Abstract
The main cause of failure of Helicobacter pylori eradication therapy is resistance to clarithromycin. The resistance is due to three point mutations in two positions on the 23S rRNA (A2142C, A2142G, and A2143G). Our aim was to develop a rapid and accurate method to detect these mutations directly on biopsy specimens. We developed a real-time PCR that included a simultaneous detection of the amplicons by hybridization of two probes labeled with LC-Red and fluorescein by using the fluorescence resonance energy transfer (FRET) technology and melting curve analysis with the LightCycler thermocycler. The assay was first applied successfully on reference strains, reference plasmids, and H. pylori-negative biopsies. Biopsies from 200 patients having failed a first eradication attempt and for whom the H. pylori strain was available were then tested with the new assay. A result was obtained in 199 cases; a single genotype was detected in 157 cases, two genotypes were detected in 41 cases, and three genotypes were detected in one case. There were, in total, seven discrepancies between the real-time PCR and the phenotypic method of determination of clarithromycin susceptibility, and in an additional four cases the two phenotypic methods were in disagreement. PCR-restriction fragment length polymorphism was applied to a sampling of biopsies, including all of the cases with multiple genotypes and all the cases with discrepant results. Finally, in four cases with discrepant results, the real-time PCR detected the resistant population at a concentration so low that it could not be detected by the phenotypic method, while in three cases other mutations could be involved. This assay had an accuracy at least as satisfactory as that of the phenotypic tests and could be performed within 2 h, allowing it to be used before the administration of therapy in the case of a first H. pylori eradication.
Helicobacter pylori infection is one of the most common chronic bacterial infections in the world and has been established as a major cause of gastritis, peptic ulcer disease, and gastric cancer (1).
Eradication therapy is recommended for patients with peptic ulcer disease (5, 10). The first-line regimen consists mainly of a triple therapy, and clarithromycin is one of the most widely used components in these treatments. However, the prevalence of primary and acquired clarithromycin resistance is increasing worldwide, jeopardizing the success of these treatments (3, 7, 14, 19). Several reports have shown that the cure rate is between 0 and 50% when the H. pylori strain is resistant to clarithromycin, whereas it is around 90% when the strain is susceptible (2, 8). Resistance to this antibiotic is due to point mutations within the peptidyltransferase-encoding region of the 23S rRNA (25). Three major point mutations in two positions on the 23S rRNA (equivalent to Escherichia coli coordinates 2058 and 2059) have been described in which an adenine residue is replaced by a guanine or a cytosine residue in different positions: A2142C, A2142G, and A2143G (18, 21, 22, 25). Mutations A2142G and A2143G are the most frequently reported, whereas mutation A2142C is less common (24). Point mutations at two additional sites, A2115G and G2141A, have been described as occurring in the same strain (9), although these mutations have never been subsequently reported.
In routine practice the detection of clarithromycin resistance is mainly based on phenotypic methods performed after culture: agar diffusion for the ɛ-Test or the agar dilution method, which is considered the reference; however, these methods are time-consuming (13). Detection of point mutations conferring resistance to clarithromycin by molecular methods may constitute a more reliable approach. Numerous PCR-based techniques have been developed to detect these mutations, such as PCR-restriction fragment length polymorphism (RFLP) (18, 25), PCR-DNA-enzyme immunoassay (11, 20), and reverse hybridization line probe assay (23). More recently, real-time PCR methods were developed that were based on amplification of a fragment of the 23S rRNA gene of H. pylori followed by melting curve analysis. The first attempt was performed by Gibson et al. on H. pylori strains (6). The studies performed on biopsies, however, included few cases with resistant strains (4) or did not include cases with the mutation A2142C (12).
The aim of the present study was to develop a rapid and reliable single-step method to detect the three most frequent clarithromycin resistance-associated gene mutations in H. pylori directly on gastric biopsies, based on a PCR assay and simultaneous detection of the amplicon by probe hybridization and thermal analysis by using fluorescence resonance energy transfer (FRET) technology with a LightCycler thermocycler.
MATERIALS AND METHODS
Bacterial strains and gastric biopsies.
Four H. pylori strains, one reference strain (CIP 101260) with the wild-type genotype and three strains (825, 683, and 677) with known mutations in the 23S rRNA gene (mutations A2142C, A2142G, and A2143G, respectively) were used as positive controls (18). Seventeen gastric biopsies obtained from patients for which all diagnostic tests for H. pylori were negative (culture, histology, and serology) were used as negative controls.
