50 nondiabetics.
The diabetic subjects reported having a higher incidence of prior fungal infections, a higher level of prior use of antifungal medications, and a larger number of oral symptoms (from self-reports). The diabetic subjects also had a larger number of oral examinations positive for Candida infections. The examinations revealed the following pathologies associated with Candida: median rhomboid glossitis, atrophic tongue papilla, denture stomatitis, angular chelitis, and pseudomembranous candidiasis. Other oral abnormalities such as lichen planus, gingival pathology (hyperplasia or gingivitis), aphthous ulcers, fissured tongue and/or parotid enlargement, and oral squamous cell carcinoma (secondary to betel nut use) were also detected.
Candida carriage status in diabetic and nondiabetic subjects.
The diabetic population had a higher level of Candida carriage (mean, 1,349 ± 246 CFU/ml) than the nondiabetic subjects (mean, 471± 134 CFU/ml) (P < 0.05). The proportion of diabetic subjects with >350 CFU/ml was 44%, and that for nondiabetic subjects was 28%. Only 5 (12%) of diabetic subjects had no detectable CFU, while 15 (30%) nondiabetic subjects had no detectable CFU.
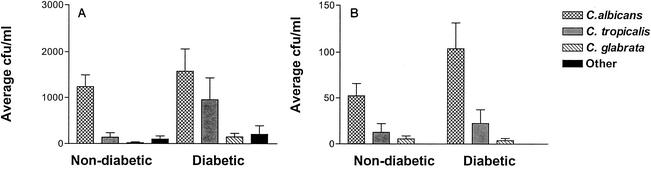
The Candida species distributions for both groups are shown in Fig. 1A and B. The difference in species identified between the diabetic and nondiabetic subjects was statistically significant (P < 0.05), with more Candida species other than C. albicans detected in the diabetic population. Although only 16% of the diabetic subjects stated a history of fungal infection and antifungal medication usage, the larger numbers of candidal species other than C. albicans might be attributed to nonreporting of previous medication usage. Colonization and species variation did not increase with respect to age in our study (data not shown), as has been reported previously (24). Among the demographic variables, only smoking had a statistically significant confounding effect. Smoking had an associative effect with elevated numbers of CFU in both groups. The effects of other variables including ethnicity, the presence of dentures, and age were not statistically significant.
FIG. 1.
Candida species distribution in type I diabetic subjects and nondiabetic subjects with high levels (>350 CFU/ml) (A) and low levels (<350 CFU/ml) (B) of Candida carriage. (A) The mean numbers of CFU per milliliter are indicated. Note that the diabetic group had significantly larger numbers of CFU of C. tropicalis and C. glabrata per milliliter than the nondiabetic group (P < 0.01). The mean numbers of CFU of C. albicans per milliliter between the diabetic subjects and the nondiabetic subjects were not significantly different. (B) The mean numbers of CFU per milliliter are indicated on an expanded scale. The C. albicans distribution in individuals with low levels of carriage differed between diabetic subjects and nondiabetic subjects (P < 0.05).
SNPs in hBD-1 in diabetic and nondiabetic subjects.
The TaqMan allelic discrimination 5′ nuclease assay was used to determine the SNPs previously reported in hBD-1 (7, 18, 39). The SNP sites included 322 (A→T at position −379), 668 (C→G at position −44), 692 (G→A at position −20), 1654 (G→A; V37I), 1741 (T→A; C66S), and 1836 (a polyadenylation site; A→G). Site 322 (−379) is located in the promoter region of hBD-1, and its frequency varies between different ethnic groups (18). Sites 668 (−44) and 692 (−20) are both located in the 5′ untranslated region of hBD-1. The changes at sites 1654 and 1741 are nonsynonymous changes in the coding region of exon 2 and have been reported earlier but occur at very low frequencies (18, 39). The resultant changes of V37I and C66S may be associated with changes in the peptide structure and function, and this was our rationale for developing the high-throughput assay for these nonsynonymous changes. However, it has recently been reported that the C66S change in the amino acid did not alter the peptide structure or function in antimicrobial assays (6). Site 1836 was also an interesting site for an SNP because it is one of two possible polyadenylation sites and may have an influence on transcription and/or translation.
The SNP frequencies for each group are shown in Table 3. The diabetic group had a higher frequency of SNPs at sites 1654 and 1836 than the nondiabetic group. We previously reported that the frequency of SNPs at site 1654 is 0 in the Coriell Northern European sample set and 0.10 in the Coriell African American sample set; in these two sample sets the frequencies of SNPs at site 1836 are 0.05 and 0.20, respectively (18). Thus, the difference in SNP frequency observed here is most likely due to ethnic differences between the diabetic and nondiabetic groups because of the greater representation of African Americans in the diabetic group (Table 2).
