Abstract
Vibrio vulnificus exhibits considerable strain-to-strain variation in virulence. Attempts to associate phenotypic or genotypic characteristics with strain virulence have been largely unsuccessful. Based on a 17-nucleotide difference throughout the sequence of the small subunit 16S rRNA gene, there are two major groups of V. vulnificus designated types A and B. In a survey of the 16S rRNA genotype in 67 V. vulnificus human clinical and nonclinical strains, we determined that the majority of nonclinical isolates are type A (31 of 33) and that there is a statistically significant association between the type B genotype and human clinical strains (26 of 34).
Vibrio vulnificus is a gram-negative halophilic bacterium common to estuarine and marine waters, especially in tropical and subtropical climates (20). At least two distinct biotypes of this organism have been identified based on lipopolysaccharide composition (4). Biotype 1 strains are most often found in association with shellfish and in the intestinal contents of fish and are a common cause of human infection, either through ingestion of raw or undercooked shellfish or by wound exposure to the organism (20). V. vulnificus biotype 2 commonly infects marine vertebrates, although infections in humans have been reported (24). The existence of a third biotype causing wound infections and bacteremia in people handling cultured tilapia in Israel has recently been proposed (5).
Among V. vulnificus biotype 1 strains, it has long been recognized that there is a wide range of virulence as measured in various animal models (18). Moreover, clinical infections of humans with this pathogen usually arise from a single strain (11), even though a single shellfish can contain hundreds to thousands of strains as determined by contour-clamped homogeneous electric field gel electrophoresis (7). Consequently, there is widespread interest in developing a method to distinguish virulent strains of V. vulnificus from those less capable of causing human disease.
Phenotypic characteristics, including cytolysin and cytotoxin titers (22) and utilization of transferrin-bound iron and production of phenolate siderophore (18), have been studied as a possible means of predicting strain virulence. To date, no single expressed factor has been identified that distinguishes naturally occurring virulent and avirulent isolates (20). All defined virulence determinants, such as the exopolysaccharide capsule (28), a combination of exotoxins and proteases (16), and a type IV pilin (R. N. Paranjpye and M. S. Strom, unpublished data), also appear to be highly conserved among all strains examined.
Distinguishing clinical or more virulent isolates from environmental isolates has often defied genotypic analysis (13). Ribotyping V. vulnificus has demonstrated considerable interstrain genetic heterogeneity and is a useful epidemiological tool but is of limited use in distinguishing virulent from less virulent isolates (1, 2, 10, 21). Contour-clamped homogeneous electric field gel electrophoresis and pulsed-field gel electrophoresis (7, 21) and random amplified polymorphic DNA analysis (2, 10, 17, 25, 26) also have generally failed to differentiate virulent from avirulent strains. In one exception, Warner and Oliver (27) applied random amplified polymorphic DNA analysis to 70 V. vulnificus strains and reported the presence of a ca. 200-bp band in 31 of 31 clinical isolates that was absent in 36 of 39 environmental isolates. However, no further information is yet available concerning the nature of this fragment. With this possible exception, these studies have generally concluded that the genetic heterogeneity that appears to be characteristic of V. vulnificus strains precludes use of these genotypic techniques for predicting the virulence of a given isolate.
Analysis of V. vulnificus strains by terminal restriction fragment length polymorphism (T-RFLP) of the gene encoding the 16S rRNA subunit (15), shows that, in the case of V. vulnificus strains, two distinctly different sets of terminal fragment sizes were observed when a portion of the 16S rRNA gene amplified by PCR was digested with the restriction endonucleases HaeIII and AluI. A subsequent survey of GenBank submissions for the 16S rRNA gene of V. vulnificus revealed that there are indeed at least two sequence variants, both originally submitted by Aznar et al. (3), and designated 16S rRNA types A and B. Alignment of the two sequence variants revealed that they differ by a total of 17 out of 1,536 bases, with most of the polymorphism centered near helix 10 of the secondary structure for bacterial 16S rRNA (23), one of several known variable regions in prokaryotic 16S rRNA genes (8). With this information in mind, we decided to screen a panel of clinical and environmental isolates of V. vulnificus to determine if there was any association between more virulent strains and a particular 16S rRNA genotype.