Two hundred successive gastric biopsies from patients included in a randomized clinical trial aiming to test second-line regimens (Strathegy) were used (H. Lamouliatte, F. Mégraud, J. C. Delchier, J. F. Bretagne, A. Courillon-Mallet, J. D. de Korwin, J. L. Fauchère, A. Labigne, J. F. Fléjou, and P. Barthélémy, submitted for publication). The first regimen used included a proton pump inhibitor, with clarithromycin and amoxicillin in 85% of the cases. Patients were included in the study if they had a positive [13C]urea breath test. They were submitted to endoscopy, and biopsies were obtained for culture from the antrum and corpus.
Gastric biopsies were ground with a tissue homogenizer (UltraTurax; LaboModerne, Paris, France), and H. pylori was cultured as described before (15). The suspension used for culture was also used for DNA isolation. DNA was isolated by using a QIAamp DNA mini kit (Qiagen SA, Courtaboeuf, France) according to the manufacturer's instructions.
Determination of the susceptibility to clarithromycin by phenotypic methods.
Two methods were used to determine the MIC of clarithromycin for H. pylori: the ɛ-Test method (AB Biodisk, Solna, Sweden) was performed just after isolation, and an agar dilution method was carried out at the end of the study. Both methods were used according to a previously described protocol (15). A strain was considered resistant to clarithromycin when the MIC was ≥1 mg/liter (17).
Detection of point mutations in the 23S rRNA gene of H. pylori by real-time PCR.
A real-time PCR-based PCR-hybridization assay was used directly on DNA obtained from gastric biopsies to detect point mutations conferring resistance to clarithromycin. The method included amplification of a fragment of the 23S rRNA gene of H. pylori coupled with simultaneous detection of the product by probe hybridization and analysis of the melting curve by using real-time PCR (26). A 267-bp fragment of the 23S rRNA gene of H. pylori was amplified by using primers HPYS and HPYA as previously described (16). The primers were analyzed for 3′-terminal specificity to assure that they were specific to H. pylori. The amplified product was detected with two probes: the sensor probe, 5′ labeled with LC-Red 640 and 3′ phosphorylated (5′-GGCAAGACGGAAAGACC-3′; nucleotides 2504 to 2520), which hybridized with the region containing the mutation sites, and the anchor probe, which hybridized three bases upstream from the former and was 3′ labeled with fluorescein (5′-TGTAGTGGAGGTGAAAATTCCTCCTACCC-3′; nucleotides 2473 to 2501) (GenBank accession number U27270). Primers and probes (Tib MolBiol Syntheselabor, Berlin, Germany) were purified by high-performance liquid chromatography.
By using the LightCycler thermocycler (Roche Diagnostics, Neuilly sur Seine, France) the PCR and hybridization reactions were carried out in glass capillaries in a volume of 20 μl containing 3 μl of template DNA, 1.6 μl of MgCl2 (25 mM), 0.4 μl of forward and reverse primers (20 μM each), 0.2 μl of sensor and anchor probes (20 μM each), and 2 μl of FastStart DNA Master Hybridization Probes (Roche Diagnostics). PCR amplification comprised an initial denaturation cycle at 95°C for 10 min, followed by 50 amplification cycles (with a temperature transition rate of 20°C/s) consisting of 95°C for 0 s, annealing at 60°C for 10 s, and extension at 72°C for 17 s. After amplification a melting step was performed, consisting of 95°C for 0 s, cooling to 45°C for 30 s (with a temperature transition rate of 20°C/s), and finally a slow rise in the temperature to 85°C at a rate of 0.1°C/s with continuous acquisition of fluorescence decline.
Determination of the sensitivity of the real-time PCR.
In order to evaluate the sensitivity of the real-time PCR regarding the detection of multiple strains in the same biopsy sample, DNA from the wild-type and A2142G and A2142C mutant strains was amplified by PCR with the primers HPYS and HPYA deduced from the 23S rRNA gene of H. pylori (GenBank accession number U27270) (16) to generate the 267-bp fragment of the 23S rRNA gene. The three PCR products were mixed in equal proportions, and the PCR product of the A2142G mutant was mixed with that of the wild-type strain in proportions of 50, 25, 10, 5, and 1%. These mixtures were analyzed with the real-time PCR.
