Abstract
Of 120 erythromycin-resistant pneumococci isolated in Italian hospitals, 39 (32.5%) were M-type isolates, carrying the mef gene alone. The mef gene was also detected, together with erm(AM), in one constitutively resistant isolate and in five isolates of the partially inducible phenotype. Among the 45 mef-positive isolates, 25 (55.6%) carried mef(A) and 20 (44.4%) carried mef(E) as observed from PCR-restriction fragment length polymorphism analysis of a 1,743-bp amplicon. The same result was obtained by a similar method applied to a more common 348-bp amplicon.
Macrolide resistance in Streptococcus pneumoniae is due to either target site modification—generally depending on a posttranscriptional modification of 23S rRNA mediated by erm-class methylases (39) or less often on mutations in 23S rRNA or ribosomal proteins (7, 37, 38)—or active efflux. In this study, 120 erythromycin-resistant pneumococci were investigated for macrolide resistance phenotypes and genotypes, with emphasis on the discrimination between mef(A) and mef(E).
Efflux-mediated resistance.
The efflux mechanism, which reduces the intracellular antibiotic concentration to subtoxic levels (40), is associated with a resistance pattern (M phenotype) characterized by resistance, among macrolides-lincosamides-streptogramin B (MLS), only to 14- and 15-membered macrolides, usually at a low level (34). M-type resistance is mediated by the mef gene, two variants of which are conventionally described: one, mef(A), originally discovered in Streptococcus pyogenes (5), and the other, mef(E), originally discovered in S. pneumoniae (36). Considering that the mef genes are detected mostly by a PCR method unable to distinguish between the two variants (33) and that mef(A) and mef(E) have 90% identity (36), they are regarded as a single gene class, designated mef(A) (28). However, mef(A) and mef(E) have recently been shown to be carried by different genetic elements in S. pneumoniae, which are inserted at different sites in the chromosome. Both elements carry, adjacent to mef, an open reading frame showing homology to the msr(A) gene associated with macrolide efflux in Staphylococcus aureus (6, 12, 29). Due to a number of important differences in the properties of mef(A)- and mef(E)-carrying pneumococci, it has been recommended that the distinction between the two genes be maintained (6).
Bacterial strains and typing.
The 120 erythromycin-resistant pneumococci studied (for which the MIC of erythromycin was ≥1 μg/ml) were independent isolates, collected from several laboratories throughout Italy between 1999 and 2002. All were clinical strains, isolated from a variety of clinical specimens (upper respiratory tract material, sputum, blood, cerebrospinal fluid). Multiple isolates from the same patient were excluded. Strain identification was confirmed in our laboratory by conventional tests such as susceptibility to optochin and solubility in bile and by employing the API system (BioMérieux, Marcy-l'Etoile, France). Serotyping, done by the capsular swelling test using specific antisera from the Statens Seruminstitut, Copenhagen, Denmark, showed that the 120 isolates were distributed over 24 serotypes, with the majority (80 isolates) belonging to five serotypes with more than 10 isolates each (23F, 14, 19A, 19F, and 6B). Typing by random amplified polymorphic DNA (RAPD) analysis as described previously (15) showed that the 120 isolates were distributed over 59 and 53 RAPD types by using primers M13 and ERIC1 (18), respectively.
Macrolide resistance phenotypes.
The M phenotype associated with efflux-mediated resistance has been described above. Methylase-mediated coresistance to MLS antibiotics can be expressed either constitutively, with high-level resistance to all MLS antibiotics (cMLS phenotype), or inducibly (iMLS phenotype); most often, however, inducibility is in regard to macrolides, particularly 16-membered ones, but not lincosamides, to which these strains are usually resistant without induction (iMcLS phenotype) (23). The macrolide resistance phenotype was determined for all isolates by the triple-disk (erythromycin plus clindamycin and rokitamycin) test and macrolide MIC induction tests as described previously (23) (Table 1). MICs were determined by the broth microdilution method according to the procedure recommended by the National Committee for Clinical Laboratory Standards (25). Compared with the distribution of strains into macrolide resistance phenotypes reported in previous studies, the rate of cMLS-type isolates rose further, from <2% among the erythromycin-resistant pneumococci isolated from 1996 to 1999 (13) and 12% among those isolated from 1998 to 2000 (23) to 17% in this study of strains isolated from 1999 to 2002. M-type isolates, accounting for approximately one-third of the erythromycin-resistant isolates in this study, were also apparently on the increase compared with rates reported in previous Italian studies and ranging from <10% (21) to 20 to 26% (6, 13, 20, 23). Moreover, we observed (for the first time in our experience) two true iMLS-type isolates, i.e., isolates showing inducibly expressed resistance not only to macrolides but also to clindamycin.