The panel of strains examined included 33 nonclinical isolates (32 oyster isolates and 1 cultured from water) and 34 clinical isolates, most from patients with V. vulnificus primary septicemia acquired through ingestion of raw oysters (Table 1). Eighteen of the clinical isolates were from patients who died as a result of their infection. The strains were grown at 30°C on Luria-Bertani agar or broth supplemented with 5 U of polymyxin B/ml. Chromosomal DNA for each strain was prepared either from a single colony using the QIAamp DNA Mini Kit (Qiagen, Valencia, Calif.) according to the manufacturer's protocol for cultured cells or from broth culture according to the protocol of Strom and Lory (19). A 492-bp segment of the 16S rRNA gene of V. vulnificus was targeted for amplification using primers UFUL and URUL (Great American Gene Co., Ramona, Calif.) (Table 2). These primers target two highly conserved regions of the prokaryotic 16S rRNA gene that flank a number of regions known to be variable among bacterial species (8, 23). This segment corresponds to deoxynucleotides (nt) 46 to 537 in the same gene of Escherichia coli (data not shown). Each 25-μl reaction contained 10 mM Tris-HCl (pH 9.0), 50 mM KCl, 0.1% Triton X-100, 2.0 mM MgCl2, a 100 nM concentration of each deoxynucleotide triphosphate, a 400 nM concentration of each primer, and 1 U of Taq polymerase (Promega, Madison, Wis.). PCR amplification was performed using a Progene thermocycler (Techne Ltd., Princeton, N.J.), equipped with a heated lid. The initial cycle consisted of 3 min at 94°C, 30 s at 57°C, and 30 s at 72°C and was followed by 30 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 30 s. The final cycle consisted of 94°C for 30 s and 57°C for 30 s and an extension at 72°C for 10 min. The resulting amplicon was purified for subsequent restriction endonuclease digestions using the Ultraclean PCR Clean-up Kit (Mo Bio Laboratories, Inc., Solana Beach, Calif.).
TABLE 1.
Strains used in this study along with their respective origins and 16S rRNA types
| Straina,b | Isolate origin | Harvest state | Harvest date (mo/day/yr or mo/yr-mo/yr) | 16S rRNA type |
|---|---|---|---|---|
| FDA2 | Oyster | Fla. | NAf | A |
| FDA4 | Oyster | N.C. | NA | A |
| FDA7 | Oyster | Fla. | NA | A |
| FDA8 | Oyster | La. | NA | A |
| FDA9 | Oyster | La. | NA | A |
| FDA11 | Oyster | La. | NA | A |
| FDA18 | Oyster | N.C. | NA | A |
| FDA29 | Oyster | Fla. | NA | A |
| 99-624 DP-C10 | Oyster | Tex. | 1/5/99 | A |
| 99-779 DP-D2 | Oyster | La. | 4/16/99 | A |
| 99-736 DP-C7 | Oyster | Fla. | 4/5/99 | A |
| 99-645 DP-C4 | Oyster | Tex. | 5/1/99 | A |
| 99-581 DP-C7 | Oyster | La. | 12/8/98 | A |
| 99-796 DP-E7 | Oyster | Fla. | 4/6/99 | A |
| 99-584 DP-B12 | Oyster | Tex. | 3/31/99 | A |
| 98-640 DP-E9 | Oyster | La. | 8/23/98 | A |
| 99-743 DP-B6 | Oyster | Tex. | 5/8/99 | B |
| 98-783 DP-A1 | Oyster | La. | 5/1/99 | A |
| 99-780 DP-E1 | Oyster | La. | 4/14/99 | A |