PCR-RFLP analysis.
A PCR-RFLP analysis was performed on all of the 42 biopsies with more than one genotype detected in the real-time PCR and on 30 randomly selected samples with a single genotype (10 wild type, 19 with one of the A→G mutations, and the biopsy with the A2142C mutation). The enzymes BbsI and BsaI (New England Biolabs, Beverly, Mass.) were used in all of the cases where the wild-type genotype and/or an A→G mutant sequence was detected, and the enzyme BceAI (New England Biolabs) was used in the cases where the mutation A2142C was detected by the real-time PCR. The four H. pylori control strains reported above were used as controls for the digestion assays. The PCR-RFLP procedures were performed as previously described (16).
Plasmid construction.
DNA from the four reference strains was amplified by PCR with the primers HPYA and HPYS containing SacI and KpnI restriction sites (5′-TAAGAGCTCAGGTTAAGAGGATGCGTCAGTC-3′ and 5′-TATGGTACCCGCATGATATTCCCATTAGCAGT-3′, respectively, where italics indicate restriction sites) as previously reported (16). The amplified PCR products were precipitated with 95% (vol/vol) ethanol, washed twice with 70% (vol/vol) ethanol, and digested overnight with the restriction enzymes SacI and KpnI according to the manufacturer's instructions (Roche Diagnostics). The phagemid vector pBluescriptII SK(+) (Stratagene, La Jolla, Calif.) was also digested by SacI and KpnI. Digested products were precipitated with ethanol and were washed twice. PCR amplicons were directionally ligated into pBluescriptII SK(+) by using the T4 DNA ligase (Roche Diagnostics), and transformation was performed by using the JM109 cells according to the manufacturer's instructions (Promega, Madison, Wis.). The construction was confirmed by restriction analysis, real-time PCR (as reported above), and sequencing in two senses by using the pBluescriptII SK(+) primers 5′-GCTATGACCATGATTACGCCA-3′ and 5′ CCCAGTCACGACGTTGTAAAA-3′. The GenBank accession numbers of these plasmids corresponding to the wild-type and to the mutations A2142C, A2142G, and A2143G are AF550406, AF550407, AF550408, and AF550409, respectively.
RESULTS
Detection of point mutations conferring resistance to clarithromycin in the control specimens.
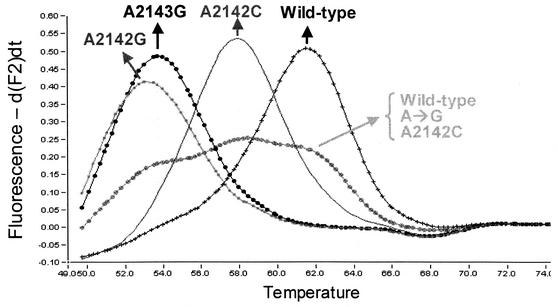
Melting curve analysis of DNA from the positive control strains produced four different melting curves, with melting temperatures (Tm) of approximately 61.5, 58.0, 53.0, and 53.6°C for the wild-type strain and mutants A2142C, A2142G, and A2143G, respectively (Fig. 1). As was expected, the existence of a nucleotide mismatch between the sequence and the hybridization probe produced a Tm lower than the Tm of the wild-type sequence when there was 100% homology between the probe and the target. A difference of 1°C maximum in the melting peak temperatures could be observed between different runs, probably resulting from variations in the temperature profile created by the LightCycler. The occurrence of close Tms between the mutations A2142G and A2143G rendered their differentiation difficult.
FIG. 1.
Melting curve analysis of the 267-bp fragment of the 23S rRNA gene obtained with real-time PCR for the wild-type and mutant strains with the A2142C, A2142G, and A2143G mutations. The melting curve corresponding to a biopsy containing the wild type and the A2142C, A2142G, and A2143G mutations is also shown, and three melting peaks can be observed with the Tms corresponding to those of the wild type, the A2142C mutation, and the A224G transition. Values on the y axis represent the ratio of the first negative derivative of the change in fluorescence (dF) to the variation in temperature (temperature values are given in degrees Celsius). A→G, A to G mutation.