TABLE 3.
SNP analysis with diabetics and nondiabetics by Candida carriage status
| SNP site | Nondiabetic subjects (n = 36) with low-level carriage
|
Nondiabetic subjects (n = 14) with high-level carriage
|
% Nondiabetics with SNP | Diabetics (n = 24) with low-level carriage
|
Diabetics (n = 19) with high-level carriage
|
% Diabetics with SNP | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of subjects with genotypea:
|
SNP frequency | % Subjects with SNP | No. of subjects with genotypea:
|
SNP frequency | % Subjects with SNP | No. of subjects with genotypea:
|
SNP frequency | % Subjects with SNP | No. of subjects with genotypea:
|
SNP frequency | % Subjects with SNP | |||||||||||
| WT | Het | MUT | WT | Het | MUT | WT | Het | MUT | WT | Het | MUT | |||||||||||
| 322 (−379)b | 12 | 17 | 7 | 0.43 | 67 | 2 | 9 | 3 | 0.54 | 86 | 72 | 11 | 10 | 3 | 0.33 | 54 | 4 | 10 | 5 | 0.53 | 79 | 65 |
| 668 (−44)d | 26 | 8 | 2 | 0.17 | 28c | 13 | 1 | 0 | 0.04 | 7c | 22 | 10 | 13 | 1 | 0.31 | 58c | 18 | 1 | 0 | 0.03 | 5c | 35 |
| 692 (−20) | 7 | 15 | 14 | 0.60 | 81 | 1 | 10 | 3 | 0.57 | 93 | 84e | 3 | 11 | 10 | 0.65 | 88 | 3 | 10 | 6 | 0.58 | 84 | 86 |
| 1654 (V37I) | 35 | 1 | 0 | 0.01 | 3 | 14 | 0 | 0 | 0 | 0 | 2e | 22 | 2 | 0 | 0.04 | 8 | 17 | 2 | 0 | 0.05 | 11 | 9e |
| 1741 (C66S) | 36 | 0 | 0 | 0 | 0 | 14 | 0 | 0 | 0 | 0 | 0 | 24 | 0 | 0 | 0 | 0 | 19 | 0 | 0 | 0 | 0 | 0 |
| 1836 [poly(A)] | 34 | 2 | 0 | 0.03 | 6 | 13 | 1 | 0 | 0.04 | 7 | 6e | 19f | 3 | 1 | 0.11 | 17 | 14 | 4 | 1 | 0.16 | 26 | 21e |
WT, wild type; Het, heterogeneous; MUT, mutant.
The information in parentheses represents the position relative to the beginning of the protein sequence start site or to the altered amino acid.
Significant difference between high- and low-level Candida carriage groups.
Boldface numbers indicate data for the site for which significant differences in occurrence between low and high Candida carriage were observed.
The difference in frequencies of SNPs at positions 1654 and 1836 is due to ethnic differences between the diabetic and nondiabetic groups and is not associated with Candida carriage.
One genotype for this group is unknown.
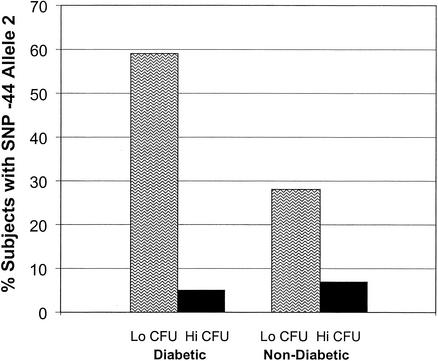
The relationship of Candida CFU, the SNP site, and the resultant alleles are also indicated in Table 3. The SNP at site 668 (C→G at position −44) exhibits a significant association with Candida carriage status in both groups (Fig. 2). The SNP 668 (−44) allele 1 was defined as a C, and 668 allele 2 was defined as a G. The frequencies of SNPs in allele 2 in diabetic subjects and nondiabetic subjects with low CFU were 58 and 28%, respectively (Fig. 2). The frequencies of SNP allele 2 in the diabetic and nondiabetic subjects with high CFU were 6 and 7%, respectively. The resultant odds ratios for relative risk are 25 and 8.5, respectively, with 95% confidence intervals of 2.8 (lower) and 221 (upper) (chi-squared = 0.007) and 0.999 (lower) and 73.5 (upper) (chi-squared = 0.114), respectively. Thus, the SNP at site 668 (−44) in allele 2 is associated with a protective effect in the diabetic and nondiabetic populations. Diabetic individuals carrying SNP allele 2 (either one or two copies) are 25 times more likely to not develop high levels of Candida carriage than individuals carrying only allele 1, and nondiabetic individuals carrying SNP allele 2 are 8.5 times more likely to not have high levels of Candida carriage than individuals carrying only allele 1. The other SNP sites were not associated with changes in the numbers of Candida CFU status and were not linked to the SNP at site −44. The changes seen in the SNPs at site −44 were independent of age, sex, smoking, dentures, previous fungal infections, and antifungal therapies.