| 99-625 DP-D8 | Oyster | Tex. | 1/5/99 | A |
| 99-738 DP-B5 | Oyster | Fla. | 4/19/99 | A |
| 99-537 DP-G7 | Oyster | Md. | 11/9/98 | A |
| 99-540 DP-B6 | Oyster | Tex. | 11/21/98 | A |
| 99-742 DP-A9 | Oyster | Miss. | 5/11/99 | A |
| 99-578 DP-B1 | Oyster | La. | 11/5/98 | B |
| 99-623 DP-F5 | Oyster | Fla. | 12/2/98 | A |
| 99-520 DP-B8 | Oyster | RI. | 12/29/98 | A |
| 99-505 DP-C8 | Oyster | Tex. | 11/9/98 | A |
| 99-609 DP-A4 | Oyster | Oreg. | 2/1/99 | A |
| 98-641 DP-G8 | Oyster | La. | 8/23/98 | A |
| 99-622 DP-E4 | Oyster | Tex. | 12/9/98 | A |
| 99-509 DP-A6 | Oyster | Tex. | 1/8/99 | A |
| PAC1c | Water | NA | NA | A |
| 9149-95 | Clinical, recovery | Fla./La. | 5/23/95 | A |
| ATL-9579 | Clinical, recovery | Tex. | 8/23/94 | A |
| ATL-9824 | Clinical, recovery | Tex. | 11/6/94 | B |
| 9070-96 | Clinical, recovery | Tex. | 10/3/96c | B |
| 9348-95 | Clinical, recovery | Fla. | 5/23/95 | A |
| 9076-96 | Clinical, recovery | La. | 10/29/96 | B |
| 9005-97 | Clinical, recovery | La. | 5/14/97 | A |
| ATL-9572 | Clinical, recovery | Fla. | 6/30/94 | A |
| 9053-96 | Clinical, recovery | Tex. | 8/16/96e | B |
| 9342-95 | Clinical, recovery | Tex./La. | 7/24/95 | B |
| 9031-96 | Clinical, recovery | Fla. | 4/30/96 | A |
| 9030-95 | Clinical, recovery | Fla. | 5/95-8/95 | A |
| 9032-95 | Clinical, recovery | Tex. | 5/13/95 | B |
| 9029-95 | Clinical, recovery | Fla. | 5/3/95 | B |
| 9349-95 | Clinical, fatality | Ala. | 7/16/95e | B |
| 9345-95 | Clinical, fatality | La. | 9/30/95 | B |
| 9075-96 | Clinical, fatality | Fla. | 10/24/96 | A |
| 9352-94 | Clinical, fatality | La. | 10/23/94 | B |
| 9340-95 | Clinical, fatality | Fla./La. | 8/7/95 | B |
| ATL-9580 | Clinical, fatality | Tex./La. | 9/2/94e | B |
| 9060-96 | Clinical, fatality | Tex. | 8/28/96 | B |
| 9003-97 | Clinical, fatality | La. | 4/29/97 | B |
| 9062-96 | Clinical, fatality | La. | 5/18/96e | B |
| 9038-96 | Clinical, fatality | Tex. | 4/27/96 | B |
| 9067-96 | Clinical, fatality | Tex. | 9/23/96 | B |
| 9074-96 | Clinical, fatality | Tex./La. | 10/9/96 | B |
| 9047-96 | Clinical, fatality | La. | 6/21/96e | B |
| 9057-96 | Clinical, fatality | Tex. | 6/18/96 | B |
| 9056-96 | Clinical, fatality | La. | 8/27/96 | B |
| 9049-96 | Clinical, fatality | Tex. | 3/20/96 | B |
| 9039-96 | Clinical, fatality | La. | NA | B |
| 9346-95 | Clinical, fatality | Fla. | 9/14/95 | B |
| MO6-24d | Clinical, outcome NA | NA | NA | B |
| C7184d | Clinical, outcome NA | NA | NA | B |
Where histories are known, all clinical isolates were from patients with typical V. vulnificus septicemia following ingestion of oysters.
Unless otherwise noted, all strains were from Gulf Coast Seafood Laboratory, the Food and Drug Administration, Dauphin Island, Ala.
Courtesy of Marie Coyle, Department of Microbiology, University of Washington, Seattle.
Courtesy of James Oliver, Department of Biology, University of North Carolina, Charlotte.
Date given is that of shellfish consumption rather than harvest date.