Biopsies from patients for which no H. pylori was detected by culture all remained negative. The plasmids containing the different sequences gave the expected Tm. To prove the specificity of the primers, various bacterial species, including E. coli (clinical isolates), Campylobacter jejuni (clinical isolates), Campylobacter coli (clinical isolates), Flexispira rappini (CCUG 29176), and Helicobacter spp. such as H. felis (CCUG 28539 T), H. bilis (CCUG 38995 B T), H. hepaticus (CCUG 33637 T), H. muridarum (CCUG 29262 T), and H. pullorum (CCUG 33842 and CCUG 33839) were tested by this new assay, and no amplification was observed (data not shown).
Detection of point mutations conferring resistance to clarithromycin in gastric biopsies by real-time PCR.
Real-time PCR, described above and applied directly to gastric biopsies, was able to detect the genotype associated with clarithromycin susceptibility or resistance in 199 of the 200 biopsies (99.5%) tested. One biopsy with a clarithromycin-susceptible strain produced a melting peak with a Tm of approximately 56°C, which was different from the expected Tm for the wild-type or the three mutant genotypes. Sequencing of a 267-bp fragment from domain V of the 23S rRNA gene by using HPYS and HPYA showed a transition from G to A in position 2141 (GenBank accession number AF550410).
In 157 cases (79%), one single genotype was detected, corresponding either to the wild type or to the mutation A→G or A→C. In 152 biopsies from this group, the results of the real-time PCR correlated perfectly with the clarithromycin susceptibility profile determined by the ɛ-Test (Table 1). Of the 64 biopsies with a matched clarithromycin-susceptible strain, 64 produced a melting peak characteristic of the wild-type genotype. Among resistant strains, mutation A→G, either in position 2142 or 2143, was detected in 87 samples, and the transversion A→C in position 2142 was detected in one biopsy. In three biopsies a wild type was present, while the phenotype was resistant. In two additional cases, the real-time PCR detected a wild-type genotype while there was discrepancy between the ɛ-Test and the agar dilution method (Table 1).
TABLE 1.
Results of the detection of point mutations conferring resistance to clarithromycin by real-time PCR on 199 gastric biopsies compared with those of ɛ-Test performed on matched cultures
In 42 cases (21%) the simultaneous presence of at least two different genotypes in the same biopsy was detected. Of these samples, 30 produced melting peaks corresponding to the wild-type and A→G sequences and two produced melting peaks corresponding to the wild-type and A→C genotypes; in three cases the two types of mutants (A→G and A→C) and in one case the wild type and the two types of mutants were detected (Fig. 1). In all of these cases, the corresponding H. pylori isolates were resistant to clarithromycin (Table 1). However, in four biopsies with a matched susceptible strain, the presence of both the wild-type and a mutant A→G genotype was observed as well as in two others where there was a discrepancy between the ɛ-Test and the agar dilution method (Table 1).
To confirm that the real-time PCR was able to detect the simultaneous presence of these different genotypes, DNA from the wild-type and A2142C and A2142G mutant strains were mixed in different proportions. In the mixture the real-time PCR was able to detect the mutant at a concentration as low as 10% (data not shown).
In total, the two phenotypic methods agreed in 195 cases (98%), and among them the real-time PCR gave concordant results in 188 cases (96.4%). When the agar dilution was regarded as the gold standard, the sensitivity and specificity of the real-time PCR were 98.4 and 94.1%, respectively.
Comparison between real-time PCR and PCR-RFLP.
In order to confirm the results of the real-time PCR, PCR-RFLP analysis was performed on each of the 42 biopsies which showed more than one genotype in the real-time PCR and on 30 randomly selected samples presenting a single genotype and distributed as follows: 10 wild-type samples, 19 samples with the A2142G or A2143G mutation, 1 sample with the A2142C mutation, and all the cases with discrepant results. Of the single-genotype group, the 23S rRNA fragment amplified from the 10 biopsies with the wild-type genotype showed no restriction with the enzymes BbsI and BsaI (Table 2). Of the 19 samples with the A→G mutation, four were restricted with BbsI and 15 were restricted with BsaI, and the only biopsy with the A→C mutation was restricted with BceAI (Table 2).
TABLE 2.