TABLE 1.
Macrolide resistance phenotypes and correlations with erythromycin resistance genes and serotypes in 120 clinical isolates of erythromycin-resistant S. pneumoniae
| Phenotype of macrolide resistance | No. (%) of strains | Antibiotica | MIC (μg/ml)b
|
No. of strains with gene:
|
Serotype (no. of strains) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Range | 50% | 90% | erm(AM) | mef(A) | mef(E) | ||||
| cMLS | 20 (16.7) | Erythromycin | 64->128 | 128 | >128 | 20 | 1 | 23F (5), 19F (4), 6B (3), 14 (3), 3 (2), 9A (1), 15A (1), 19A (1) | |
| Clindamycin | 128->128 | >128 | >128 | ||||||
| Clindamycin (ind.) | >128 | >128 | >128 | ||||||
| Rokitamycin | 4->128 | 32 | >128 | ||||||
| Rokitamycin (ind.) | >128 | >128 | >128 | ||||||
| iMLS | 2 (1.7) | Erythromycin | 64->128 | 2 | 7F (1), 23F (1) | ||||
| Clindamycin | 0.06-0.12 | ||||||||
| Clindamycin (ind.) | >128 | ||||||||
| Rokitamycin | ≤0.03-0.25 | ||||||||
| Rokitamycin (ind.) | >128 | ||||||||
| iMcLS | 59 (49.2) | Erythromycin | 4->128 | 128 | >128 | 59 | 1 | 4 | 19A (11), 23F (10), 19F (9), 6B (6), 3 (5), 6A (3), 10F (3), 14 (3), 10A (2), 23A (2), 9V (1), 15A (1), 20 (1), 33F (1), 36 (1) |
| Clindamycin | 2->128 | 64 | >128 | ||||||
| Clindamycin (ind.) | 32->128 | >128 | >128 | ||||||
| Rokitamycin | ≤0.03-1 | 0.25 | 1 | ||||||
| Rokitamycin (ind.) | 4->128 | >128 | >128 | ||||||
| M | 39 (32.5) | Erythromycin | 2-16 | 8 | 16 | 23 | 16 | 14 (14), 23F (2), 19A (2), 3 (2), 6B (1), 15B (1), 33A (1)c | |
| Clindamycin | ≤0.03-0.12 | ≤0.03 | 0.12 | ||||||
| Clindamycin (ind.) | ≤0.03-0.25 | 0.06 | 0.12 | 23A (3), 23F (3), 1 (2), 6B (1), 9V (1), 10F (1), 11A (1), 12F (1) 18F (1), 19A (1), 35 (1)d | |||||
| Rokitamycin | ≤0.03-0.25 | ≤0.03 | 0.25 | ||||||
| Rokitamycin (ind.) | ≤0.03-0.25 | ≤0.03 | 0.25 | ||||||
ind., after induction by pregrowth in 0.05 μg of erythromycin per ml.
50% and 90%, MICs at which 50 and 90% of isolates, respectively, are inhibited.
Isolates carrying the mef(A) gene.
Isolates carrying the mef(E) gene.
Resistance genes.
While efflux-mediated resistance is encoded by the above-mentioned mef genes, methylase-mediated resistance is usually encoded by the conventional erm(AM) gene, which belongs to gene class erm(B) (28). Another methylase, mediated by the erm(TR) gene, belonging to gene class erm(A) (28), was first described (31) and then found to be extensively present (14, 19) in S. pyogenes, whereas its presence in S. pneumoniae has been reported only occasionally (4, 35). The presence of erythromycin resistance genes was investigated by PCR. Primer pairs specific for the detection of erm(AM) and erm(TR) were as reported by Sutcliffe et al. (expected amplicon size, 639 bp) (33) and by Seppälä et al. (primers III8 and III10; expected amplicon size, 208 bp) (31), respectively. Two primer pairs were used to detect the mef gene: the one described by Sutcliffe et al. (expected amplicon size, 348 bp) (33), and the one described by Del Grosso et al. [primers MEF3 and MEF4, derived from Tait-Kamradt et al. (36); expected amplicon size, 1,743 bp] (6). All cMLS, iMLS, and iMcLS isolates had the erm(AM) gene, and all M-type isolates had the mef gene. The latter gene was also detected, besides erm(AM), in one cMLS isolate and in five iMcLS isolates (Table 1). No isolate had the erm(TR) gene.