NA, not available.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Nucleotide sequence | Positiona | Reference or source |
|---|---|---|---|
| UFUL | GCCTAACACATGCAAGTCGA | 39-58 | 15 |
| URUL | CGTATTACCGCGGCTGCTGG | 530-511 | 15 |
| Vvu16SF | GATCATGGCTCAGATTGAACG | 8-28 | This work |
| Vvu16SR | GTGATCCAGCGCCAGGTTC | 1529-1511 | This work |
| Vvu511F | CCAGCAGCCGCGGTAATACG | 511-530 | This work |
| Vvu977F | CCTACTCTTGACATCCAGAG | 977-996 | This work |
| Vvu996R | CTCTGGATGTCAAGAGTAGG | 996-977 | This work |
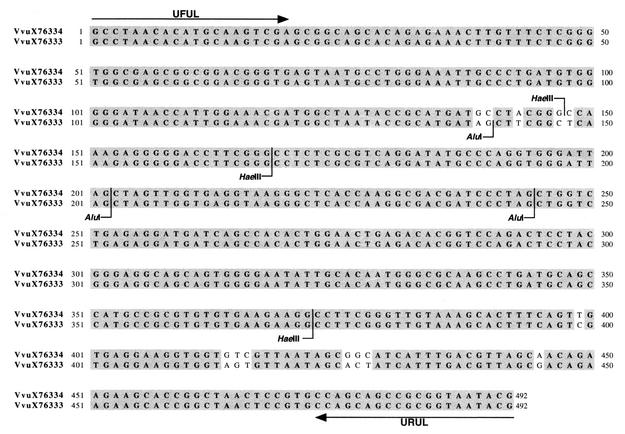
Figure 1 shows the alignment of the 492-bp amplified region for both 16S rRNA types (accession numbers X76333 [16S rRNA type A] and X76334 [16S rRNA type B]) from the original GenBank submissions (3). Both GenBank entries show AluI cleavage sites after nt 202 and 244 and HaeIII cleavage sites after nt 168 and 372. V. vulnificus X76333 (rRNA type A) contains an additional AluI cleavage site after nt 140, while V. vulnificus X76334 (16S rRNA type B) contains an additional HaeIII site after nt 147. As a result, the AluI digest of an amplicon from a given strain of V. vulnificus is predicted to have fragments of 140, 62, 42, and 248 bp if it is of 16S rRNA type A and 202, 42, and 248 bp if it is of 16S rRNA type B. Similarly, a digest of the amplicon with HaeIII is expected to give fragments of 147, 21, 204, and 120 bp for those of type B, while type A isolates should give fragments of 168, 204, and 120 bp. The 16S rRNA type for each of the 67 strains was determined by digestion of the amplicons with AluI and HaeIII (New England Biolabs, Beverly, Mass.) and analysis by electrophoresis on either a 2% agarose gel with staining by ethidium bromide or a 4% agarose gel composed of a 3:1 mixture of NuSieve GTG agarose (BMA, Rockland, Maine) and Agarose MB (Midwest Scientific, St. Louis, Mo.) with staining by the GelStar nucleic acid stain (BMA). AluI digests of the amplicons of type A and B strains can be readily distinguished on these gels (data not shown). The HaeIII digest of the same amplicon also differentiates the two 16S rRNA types, although more careful inspection is necessary due to the smaller differences in size of the fragments for the two types (data not shown). Assignments of 16S rRNA type based on RFLP results were verified by performing T-RFLP analyses (15) on the HaeIII and AluI digests of all strains (data not shown).
FIG. 1.
An alignment of the partial gene sequences for the 16S rRNA genes for V. vulnificus (Vvu) types A and B (GenBank accession numbers X76333 and X76334, respectively). Target sequences for primers used to generate the fragments are indicated. Polymorphic regions are unshaded, and endonuclease recognition sites for both sequences as well as sites present in only one of the sequences are indicated.
Table 1 provides a summary of the 16S rRNA types of all 67 strains. Of the 34 V. vulnificus isolates from human clinical cases, 26 are classified as type B while 31 of 33 environmental isolates belong to type A. The nucleotide sequence of the 16S rRNA gene for representative type A and B strains was determined to verify the RFLP analysis. Sequencing was performed using primers listed in Table 2 and the ABI BigDye Terminator Cycle Sequencing Ready Reaction Kit, version 3.0 (Applied Biosystems, Foster City, Calif.), with the resulting products being analyzed on an ABI 3100 Genetic Analyzer (Applied Biosystems). V. vulnificus strain FDA2 showed 100% identity with the type A strain (GenBank accession number X76333) as originally submitted by Aznar et al. (3) (data not shown). Similarly, the sequence of the 16S rRNA gene from strain C7184 revealed 100% identity with that of the type B strain (GenBank accession number X76334).