Results of the PCR-RFLP analysis on 30 biopsies with a single genotype (10 wild type, 19 with the A2142/43G mutation, and 1 with the A2143C mutation) and on all 42 samples with more than one genotype detected by real-time PCR
| Genotype detected by real-time PCR | PCR-RFLP status (no. of samples) |
|---|---|
| Wild type | NR (10)a |
| A→G | BbsI (4), BsaI (15) |
| A→C | BceAI (1) |
| Wild type and A→G | BbsI and NR (15), BsaI and NR (20) |
| Wild type and A→G | BbsI and BsaI and NR (1) |
| Wild type and A→C | BceAI and NR (2) |
| A→G and A→C | BbsI and BceAI (2) |
| A→G and A→C | BsaI and BceAI (1) |
| Wild type and A→G and A→C | BbsI and BsaI and BceAI and NR (1) |
NR, not restricted.
The melting peaks of this biopsy are represented in Fig. 1.
Of the 42 biopsies with more than one genotype, 36 presented a mixture of wild-type and A→G genotypes. Among these, 15 presented the uncut PCR product and the products resulting from the restriction with BbsI, 20 presented the uncut fragment and the BsaI digestion products, and one biopsy was restricted with both enzymes in addition to showing the presence of the uncut fragment (Table 2).
The two biopsies with a mixture of wild-type and A→C genotypes were restricted with BceAI, and the uncut fragment was also visible. The presence of both A→G and A→C genotypes in three biopsies was confirmed by restriction with BbsI and BceAI in two cases and with BsaI and BceAI in one case (Table 2). Finally, the biopsy presenting three melting peaks corresponding to the wild-type, A→G, and A→C genotypes was restricted with BbsI, BsaI, and BceAI in addition to showing the presence of the uncut fragment in the digestion profiles (Table 2).
There were, altogether, 11 discrepant results (Table 3). In four cases, both phenotypic methods indicated a susceptible strain while the real-time PCR as well as the PCR-RFLP indicated a mixture of the wild-type genotypes and genotypes restricted by BbsI or BsaI.
TABLE 3.
Discrepant results of susceptibility testing for clarithromycin determined by phenotypic methods (MIC by ɛ-Test and agar dilution), real-time PCR, and PCR-RFLPa
| Biopsy no. | ɛ-Test | Agar dilution | Real-time PCR
|
PCR-RFLP
|
|||
|---|---|---|---|---|---|---|---|
| WT | A2142/43G | WT | BbsI | BsaI | |||
| B18 | S | S | + | + | + | + | |
| B37 | S | S | + | + | + | + | |
| B92 | S | S | + | + | + | + | |
| B260 | S | S | + | + | + | + | |
| B128 | R | R | + | + | |||
| B130 | R | R | + | + | |||
| B250 | R | R | + | + | |||
| B73 | S | R | + | + | |||
| B55 | S | R | + | + | + | + | |
| B294 | S | R | + | + | + | + | |
| B202 | S | R | + | + | + | ||
WT, wild type. R, resistant; S, susceptible.
In three cases the reverse situation was observed; i.e., the samples were determined to be resistant by phenotypic methods (agar dilution and Etest) and were determined to have the wild-type genotype by genotypic methods (PCR-RFLP and real-time PCR).
Finally, in four cases there was a discrepancy between ɛ-Test and agar dilution method results: one sample appeared to have a wild-type genotype and two samples had a mixture of wild-type and mutant genotypes, and in only one sample did PCR-RFLP show a mixture of wild-type and mutant genotypes while the real-time PCR indicated a wild-type genotype.
DISCUSSION
In this study we describe a real-time PCR-based method for detection of the most common point mutations occurring in the 23S rRNA gene of H. pylori that confer resistance to clarithromycin (24). This method is applied directly to gastric biopsies and consists of an amplification of a fragment of the 23S rRNA gene of H. pylori and subsequent detection of the mutations by probe hybridization and melting curve analysis with real-time PCR technology. The primers used have been proven to be specific for H. pylori, since no positive reaction was obtained with negative gastric biopsies or with a range of bacteria which could be present in gastric biopsies.
The assay described above permits the detection of the transition A→G, in position 2142 or 2143, the transversion A→C in position 2142, and the wild-type genotype. The distinction between genotypes is based on the Tm of the corresponding melting curves, and in this assay the Tms were well established and reproducible for each genotype. Due to the close proximity of the melting Tm between the A2142G and A2143G mutations and to the fact that the melting peaks present slight variations between different runs, the distinction between these two genotypes is sometimes difficult. Therefore, it was considered that the assay described in this study can be applied to detect and distinguish the two transitions A→G and A→C and the wild-type genotype but cannot distinguish between A2142G and A2143G mutations, in contrast to some other methods (11, 23). However, we think that the clinical relevance of this distinction has limited value.