Discrimination between mef(A) and mef(E).
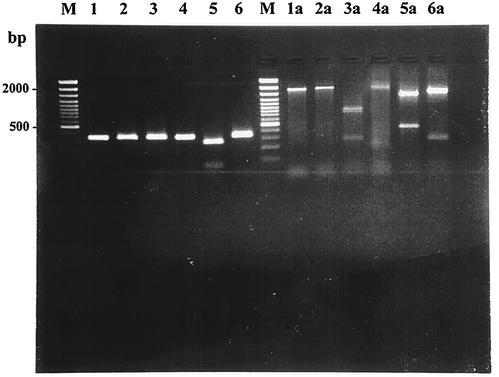
mef(A) and mef(E) were distinguished by PCR-restriction fragment length polymorphism (RFLP) analysis as suggested by Del Grosso et al. (6), i.e., by digesting the 1,743-bp amplicon with restriction endonucleases (New England Biolabs, Beverly, Mass.) BamHI [which has no restriction site in mef(E) and one in mef(A), generating two fragments of 1,340 and 403 bp] and DraI [which has two restriction sites in mef(E), generating three fragments of 782, 711, and 250 bp, and one in mef(A) generating two fragments of 1,493 and 250 bp]. In this way, of the 39 M-type isolates, 23 were found to carry mef(A) and 16 were found to carry mef(E); of the five mef-positive iMcLS-type isolates, four carried mef(E) and one carried mef(A); and the mef-positive cMLS-type isolate carried mef(A) (Table 1). In additional experiments, we attempted to apply PCR-RFLP analysis to the 348-bp amplicon yielded by the primers described by Sutcliffe et al. (33). Complete overlap with the above-reported results of the discrimination between mef(A) and mef(E) was obtained by digesting the 348-bp amplicon with BamHI, which had no restriction site in mef(E) and one in mef(A) generating two fragments of 284 and 64 bp; DraI cut neither mef(A) nor mef(E) (Fig. 1). Aliquots of 10 μl of the PCR product were digested with 1 U of enzyme following the instructions of the manufacturer.
FIG. 1.
PCR-RFLP analysis of two M-type erythromycin-resistant pneumococci, one carrying the mef(E) gene (lanes 1 to 3 and 1a to 3a) and the other carrying the mef(A) gene (lanes 4 to 6 and 4a to 6a). Lanes 1 and 4, PCR amplicon (348 bp) from the mef primers designed by Sutcliffe et al. (34); lanes 1a and 4a, PCR amplicon (1,743 bp) from primers MEF3 and MEF4 designed by Del Grosso et al. (6); lanes 2, 5, 2a, and 5a, restriction patterns from BamHI enzyme; lanes 3, 6, 3a, and 6a, restriction patterns from DraI enzyme; lanes M, 100-bp DNA ladder.
Correlations between mef genes and serotypes.
Fourteen of the 23 isolates carrying mef(A) belonged to serotype 14, the remaining 9 being distributed over six other serotypes (Table 1). The 16 isolates carrying mef(E) were more scattered (over 11 serotypes), with no more than 3 isolates in each serotype (23A and 23F). In another recent Italian study of 20 M-type pneumococci, all of the 17 isolates carrying mef(A) belonged to serotype 14, whereas the 3 isolates carrying mef(E) belonged to different serotypes (6).
Distribution of M-type pneumococci.
The distribution of phenotypes and genotypes of erythromycin-resistant pneumococci may vary considerably from area to area. As regards M-type isolates, recent Italian rates ranging between one-third (in the present study) and one-fifth (6, 13, 20, 23) of erythromycin-resistant isolates contrast with the complete absence of M-type pneumococci in recent surveys carried out in a country as close as France (1, 10). Rates of M-type isolates similar to those found in Italy have been reported in Greece (35), in a multinational European study (30), in Georgia (United States) (11), and in Taiwan (17); lower rates have been reported in Spain (32), Belgium (8), Central and Eastern European countries (24), and South Africa (22); higher rates have been reported in Germany (27), Japan (26), Canada (16), and in a nationwide study in the United States (9).
mef(A) and mef(E) genes in M-type pneumococci.