Statistical treatment using chi-square analysis of the results comparing the human clinical strains and those isolated from nonclinical sources indicates a highly significant association between strains possessing 16S rRNA type B and those that caused illness or death (Table 3, P < 0.0001, as calculated with Fisher's exact test at α = 0.05; Graphpad Prism, GraphPad Software, Inc.). The results of this study suggest a possible association between the type B 16S rRNA allele (3) and the potential virulence of a given V. vulnificus strain. By exploiting the nucleotide differences in the 16S rRNA gene sequence that result in different AluI and HaeIII restriction patterns between the types, we show that strains can be quickly differentiated by first using universal 16S rRNA gene primers to amplify a 492-bp DNA fragment, followed by digestion with these restriction enzymes. The characterization of the 16S rRNA type is then readily determined by visualization of an AluI or HaeIII digest of the amplicon on a 2% agarose gel. Alternatively, both the AluI- and HaeIII-digested amplicons can be analyzed by T-RFLP (15) using an ABI Prism 310 Genetic Analyzer (data not shown). Although T-RFLP analysis requires specialized equipment and primers labeled with fluorescent dyes, this technique is more automated than simple agarose gel analysis and is fully capable of detecting even smaller differences in fragment size.
TABLE 3.
Summary of the 16S rRNA types and the strain origin for all strains included in this studya
| Source | No. of isolates from:
|
Total no. of strains | |
|---|---|---|---|
| 16S rRNA type A | 16S rRNA type B | ||
| Nonclinical | 31 | 2 | 33 |
| Clinical, recovery | 7 | 7 | 14 |
| Clinical, outcome unknown | 2 | 2 | |
| Clinical, fatality | 1 | 17 | 18 |
| Total | 39 | 28 | 67 |
Statistical analysis (see Results) demonstrates a strong association between strains carrying 16S rRNA type B and those causing illness or death (P < 0.0001; α = 0.05).
It is especially notable that 17 of 18 isolates (94%) from clinical fatalities were of rRNA type B, while only 2 of 33 (6%) nonclinical isolates were of type B. Since type B strains appear to be relatively rare in nonclinical samples and were much more commonly isolated from clinical fatalities, the data strongly suggest that type B strains are significantly more virulent. On the other hand, type A strains are certainly not a priori avirulent, since isolates from 1 of 18 fatalities and 7 of 14 of the clinical but nonfatal illnesses were of type A. A survey of available medical history information for individuals from which strains were cultured reveals that most exhibited classical predisposing factors, e.g., liver disease, diabetes mellitus, or immune deficiency. This trend appeared to hold whether the individual eventually recovered or whether the strain isolated was of either 16S rRNA type. Nonetheless, the data appear to suggest that the 16S rRNA type might be one important indicator of the potential virulence of a given V. vulnificus strain.
Kim and Jeong (12) recently described a triprimer PCR assay that appears to be yet another method for differentiating type A and B strains. The assay was applied to 40 environmental strains isolated from oysters, sediment, and seawater off of the southern coast of Korea. This survey reported that the majority of the Korean isolates (65%) were type B, in contrast to our findings that the vast majority of environmental isolates (31 of 33) from the Gulf of Mexico and the U.S. Atlantic coast were type A. Whether these divergent findings can be explained solely on the basis of geographical considerations is impossible to say and may warrant further study.
There are a few reports in the literature that demonstrate a similar relationship between 16S rRNA polymorphisms and a virulent phenotype. In studies of Fusobacterium necrophorum (14) and Moraxella catarrhalis (6), data were presented suggesting that subpopulations of each organism that could be distinguished by phenotypic differences also differed in 16S rRNA sequence. Our finding with V. vulnificus suggests that there may exist a similar set of subpopulations of this species differing in virulence that can be distinguished by 16S rRNA type. However, known virulence determinants such as the exopolysaccharide capsule, a type IV pilin (Paranjpye and Strom, unpublished), and a possible combination of type IV pili and exoenzymes (16) appear to be present in all strains examined, with no single virulence determinant that is present in virulent but not in less virulent or avirulent isolates. The division of V. vulnificus biotype 1 strains into two types by 16S rRNA polymorphism analysis might be a good starting point for selecting strains for further study of differences in phenotypic traits that correlate with V. vulnificus virulence using techniques such as suppression subtractive hybridization (9, 29). Finally, although we used universal eubacterial primers that flank regions of polymorphism in the 16S rRNA gene that serve to distinguish the two types, species-specific primers presumably could be designed for the same purpose. It therefore seems possible that polymorphism in the 16S rRNA gene could be exploited for developing screening methods such as type-specific DNA probes or real-time quantitative PCR that could be used to screen for the presence of more virulent V. vulnificus strains in shellfish.