FIG. 2.
Frequency of SNP at site −44 (C224G) in type I diabetic subjects and nondiabetic controls with high and low levels of Candida carriage. The groups with the low levels of Candida carriage had a high frequency of SNP 668 (−44), suggesting an association of this SNP with protection from Candida carriage (P < 0.01).
DISCUSSION
Candidiasis is a common oral fungal infection whose effects range from non-life-threatening mucocutaneous lesions to invasive disease. In the present study we examined the association of Candida carriage status and genetic polymorphisms in the DEFB1 gene encoding constitutively expressed hBD-1. The β-defensins are expressed by epithelial cells and convey potent antimicrobial properties. The antimicrobial action of β-defensins is by pore formation in the microbial cell membrane and necessitates direct contact between the peptides and microbe and the resultant membrane disruption (27). The major mechanism for candidal invasion is by adhesion and invasion of the host epithelial surface. Thus, a plausible role for the β-defensin peptides in the innate immune defense is to destroy fungal species when adhesion to the cell surface occurs. This mechanism may be an effective backup system in the event of failure of the antiadhesion or agglutination properties of saliva or the fungicidal actions of histatins (36). Our results concur with those of others that rate of Candida carriage is greater in diabetic subjects than in nondiabetic subjects (12, 13, 15, 17, 35). In our study the mean level of candidal carriage in diabetic subjects was somewhat lower than that detected by other investigators but may reflect differences in collection, sample handling, and/or colony counting. Smoking was the only confounding variable that had a statistically significant relationship with Candida carriage status. Smoking was associated with an increase in the level of candidal carriage in both diabetic subjects and controls. Other variables (age, gender, dentures) were tested by logistic and linear regression analyses for confounding effects but were found to not exhibit significant effects. Although dentures have been associated with Candida infection (4), this was not observed in this population, in which salivary Candida carriage rather than denture-associated Candida carriage was measured.
A statistically significant difference in the proportions of the candidal species carried between diabetic and nondiabetic subjects with high levels of Candida carriage was seen. The mean values for the numbers of CFU of C. glabrata and C. tropicalis per milliliter were significantly greater (P < 0.05) in the diabetic group, whereas the mean values for C. albicans were not statistically different between groups. This observation may have clinical significance. Several studies have shown that non-C. albicans species are less sensitive to certain antifungals and, consequently, are more difficult to treat. This can contribute to a greater risk for systemic infection (10, 16, 28, 42). Although only 16% of the diabetic subjects stated a history of fungal infection and antifungal medication usage, the larger numbers of other candidal species might be attributed to the unreported use of antifungal medications. It has been shown that prior antifungal medication can alter the Candida species distribution, with resistant strains becoming more prevalent (41). This shift in species was exaggerated in the diabetic population and warrants further investigation.
A single SNP in the DEFB1 gene encoding hBD-1 showed an association with Candida carriage. This SNP at position −44 (C→G) was associated with a protective effect in both groups. The resultant odds ratios of risk were calculated to be 25 and 8.5 for diabetic subjects and nondiabetic subjects with Candida carriage levels of ≤350 CFU/ml, respectively. This protective effect means that individuals carrying either one or two copies of the SNP allele 2 (G) are 25 times more likely (or 8.5 times more likely for the nondiabetic group) of not having elevated levels of Candida carriage compared to the likelihood for those individuals with only SNP allele 1 (C) at this position. The location of the SNP is in the 5′ untranslated region of hBD-1, a region that is highly conserved in other primate species The resultant change does not confer a change in the amino acid composition of the peptide but may be associated with a variation in translation or transcription of hBD-1 or another linked gene.