Far fewer data are available to assess the actual and different contributions of the mef(A) and mef(E) genes to M-type erythromycin resistance in pneumococci. Our finding of a prevalence of mef(A) over mef(E) is consistent with the results of another recent investigation in Italy (6): in our study, however, the mef(A)-to-mef(E) ratio was lower, and the mef(A)-positive strains were distributed over seven serotypes rather than belonging to serotype 14 only (6).
Conclusions.
The custom of designating the mef gene as mef(A) or mef(E) depending on its detection in S. pyogenes or S. pneumoniae rather than on the basis of specific, molecular differentiation should be reconsidered. The discrimination of mef(A) from mef(E) by RFLP analysis of the 348-bp PCR fragment (33) using a single endonuclease (BamHI) suggested herein is simple and rapid and should be recommended for future studies of M-type pneumococci. It would be interesting to see whether a similar mixed occurrence of mef(A) and mef(E) is also found in the M-type strains of S. pyogenes, which are usually considered to be associated with the mef(A) gene but without any specific attempt at discriminating between the two mef variants. In at least one study (2), both mef(A) and mef(E) have been separately detected in M-type strains of S. pyogenes. The same investigators also separately detected both mef(A) and mef(E) in M-type strains of Streptococcus agalactiae (3).
Acknowledgments
This work was supported in part by a grant from the Italian Ministry of Education, University and Research.
REFERENCES
- 1.Angot, P., M. Vergnaud, M. Auzou, R. Leclercq, and Observatoire de Normandie du Pneumocoque. 2000. Macrolide resistance phenotypes and genotypes in French clinical isolates of Streptococcus pneumoniae. Eur. J. Clin. Microbiol. Infect. Dis. 19:755-758. [DOI] [PubMed] [Google Scholar]
- 2.Arpin, C., M. H. Canron, P. Noury, and C. Quentin. 1999. Emergence of mefA and mefE genes in beta-haemolytic streptococci and pneumococci in France. J. Antimicrob. Chemother. 44:133-138. [DOI] [PubMed] [Google Scholar]
- 3.Arpin, C., H. Daube, F. Tessier, and C. Quentin. 1999. Presence of mefA and mefE genes in Streptococcus agalactiae. Antimicrob. Agents Chemother. 43:944-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betriu, C., M. Redondo, M. L. Palau, A. Sánchez, M. Gómez, E. Culebras, A. Boloix, and J. J. Picazo. 2000. Comparative in vitro activities of linezolid, quinupristin-dalfopristin, moxifloxacin, and trovafloxacin against erythromycin-susceptible and -resistant streptococci. Antimicrob. Agents Chemother. 44:1838-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clancy, J., J. Petitpas, F. Dib-Hajj, W. Yuan, M. Cronan, A. V. Kamath, J. Bergeron, and J. A. Retsema. 1996. Molecular cloning and functional analysis of a novel macrolide resistance determinant, mefA, from Streptococcus pyogenes. Mol. Microbiol. 22:867-879. [DOI] [PubMed] [Google Scholar]
- 6.Del Grosso, M., F. Iannelli, C. Messina, M. Santagati, N. Petrosillo, S. Stefani, G. Pozzi, A. Pantosti. 2002. Macrolide efflux genes mef(A) and mef(E) are carried by different genetic elements in Streptococcus pneumoniae. J. Clin. Microbiol. 40:774-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Depardieu, F., and P. Courvalin. 2001. Mutation in 23S rRNA responsible for resistance to 16-membered macrolides and streptogramins in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:319-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Descheemaeker, P., S. Chapelle, C. Lammens, M. Hauchecorne, M. Wijdooghe, P. Vandamme, M. Ieven, and H. Goessens. 2000. Macrolide resistance and erythromycin resistance determinants among Belgian Streptococcus pyogenes and Streptococcus pneumoniae isolates. J. Antimicrob. Chemother. 45:167-173. [DOI] [PubMed] [Google Scholar]