Acknowledgments
We gratefully acknowledge the support of the Northwest Fisheries Science Center, National Marine Fisheries Service for funding this project.
The Centers for Disease Control and Prevention and the Food and Drug Administration Southeast Regional Laboratory provided clinical and oyster strains, respectively. We also thank Michael Vickery and Cyndy Masada for reviewing the manuscript.
REFERENCES
- 1.Arias, C. R., M. J. Pujalte, E. Garay, and R. Aznar. 1998. Genetic relatedness among environmental, clinical, and diseased-eel Vibrio vulnificus isolates from different geographical regions by ribotyping and randomly amplified polymorphic DNA PCR. Appl. Environ. Microbiol. 64:3403-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aznar, R., W. Ludwig, and K.-H. Schleifer. 1993. Ribotyping and randomly amplified polymorphic DNA analysis of Vibrio vulnificus biotypes. Syst. Appl. Microbiol. 16:303-309. [Google Scholar]
- 3.Aznar, R., W. Ludwig, R. I. Amann, and K. H. Schleifer. 1994. Sequence determination of rRNA genes of pathogenic Vibrio species and whole-cell identification of Vibrio vulnificus with rRNA-targeted oligonucleotide probes. Int. J. Syst. Bacteriol. 44:330-337. [DOI] [PubMed] [Google Scholar]
- 4.Biosca, E. G., J. D. Oliver, and C. Amaro. 1996. Phenotypic characterization of Vibrio vulnificus biotype 2, a lipopolysaccharide-based homogenous O serogroup within Vibrio vulnificus. Appl. Environ. Microbiol. 62:918-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bisharat, N., V. Agmon, R. Finkelstein, R. Raz, G. Ben-Dror, L. Lerner, S. Soboh, R. Colodner, D. N. Cameron, D. L. Wykstra, D. L. Swerdlow, J. J. Farmer III, et al. 1999. Clinical, epidemiological, and microbiological features of Vibrio vulnificus biogroup 3 causing outbreaks of wound infection and bacteraemia in Israel. Lancet 354:1421-1424. [DOI] [PubMed] [Google Scholar]
- 6.Bootsma, H. J., H. G. J. van der Heide, S. van de Pas, L. M. Schouls, and F. R. Mooi. 2000. Analysis of Moraxella catarrhalis by DNA typing: evidence for a distinct subpopulation associated with virulence traits. J. Infect. Dis. 181:1376-1387. [DOI] [PubMed] [Google Scholar]
- 7.Buchrieser, C., V. V. Gangar, R. L. Murphree, M. L. Tamplin, and C. W. Kaspar. 1995. Multiple Vibrio vulnificus strains in oysters as demonstrated by clamped homogeneous electric field gel electrophoresis. Appl. Environ. Microbiol. 61:1163-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Rijk, P., J.-M. Neefs, Y. Van de Peer, and R. De Wachter. 1992. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 20:2075-2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diatchenko, L., Y.-F. C. Lau, A. P. Campbell, A. Chenchik, F. Moqadam, B. Huang, S. Lukyanov, K. Lukyanov, N. Gurskaya, E. D. Sverdlov, and P. D. Siebert. 1996. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. USA 93:6025-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Høi, L., A. Dalsgaard, J. L. Larsen, J. M. Warner, and J. D. Oliver. 1997. Comparison of ribotyping and randomly amplified polymorphic DNA PCR for characterization of Vibrio vulnificus. Appl. Environ. Microbiol. 63:1674-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson, J. K., R. L. Murphree, and M. L. Tamplin. 1997. Evidence that mortality from Vibrio vulnificus infection results from single strains among heterogeneous populations in shellfish. J. Clin. Microbiol. 35:2098-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim, M. S., and H. D. Jeong. 2001. Development of 16S rRNA targeted PCR methods for the detection and differentiation of Vibrio vulnificus in marine environments. Aquaculture 193:199-211. [Google Scholar]
- 13.Maslow, J. N., M. E. Mulligan, and R. D. Arbeit. 1993. Molecular epidemiology: application of contemporary techniques to the typing of microorganisms. Clin. Infect. Dis. 17:153-164. [DOI] [PubMed] [Google Scholar]