The two mutations occurring simultaneously in the 23S rRNA gene of H. pylori (in the positions A2115G and G2141A) and described by Hultén et al. (9) as being associated with clarithromycin resistance seem to be very rare, as they were not detected in other studies (15, 24). In this study, the G2141A change was detected by the real-time PCR with a susceptible strain, proving that this assay is able to detect and identify this genotype as long as a control DNA is used in parallel. The A2115G mutation is localized within the annealing region of the anchor probe. Only testing a control strain containing this mutation would show if it could be detected by this assay. However, in theory the A2115G mutation should not be detected by the system of probes described in this study because the Tm difference between the sensor and the anchor probes is approximately 10°C, and one mutation occurring in the region of the anchor probe presumably will not reduce the Tm by more than 10°C. The only way to be sure will be to include a control strain, which we are presently lacking. Sequencing showed that the A2115G mutation was not present in this biopsy. In our study, the G2141A mutation was not associated with a phenotype exhibiting resistance to clarithromycin, indicating that this mutation alone is not sufficient to induce clarithromycin resistance.
The real-time PCR described in this study has proven to be very sensitive in the detection of mixed populations, since it was able to detect the presence of a mutant among wild-type strains at a level of 10% (data not shown). It was also able to detect two mutants and the wild-type genotype in the same biopsy (Fig. 1).
The genotype responsible for the susceptibility profile to clarithromycin was successfully identified in 199 of the 200 biopsies tested. A single genotype was detected in the majority of the biopsies, whereas the presence of more than one genotype was detected in 21% of the cases. The presence of two different genotypes in the same biopsy was detected in 41 cases, with either the wild-type genotype combined with the A→G or A→C mutation or with the two mutated genotypes. One biopsy with three genotypes, wild type, A→G mutation, and A→C mutation, was also observed.
The results of the real-time PCR were compared with the results of the clarithromycin susceptibility testing by the ɛ-Test and the agar dilution method. A perfect correlation between the genotype and the susceptibility profile was found in 188 biopsies (96.4%). In four cases with a clarithromycin-susceptible H. pylori isolate, the real-time PCR was able to detect the presence of both the wild type and an A→G mutant. This discrepant result may be explained by the fact that the resistant population was present at a concentration so low that it could not be detected by the phenotypic method, showing that the genotypic method is more sensitive in detecting several populations in the same sample than the phenotypic method. The hypothesis that the two genotypes may correspond to different 23S ribosomal DNA alleles in a single strain cannot be excluded. However, since this study was performed on biopsies, this hypothesis is less probable than the hypothesis of the presence of mixed populations in the same sample.
In three cases a wild-type genotype was detected in biopsies containing a clarithromycin-resistant strain, suggesting that probably other mutations in the 23S ribosomal DNA not detected by the method or other mechanisms not related to mutations in this region may be involved.
The detection of the point mutations was also performed by PCR-RFLP on a group of biopsies, with a single genotype and in all of the samples comprised of more than one genotype, in order to confirm the results obtained by the real-time PCR. In the former group the results of the two genotypic methods were totally in agreement. With regard to the samples with mixed populations, an incomplete cleavage was observed together with the digestion products by using either BbsI, BsaI, or BceAI. The presence of an uncut fragment with the same intensity as the restriction products was considered to be indicative of a mixed population. The results of the PCR-RFLP were concordant with the results of the real-time PCR in all of the cases except one, and again, the very small amount of the missed genotype may be the reason why it was not detected.
In conclusion, we have developed a new real-time PCR assay for detection directly on gastric biopsies of the most common point mutations occurring in the 23S rRNA gene of H. pylori that confer resistance to clarithromycin. The method is rapid; in fact, the entire procedure only takes 2 h, from the reception of the samples, involving the isolation of the DNA from the biopsies, to the detection of the mutations by real-time PCR and melting curve analysis. Moreover, the system of sensor and anchor probes used in this assay is able to discriminate between all of the genotypes in one single reaction, also contributing to the rapidity of the method. The assay also proved to be reliable and more sensitive than the phenotypic methods, especially in the detection of resistant strains. Since the amplified fragment is species specific for H. pylori, the determination of an H. pylori status is performed simultaneously. Compared to the real-time PCR biopsy assays previously published, our assay proved to be able to detect all of the common mutations associated with clarithromycin resistance, while the assay described by Chisholm et al. (4) appeared to be less sensitive (4 biopsies out of 56 were found to be falsely H. pylori negative) and the evaluation of the clarithromycin resistance was only performed on four biopsies.