We have seen in our work and other investigators have found (20, 25) that hBD-1 expression varies between individuals, even though this is considered to be a constitutively expressed peptide (38). The variability of expression may be associated with changes in the portion of the gene with the SNP. An alternative possibility is that the SNP is in linkage disequilibrium with another gene, perhaps another β-defensin or an α-defensin, that has a direct effect on Candida survival. The DEFB1 gene, which encoded the hBD-1 gene, is located on chromosome 8p 22-23 in a cluster of the α-defensin genes, which are more telomeric than the other β-defensin genes (23). Thus, the effects of this SNP may be linked to the neighboring α- or β-defensins. An SNP in the coding region of hBD-1 (position 1654; V37I) was shown to be associated with chronic obstructive pulmonary disease (26); however, the SNP at position 1654 showed no association with Candida carriage in our study.
The epithelial barrier and the associated antimicrobial peptides, in concert with salivary flow and salivary proteins, masticatory forces, oral hygiene, and other innate host defense mechanisms, play pivotal roles in maintaining the commensal behavior of candidal organisms and preventing them from becoming opportunistic pathogens. We have shown that an SNP in the gene for an epithelial antimicrobial peptide, hBD-1, is associated with a protective effect against Candida carriage. With the increased rates of fungal infections and the increased numbers of resistant fungal species that are occurring as a result of nonjudicial antifungal pharmacotherapy, susceptibility to candidal carriage by populations at risk needs to be examined. We provide here a marker that may be useful for this purpose. Although this SNP may be only one of several factors that contribute to susceptibility to candidal infection, it could prove to be a valuable marker for assessment of risk for Candida infection.
Acknowledgments
This study was supported by U.S. Public Health Service grants 1 R41 DE13448-01A1, P60 DE97002, and K16 DE00161 from the National Institute for Dental and Craniofacial Research.
We gratefully acknowledge useful discussions with Deborah Nickerson and offer our sincere gratitude to the staff of the Diabetology Clinic and the Madison Clinic at Harborview Medical Center, Seattle, Wash., especially Kathleen Givens, Scott Weigle, Carol Glenn, and M. Hooton, for assistance and support. We also express our appreciation to Dwight Baker, Albany Molecular Research, for helpful discussions and support.
REFERENCES
- 1.Andriole, V. T. 1999. The 1998 Garrod lecture. Current and future antifungal therapy: new targets for antifungal agents. J. Antimicrob. Chemother. 44:151-162. [DOI] [PubMed] [Google Scholar]
- 2.Bensch, K. W., M. Raida, H. J. Magert, K.-P. Schulz, and W. G. Forssmann. 1995. hBD 1: a novel beta defensin from human plasma. FEBS Lett. 368:331-335. [DOI] [PubMed] [Google Scholar]
- 3.Boman, H. G. 1998. Gene encoded peptide antibiotics and the concept of innate immunity: an update review. Scand. J. Immunol. 48:15-25. [DOI] [PubMed] [Google Scholar]
- 4.Cannon, R. D., and W. L. Chaffin. 1999. Oral colonization by Candida albicans. Crit. Rev. Oral Biol. Med. 10:359-383. [DOI] [PubMed] [Google Scholar]
- 5.Cannon, R. D., A. R. Holmes, A. B. Mason, and B. C. Monk. 1995. Oral Candida: clearance, colonization, or candidiasis? J. Dent. Res. 74:1152-1161. [DOI] [PubMed] [Google Scholar]
- 6.Circo, R., B. Skerlavaj, R. Gennaro, A. Amoroso, and M. Zanetti. 2002. Structural and functional characterization of hBD-1(Ser35), a peptide deduced from a DEFB1 polymorphism. Biochem. Biophys. Res. Commun. 293:586-592. [DOI] [PubMed] [Google Scholar]
- 7.Dork, T. S. M. 1998. Polymorphisms of the human B-defensin gene. Mol. Cell. Probes 12:171-173. [DOI] [PubMed] [Google Scholar]
- 8.Ganz, T., and R. I. Lehrer. 1998. Antimicrobial peptides of vertebrates. Curr. Opin. Immunol. 10:41-44. [DOI] [PubMed] [Google Scholar]
- 9.Garcia, J. R., A. Krause, S. Schulz, F. J. Rodriguez-Jimenez, E. Kluver, K. Adermann, U. Forssmann, A. Frimpong-Boateng, R. Bals, and W. G. Forssmann. 2001. Human beta-defensin 4: a novel inducible peptide with a specific salt-sensitive spectrum of antimicrobial activity. FASEB J. 15:1819-1821. [PubMed] [Google Scholar]