- 14.Nicholson, L. A., C. J. Morrow, L. A. Corner, and A. L. M. Hodgson. 1994. Phylogenetic relationship of Fusobacterium necrophorum A, AB, and B biotypes based upon 16S rRNA gene sequence analysis. Int. J. Syst. Bacteriol. 44:315-319. [DOI] [PubMed] [Google Scholar]
- 15.Nilsson, W. B., and M. S. Strom. 2002. Detection and identification of bacterial pathogens of fish in kidney tissue using terminal restriction length polymorphism (T-RFLP) analysis of 16S rRNA genes. Dis. Aquat. Org. 48:175-185. [DOI] [PubMed] [Google Scholar]
- 16.Paranjpye, R. N., J. C. Lara, J. C. Pepe, C. M. Pepe, and M. S. Strom. 1998. The type IV leader peptidase/N-methyltransferase of Vibrio vulnificus controls factors required for adherence to HEp-2 cells and virulence in iron-overloaded mice. Infect. Immun. 66:5659-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryang, D. W., S. W. Cho, M. G. Shin, J. H. Shin, and S. P. Suh. 1997. Molecular typing of Vibrio vulnificus isolates by random amplified polymorphic DNA (RAPD) analysis. Jpn. J. Med. Sci. Biol. 50:113-121. [DOI] [PubMed] [Google Scholar]
- 18.Stelma, G. N., Jr., A. L. Reyes, J. T. Peeler, C. H. Johnson, and P. L. Spaulding. 1992. Virulence characteristics of clinical and environmental isolates of Vibrio vulnificus. Appl. Environ. Microbiol. 58:2776-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strom, M. S., and S. Lory. 1986. Cloning and expression of the pilin gene of Pseudomonas aeruginosa PAK in Escherichia coli. J. Bacteriol. 165:367-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strom, M. S., and R. N. Paranjpye. 2000. Epidemiology and pathogenesis of Vibrio vulnificus. Microbes Infect. 2:177-188. [DOI] [PubMed] [Google Scholar]
- 21.Tamplin, M. L., J. K. Jackson, C. Buchrieser, R. L. Murphree, K. M. Portier, V. Gangar, L. G. Miller, and C. W. Kaspar. 1996. Pulsed-field gel electrophoresis and ribotype profiles of clinical and environmental Vibrio vulnificus isolates. Appl. Environ. Microbiol. 62:3572-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tison, D. L., and M. T. Kelly. 1986. Virulence of Vibrio vulnificus strains from marine environments. Appl. Environ. Microbiol. 51:1004-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van de Peer, Y., S. Nicolaï, P. De Rijk, and R. De Wachter. 1996. Database on the structure of small ribosomal subunit RNA. Nucleic Acids Res. 24:86-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veenstra, J., P. J. Rietra, C. P. Stoutenbeek, J. M. Coster, H. H. de Gier, and S. Dirks-Go. 1992. Infection by an indole-negative variant of Vibrio vulnificus transmitted by eels. J. Infect. Dis. 166:209-210. [DOI] [PubMed] [Google Scholar]
- 25.Vickery, M. C. L., A. L. Smith, A. DePaola, D. D. Jones, R. J. Steffan, and A. K. Bej. 1998. Optimization of the arbitrarily-primed polymerase chain reaction (AP-PCR) for intra-species differentiation of Vibrio vulnificus. J. Microbiol. Methods 33:181-189. [Google Scholar]
- 26.Vickery, M. C. L., N. Harold, and A. K. Bej. 2000. Cluster analysis of AP-PCR generated DNA fingerprints of Vibrio vulnificus isolates from patients fatally infected after consumption of raw oysters. Lett. Appl. Microbiol. 30:258-262. [DOI] [PubMed] [Google Scholar]
- 27.Warner, J. M., and J. D. Oliver. 1999. Randomly amplified polymorphic DNA analysis of clinical and environmental isolates of Vibrio vulnificus and other Vibrio species. Appl. Environ. Microbiol. 65:1141-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshida, S.-I., M. Ogawa, and Y. Mizuguchi. 1985. Relation of capsular materials and colony opacity to virulence of Vibrio vulnificus. Infect. Immun. 47:446-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang, Y. L., C. T. Ong, and K. Y. Leung. 2000. Molecular analysis of genetic differences between virulent and avirulent strains of Aeromonas hydrophila isolated from diseased fish. Microbiology 146:999-1009. [DOI] [PubMed] [Google Scholar]