The application of this new method to the rapid detection of H. pylori and determination of its resistance profile to clarithromycin allows the reconsideration of the strategy of empirical therapy in H. pylori infection. We now have a tool to obtain the information on clarithromycin susceptibility or resistance without delaying the treatment, so this method should be used before the administration of a fast course of eradication treatment without having to wait for cases of failure to perform the test. This strategy will reduce the proportion of treatment failures and, therefore, will reduce the cost of treatment.
REFERENCES
- 1.Blaser, M. J. 1997. Ecology of Helicobacter pylori in the human stomach. J. Clin. Investig. 100:759-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broutet, N., S. Tchamgoué, E. Pereira, H. Lamouliatte, R. Salamon, and F. Mégraud. Results of an individual data analysis of 2751 patients. Ailment. Pharmacol. Ther., in press. [DOI] [PubMed]
- 3.Cabrita, J., M. Oleastro, R. Matos, A. Manhente, J. Cabral, R. Barros, A. I. Lopes, P. Ramalho, B. C. Neves, and A. S. Guerreiro. 2000. Features and trends in Helicobacter pylori antibiotic resistance in Lisbon area, Portugal (1990-1999). J. Antimicrob. Chemother. 46:1029-1031. [DOI] [PubMed] [Google Scholar]
- 4.Chisholm, S. A., R. J. Owen, E. L. Teare, and S. Saverymuttu. 2001. PCR-based diagnosis of Helicobacter pylori infection and real-time determination of clarithromycin resistance directly from human gastric biopsy samples. J. Clin. Microbiol. 39:1217-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gastroenterology. 1997. The report of the Digestive Health Initiative International Update Conference on Helicobacter pylori. Gastroenterology 113(Suppl.):S4-S8. [DOI] [PubMed] [Google Scholar]
- 6.Gibson, J. R., N. A. Saunders, B. Burke, and R. J. Owen. 1999. Novel method for rapid determination of clarithromycin sensitivity in Helicobacter pylori. J. Clin. Microbiol. 37:3746-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glupczynski, Y., F. Mégraud, L. P. Andersen, and M. Lopez-Brea. 1999. Antibiotic susceptibility of Helicobacter pylori in Europe in 1998: results of the third multicentre study. Gut 45(Suppl. 3):A3. [Google Scholar]
- 8.Houben, M. H., D. van Der Beek, E. F. Hensen, A. J. Craen, E. A. Rauws, and G. N. Tytgat. 1999. A systematic review of Helicobacter pylori eradication therapy—the impact of antimicrobial resistance on eradication rates. Aliment. Pharmacol. Ther. 13:1047-1055. [DOI] [PubMed] [Google Scholar]
- 9.Hultén, K., A. Gibreel, O. Skold, and L. Engstrand. 1997. Macrolide resistance in Helicobacter pylori: mechanism and stability in strains from clarithromycin-treated patients. Antimicrob. Agents Chemother. 41:2550-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malfertheiner, P., F. Mégraud, C. O'Morain, A. P. S. Hungin, R. Jones, A. Axon, D. Y. Graham, G. Tytgat, and The European Helicobacter Pylori Study Group (EHPSG). 2002. Current concepts in the management of Helicobacter pylori infection. The Maastricht Consensus Report. Aliment. Pharmacol. Ther. 16:167-180. [DOI] [PubMed] [Google Scholar]
- 11.Marais, A., L. Monteiro, A. Occhialini, M. Pina, H. Lamouliatte, and F. Mégraud. 1999. Direct detection of Helicobacter pylori resistance to macrolides by a polymerase chain reaction/DNA enzyme immunoassay in gastric biopsy specimens. Gut 44:463-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsumura, M., Y. Hikiba, K. Ogura, G. Togo, I. Tsukuda, K. Ushikawa, Y. Shiratori, and M. Omata. 2001. Rapid detection of mutations in the 23S rRNA gene of Helicobacter pylori that confers resistance to clarithromycin treatment to the bacterium. J. Clin. Microbiol. 39:691-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mégraud, F. 1997. Resistance of Helicobacter pylori to antibiotics. Aliment. Pharmacol. Ther. 11(Suppl. 1):43-53. [DOI] [PubMed] [Google Scholar]