- 9.Doern, G. V., K. P. Heilmann, H. K. Huynh, P. R. Rhomberg, S. L. Coffman, and A. B. Brueggemann. 2001. Antimicrobial resistance among clinical isolates of Streptococcus pneumoniae in the United States during 1999-2000, including comparison of resistance rates since 1994-1995. Antimicrob. Agents Chemother. 45:1721-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitoussi, F., C. Doit, P. Geslin, N. Brahimi, and E. Bingen. 2001. Mechanism of macrolide resistance in clinical pneumococcal isolates in France. Antimicrob. Agents Chemother. 45:636-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gay, K., W. Baughman, Y. Miller, D. Jackson, C. G. Whitney, A. Schuchat, M. M. Farley, F. Tenover, and D. S. Stephens. 2000. The emergence of Streptococcus pneumoniae resistant to macrolide antimicrobial agents: a 6-year population-based assessment. J. Infect. Dis. 182:1417-1424. [DOI] [PubMed] [Google Scholar]
- 12.Gay, K., and D. S. Stephens. 2001. Structure and dissemination of a chromosomal insertion element encoding macrolide efflux in Streptococcus pneumoniae. J. Infect. Dis. 184:56-65. [DOI] [PubMed] [Google Scholar]
- 13.Giovanetti, E., M. P. Montanari, F. Marchetti, and P. E. Varaldo. 2000. In vitro activity of ketolides telithromycin and HMR 3004 against Italian isolates of Streptococcus pyogenes and Streptococcus pneumoniae with different erythromycin susceptibility. J. Antimicrob. Chemother. 46:905-908. [DOI] [PubMed] [Google Scholar]
- 14.Giovanetti, E., M. P. Montanari, M. Mingoia, and P. E. Varaldo. 1999. Phenotypes and genotypes of erythromycin-resistant Streptococcus pyogenes strains in Italy and heterogeneity of inducibly resistant strains. Antimicrob. Agents Chemother. 43:1935-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruteke, P., A. van Belkum, L. M. Schouls, W. D. H. Hendriks, F. A. G. Reubsaet, J. Dokter, H. Boxma, and H. A. Verbrugh. 1996. Outbreak of group A streptococci in a burn center: use of pheno- and genotypic procedures for strain tracking. J. Clin. Microbiol. 34:114-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoban, D. J., A. K. Wierzbowski, K. Nichol, and G. G. Zhanel. 2001. Macrolide-resistant Streptococcus pneumoniae in Canada during 1998-1999: prevalence of mef(A) and erm(B) and susceptibility to ketolides. Antimicrob. Agents Chemother. 45:2147-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsueh, P. R., L. J Teng, L. N. Lee, P. C. Yang, S. W. Ho, C. H. Lue, and K. T. Luh. 2002. Increased prevalence of erythromycin resistance in streptococci: substantial upsurge in erythromycin-resistant M phenotype in Streptococcus pyogenes (1979-1998) but not in Streptococcus pneumoniae (1985-1999) in Taiwan. Microb. Drug Resist. 8:27-33. [DOI] [PubMed] [Google Scholar]
- 18.Hsueh, P. R., L. J Teng, L. N. Lee, P. C. Yang, S. W. Ho, and K. T. Luh. 1999. Dissemination of high-level penicillin-, extended-spectrum cephalosporin-, and erythromycin-resistant Streptococcus pneumoniae clones in Taiwan. J. Clin. Microbiol. 37:221-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kataja, J., P. Huovinen, the Macrolide Resistance Study Group, and H. Seppälä. 2000. Erythromycin resistance genes in group A streptococci of different geographical origins. J. Antimicrob. Chemother. 46:789-792. [DOI] [PubMed] [Google Scholar]
- 20.Latini, L., M. P. Ronchetti, R. Merolla, F. Guglielmi, S. Bajaksouzian, M. P. Villa, M. R. Jacobs, and R. Ronchetti. 1999. Prevalence of mefE, erm and tet(M) genes in Streptococcus pneumoniae strains from central Italy. Int. J. Antimicrob. Agents 13:29-33. [DOI] [PubMed] [Google Scholar]
- 21.Marchese, A., E. Tonoli, E. A. Debbia, and G. C. Schito. 1999. Macrolide resistance mechanisms and expression of phenotypes among Streptococcus pneumoniae circulating in Italy. J. Antimicrob. Chemother. 44:461-464. [DOI] [PubMed] [Google Scholar]