- 10.Goswami, R., V. Dadhwal, S. Tejaswi, K. Datta, A. Paul, R. N. Haricharan, U. Banerjee, and N. P. Kochupillai. 2000. Species-specific prevalence of vaginal candidiasis among patients with diabetes mellitus and its relation to their glycaemic status. J. Infect. 41:162-166. [DOI] [PubMed] [Google Scholar]
- 11.Greenspan, D., and J. S. Greenspan. 1996. HIV related oral disease. Lancet 348:729-733. [DOI] [PubMed] [Google Scholar]
- 12.Guggenheimer, J., P. A. Moore, K. Rossie, D. Myers, M. B. Mongelluzzo, H. M. Block, R. Weyant, and T. Orchard. 2000. Insulin-dependent diabetes mellitus and oral soft tissue pathologies. I. Prevalence and characteristics of non-candidal lesions. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 89:563-569. [DOI] [PubMed] [Google Scholar]
- 13.Guggenheimer, J., P. A. Moore, K. Rossie, D. Myers, M. B. Mongelluzzo, H. M. Block, R. Weyant, and T. Orchard. 2000. Insulin-dependent diabetes mellitus and oral soft tissue pathologies. II. Prevalence and characteristics of Candida and candidal lesions. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 89:570-576. [DOI] [PubMed] [Google Scholar]
- 14.Harder, J., J. Bartels, E. Christophers, and J. M. Schroder. 1997. A peptide antibiotic from human skin. Nature 387:861.. [DOI] [PubMed] [Google Scholar]
- 15.Hill, L. V., M. H. Tan, L. H. Pereira, and J. A. Embil. 1989. Association of oral candidiasis with diabetic control. J. Clin. Pathol. 42:502-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horowitz, S., R. Osborne, and F. DeGeorge. 1958. Caries experience in twins. Science 128:300-301. [DOI] [PubMed] [Google Scholar]
- 17.Hostetter, M. K. 1990. Handicaps to host defense. Effects of hyperglycemia on C3 and Candida albicans. Diabetes 39:271-275. [DOI] [PubMed] [Google Scholar]
- 18.Jurevic, R. J., P. Chrisman, L. Mancl, R. Livingston, and B. A. Dale. 2002. Single nucleotide polymorphisms in β-defensin genes in different ethnic populations. Genet. Test. 6:261-269. [DOI] [PubMed]
- 19.Krisanaprakornkit, K., J. R. Kimball, A. Weinberg, R. P. Darveau, B. W. Bainbridge, and B. A. Dale. 2000. Inducible expression of human β-defensin-2 by Fusobacterium nucleatum in oral epithelial cells: multiple signaling pathways and the role of commensal bacteria in innate immunity and the epithelial barrier. Infect. Immun. 68:2907-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krisanaprakornkit, S., A. Weinberg, C. N. Perez, and B. A. Dale. 1998. Expression of the peptide antibiotic human beta-defensin 1 in cultured gingival epithelial cells and gingival tissue. Infect. Immun. 66:4222-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamey, P. J., A. Darwaza, B. M. Fisher, L. P. Samaranayake, T. W. Macfarlane, and B. M. Frier. 1988. Secretor status, candidal carriage and candidal infection in patients with diabetes mellitus. J. Oral Pathol. 17:354-357. [DOI] [PubMed] [Google Scholar]
- 22.Lehrer, R. I., and T. Ganz. 1996. Endogenous vertebrate antibiotics. Defensins, protegrins, and other cysteine rich antimicrobial peptides. Ann. N. Y. Acad. Sci. 797:228-239. [DOI] [PubMed] [Google Scholar]
- 23.Liu, L., C. Zhao, H. H. Q. Heng, and T. Ganz. 1997. The human beta defensin-1 and alpha-defensins are encoded by adjacent genes: two families with differing disulfide topology share a common ancestry. Genomics 43:316-320. [DOI] [PubMed] [Google Scholar]
- 24.Lockhart, S. R., S. Joly, K. Vargas, J. Swails-Wenger, L. Enger, and D. R. Soll. 1999. Natural defenses against Candida colonization breakdown in the oral cavities of the elderly. J. Dent. Res. 78:857-868. [DOI] [PubMed] [Google Scholar]
- 25.Mathews, M., H. P. Jia, J. M. Guthmiller, G. Losh, S. Graham, G. K. Johnson, B. F. Tack, and P. B. McCray, Jr. 1999. Production of beta-defensin antimicrobial peptides by the oral mucosa and salivary glands. Infect. Immun. 67:2740-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsushita, I., K. Hasegawa, K. Nakata, K. Yasuda, K. Tokunaga, and N. Keicho. 2002. Genetic variants of human beta-defensin-1 and chronic obstructive pulmonary disease. Biochem. Biophys. Res. Commun. 291:17-22. [DOI] [PubMed] [Google Scholar]
- 27.Matsuzaki, K. 2001. Development of novel antimicrobial peptides: emerging strategies. Horizon Scientific Press, Wymondham, United Kingdom.