- 14.Mégraud, F. 1998. Epidemiology and mechanism of antibiotic resistance in Helicobacter pylori. Gastroenterology 115:1278-1282. [DOI] [PubMed] [Google Scholar]
- 15.Mégraud, F., N. Lehn, T. Lind, E. Bayerdorffer, C. O'Morain, R. Spiller, P. Unge, S. Veldhuyzen van Zanten, M. Wrangstadh, and C. F. Burman. 1999. Antimicrobial susceptibility testing of Helicobacter pylori in a large multi-center trial: the MACH 2 study. Antimicrob. Agents Chemother. 43:2747-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ménard, A., M. Oleastro, A. Santos, and F. Mégraud. 2002. PCR-RFLP can also detect the point mutation A2142C of the 23S rRNA gene associated with resistance of Helicobacter pylori to clarithromycin. Antimicrob. Agents Chemother. 46:1156-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial susceptibility testing. Sixth informational supplement M100 S9.19,1. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 18.Occhialini, A., M. Urdaci, F. Doucet-Populaire, C. M. Bébéar, H. Lamouliatte, and F. Mégraud. 1997. Macrolide resistance in Helicobacter pylori: rapid detection of point mutations and assays of macrolide binding to ribosomes. Antimicrob. Agents Chemother. 41:2724-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osato, M. S., R. Reddy, S. G. Reddy, R. L. Penland, H. M. Malaty, and D. Y. Graham. 2001. Pattern of primary resistance of Helicobacter pylori to metronidazole or clarithromycin in the United States. Arch. Intern. Med. 161:1217-1220. [DOI] [PubMed] [Google Scholar]
- 20.Pina, M., A. Occhialini, L. Monteiro, H.-P. Doermann, and F. Mégraud. 1998. Detection of point mutations associated with resistance of Helicobacter pylori to clarithromycin by hybridization in liquid phase. J. Clin. Microbiol. 36:3285-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stone, G. G., D. Shortridge, J. Versalovic, J. Beyer, R. K. Flamm, D. Y. Graham, A. T. Ghoneim, and S. K. Tanaka. 1997. A PCR-oligonucleotide ligation assay to determine the prevalence of 23S rRNA gene mutations in clarithromycin-resistant Helicobacter pylori. Antimicrob. Agents Chemother. 41:712-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor, D. E., Z. Ge, D. Purych, T. Lo, and K. Hiratsuka. 1997. Cloning and sequence analysis of two copies of a 23S rRNA gene from Helicobacter pylori and association of clarithromycin resistance with 23S rRNA mutations. Antimicrob. Agents Chemother. 41:2621-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Doorn, L. J., Y. J. Debets-Ossekopp, A. Marais, R. Sanna, F. Mégraud, J. G. Kusters, and W. G. V. Quint. 1999. Rapid detection, by PCR and reverse hybridization, of mutations in the Helicobacter pylori 23S rRNA gene, associated with macrolide resistance. Antimicrob. Agents Chemother. 43:1779-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Doorn, L. J., Y. Glupczynski, J. G. Kusters, F. Megraud, P. Midolo, N. Maggi-Solca, D. M. Queiroz, N. Nouhan, E. Stet, and W. G. Quint. 2001. Accurate prediction of macrolide resistance in Helicobacter pylori by a PCR line probe assay for detection of mutations in the 23S rRNA gene: multicenter validation study. Antimicrob. Agents Chemother. 45:1500-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Versalovic, J., D. Shortridge, K. Kibler, M. V. Griffy, J. Beyer, R. K. Flamm, S. K. Tanaka, D. Y. Graham, and M. F. Go. 1996. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 40:477-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wittwer, C. T., K. M. Ririe, R. V. Andrew, D. A. David, R. A. Gundry, and U. J. Balis. 1997. The LightcyclerTM: a microvolume multisample fluorimeter with rapid temperature control. BioTechniques 22:176-181. [DOI] [PubMed] [Google Scholar]