- 22.McGee, L., K. P. Klugman, A. Wasas, T. Capper, A. Brink, and the Antibiotics Surveillance Forum of South Africa. 2001. Serotype 19F multiresistant pneumococcal clone harboring two erythromycin resistance determinants [erm(B) and mef(A)] in South Africa. Antimicrob. Agents Chemother. 45:1595-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montanari, M. P., M. Mingoia, E. Giovanetti, and P. E. Varaldo. 2001. Differentiation of resistance phenotypes among erythromycin-resistant pneumococci. J. Clin. Microbiol. 39:1311-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagai, K., P. C. Appelbaum, T. A. Davies, L. M. Kelly, D. B. Hoellman, A. T. Andrasevic, L. Drukalska, W. Hryniewicz, M. R. Jacobs, J. Kolman, J. Miciuleviciene, M. Pana, L. Setchanova, M. K. Thege, H. Hupkova, J. Trupl, and P. Urbaskova. 2002. Susceptibilities to telithromycin and six other agents and prevalence of macrolide resistance due to L4 ribosomal protein mutation among 992 pneumococci from 10 Central and Eastern European countries. Antimicrob. Agents Chemother. 46:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 26.Nishijima, T., Y. Saito, A. Aoki, M. Toriya, Y. Toyonaga, and R. Fujii. 1999. Distribution of mefE and ermB genes in macrolide-resistant strains of Streptococcus pneumoniae and their variable susceptibility to various antibiotics. J. Antimicrob. Chemother. 43:637-643. [DOI] [PubMed] [Google Scholar]
- 27.Reinert, R. R., A. Al-Lahham, M. Lemperle, C. Tenholte, C. Briefs, S. Haupts, H. H. Gerards, and R. Lütticken. 2002. Emergence of macrolide and penicillin resistance among invasive pneumococcal isolates in Germany. J. Antimicrob. Chemother. 49:61-68. [DOI] [PubMed] [Google Scholar]
- 28.Roberts, M. C., J. Sutcliffe, P. Courvalin, L. B. Jensen, J. Rood, and H. Seppälä. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santagati, M., F. Iannelli, F., M. R. Oggioni, S. Stefani, and G. Pozzi. 2000. Characterization of a genetic element carrying the macrolide efflux gene mef(A) in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:2585-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitz, F. J., M. Perdikouli, A. Beeck, J. Verhoef, and A. C. Fluit. 2001. Molecular surveillance of macrolide, tetracycline and quinolone resistance mechanisms in 1191 clinical European Streptococcus pneumoniae isolates. Int. J. Antimicrob. Agents 18:433-436. [DOI] [PubMed] [Google Scholar]
- 31.Seppälä, H., M. Skurnik, H. Soini, M. C. Roberts, and P. Huovinen. 1998. A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob. Agents Chemother. 42:257-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seral, C., F. J. Castillo, M. C. Rubio-Calvo, A. Fenoll, C. García, and R. Gómez-Lus. 2001. Distribution of resistance genes tet(M), aph3′-III, catpC194 and the integrase gene of Tn1545 in clinical Streptococcus pneumoniae harbouring erm(B) and mef(A) genes in Spain. J. Antimicrob. Chemother. 47:863-866. [DOI] [PubMed] [Google Scholar]
- 33.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sutcliffe, J., A. Tait-Kamradt, and L. Wondrack. 1996. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob. Agents Chemother. 40:1817-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Syrogiannopoulos, G. A., I. N. Grivea, A. Tait-Kamradt, G. D. Katopodis, N. G. Beratis, J. Sutcliffe, P. C. Appelbaum, and T. A. Davies. 2001. Identification of an erm(A) erythromycin resistance methylase gene in Streptococcus pneumoniae isolated in Greece. Antimicrob. Agents Chemother. 45:342-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tait-Kamradt, A., J. Clancy, M. Cronan, F. Dib-Hajj, L. Wondrack, W. Yuan, and J. Sutcliffe. 1997. mefE is necessary for the erythromycin-resistant M phenotype in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41:2251-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tait-Kamradt, A., T. Davies, P. C. Appelbaum, F. Depardieu, P. Courvalin, J. Petipas, L. Wondrack, A. Walker, M. R. Jacobs, and J. Sutcliffe. 2000. Two new mechanisms of macrolide resistance in clinical strains of Streptococcus pneumoniae from Eastern Europe and North America. Antimicrob. Agents Chemother. 44:3395-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tait-Kamradt, A., T. Davies, M. Cronan, M. R. Jacobs, P. C. Appelbaum, and J. Sutcliffe. 2000. Mutations in 23S rRNA and L4 ribosomal protein account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob. Agents Chemother. 44:2118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weisblum, B. 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39:577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong, P., and V. D. Shortridge. 2000. The role of efflux in macrolide resistance. Drug Resist. Updat. 3:325-329. [DOI] [PubMed] [Google Scholar]