- 28.Odds, F. C. 1993. Resistance of yeasts to azole-derivative antifungals. J. Antimicrob. Chemother. 31:463-471. [DOI] [PubMed] [Google Scholar]
- 29.Odds, F. C., and R. Bernaerts. 1994. CHROMagar Candida, a new differential isolation medium for presumptive identification of clinically important Candida species. J. Clin. Microbiol. 32:1923-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samaranayake, L. P. 1997. Candida krusei infections and fluconazole therapy. Hong Kong Med. J. 3:312-314. [PubMed] [Google Scholar]
- 31.Samaranayake, L. P. 1990. Oral candidosis: an old disease in new guises. Dent. Update 17:36-38. [PubMed] [Google Scholar]
- 32.Samaranayake, Y. H., L. P. Samaranayake, E. H. Pow, V. T. Beena, and K. W. Yeung. 2001. Antifungal effects of lysozyme and lactoferrin against genetically similar, sequential Candida albicans isolates from a human immunodeficiency virus-infected southern Chinese cohort. J. Clin. Microbiol. 39:3296-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schutte, B. C., J. P. Mitros, J. A. Bartlett, J. D. Walters, H. P. Jia, M. J. Welsh, T. L. Casavant, and P. B. McCray, Jr. 2002. Discovery of five conserved beta-defensin gene clusters using a computational search strategy. Proc. Natl. Acad. Sci. USA 99:2129-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snider, J. 1999. PCR applications: protocols for functional genomics. Academic Press, Inc., New York, N.Y.
- 35.Tapper-Jones, L. M., M. J. Aldred, D. M. Walker, and T. M. Hayes. 1981. Candidal infections and populations of Candida albicans in mouths of diabetics. J. Clin. Pathol. 34:706-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai, H., and L. A. Bobek. 1998. Human salivary histatins: promising anti-fungal therapeutic agents. Crit. Rev. Oral Biol. Med. 9:480-497. [DOI] [PubMed] [Google Scholar]
- 37.Ueta, E., T. Tanida, K. Yoneda, T. Yamamoto, and T. Osaki. 2001. Increase of Candida cell virulence by anticancer drugs and irradiation. Oral Microbiol. Immunol. 16:243-249. [DOI] [PubMed] [Google Scholar]
- 38.Valore, E. V., C. H. Park, A. J. Quayle, K. R. Wiles, P. B. McCray, Jr., and T. Ganz. 1998. Human beta-defensin-1: an antimicrobial peptide of urogenital tissues. J. Clin. Investig. 101:1633-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vatta, S., M. Boniotto, E. Bevilacqua, A. Belgrano, D. Pirulli, S. Crovella, and A. Amoroso. 2000. Human beta defensin 1 gene: six new variants. Hum. Mutat. 15:582-583. [DOI] [PubMed] [Google Scholar]
- 40.Weinberg, A., S. Krisanaprakornkit, and B. A. Dale. 1998. Epithelial antimicrobial peptides: review and significance for oral applications. Crit. Rev. Oral Biol. Med. 9:399-414. [DOI] [PubMed] [Google Scholar]
- 41.White, T. C., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willis, A. M., W. A. Coulter, C. R. Fulton, J. R. Hayes, P. M. Bell, and P. J. Lamey. 2001. The influence of antifungal drugs on virulence properties of Candida albicans in patients with diabetes mellitus. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 91:317-321. [DOI] [PubMed] [Google Scholar]
- 43.Willis, A. M., W. A. Coulter, C. R. Fulton, J. R. Hayes, P. M. Bell, and P. J. Lamey. 1999. Oral candidal carriage and infection in insulin-treated diabetic patients. Diabet. Med. 16:675-679. [DOI] [PubMed] [Google Scholar]
- 44.Zhao, C., I. Wang, and R. I. Lehrer. 1996. Widespread expression of beta-defensin hBD-1 in human secretory glands and epithelial cells. FEBS Lett. 396:319-322. [DOI] [PubMed] [Google Scholar]