Abstract
One hundred thirty-two methicillin-resistant Staphylococcus aureus (MRSA) isolates recovered from patients with S. aureus infections between January 1998 and February 1999 in two hospitals, one located in Taipei, Taiwan, and another in Nanjing, People's Republic of China, were examined for antibiotic susceptibility and for clonal type by a combination of three methods: hybridization of ClaI restriction digests with mecA- and Tn554-specific DNA probes and pulsed-field gel electrophoresis of chromosomal SmaI digests. Selected isolates representing each clonal type were also analyzed by spaA typing, multilocus sequence typing, and a multiplex PCR method capable of identifying the structural type of the staphylococcal cassette chromosome mec (SCCmec) carried by the bacteria. The overwhelming majority of isolates (126 of 132 or 95%) belonged to minor variants of a single clonal type resembling the Brazilian and Hungarian epidemic MRSA clones, which showed a common spaA type and which were either sequence type 239 (ST239) or ST241 (a single-locus variant of ST239) in association with SCCmec type III or IIIA.
Methicillin-resistant Staphylococcus aureus (MRSA) is a serious problem in both Taiwan and China. The frequency of nosocomial infections caused by MRSA in Taiwan has increased rapidly during the past 10 years (4). In most major hospitals, MRSA accounts for more than 60% of the S. aureus isolates (30). According to a study from 1991 to 1996 at the Veterans General Hospital of Taipei, the prevalence of MRSA in this hospital was estimated as 88.2% (28). Furthermore, community-acquired MRSA infections seem to occur frequently in Taiwan among patients (infants and children) with no associated risk factors (31). In China the incidence of MRSA from hospital infection can be over 80% and the incidence of MRSA from community-acquired infections is close to 22% (14).
The Center for Molecular Epidemiology and International Network (CEM/NET) has been created to keep track of the movement of major multidrug-resistant clones of S. aureus and other gram-positive pathogens and to identify their reservoirs (6). Under this initiative, clinical isolates of MRSA collected in different countries were analyzed by molecular typing techniques involving ClaI-mecA polymorphisms, Tn554 insertion patterns, and pulsed-field gel electrophoresis (PFGE). These techniques have allowed us to identify so far five multiresistant MRSA clones (22). The names assigned to these five pandemic MRSA clones, Iberian (8, 25), Brazilian (27), Hungarian (7, 18), New York/Japan (1, 23), and pediatric (24), reflect the geographic area in which they were first identified or indicate some unique epidemiological property. Enright et al. (10) proposed a different nomenclature for these clones based on their sequence types (ST) (9) and staphylococcal cassette chromosome mec (SCCmec) types (I through IV) (11, 12). If subtypes IA and IIIA are included (20), the designations of the five pandemic clones are ST247-IA, ST239-IIIA, ST239-III, ST5-II, and ST5-IV, respectively (10, 19).
The aim of this study was to characterize a collection of clinical MRSA strains from Taiwan and China and to evaluate their geographic spread by comparison with information included in the CEM/NET and multilocus sequence typing (MLST) (http://www.mlst.net) databases.
MATERIALS AND METHODS
Bacterial isolates.
One hundred thirty-two clinical MRSA isolates were collected from January 1998 to February 1999 from infection sites of individual patients.
Hospitals.
Most of the isolates (n = 118) were from the Tri-Service General Hospital/National Defense Medical Center of Taipei, Taiwan, a 1,200-bed medical center. A small sample (14 isolates) was received from the First Teaching Hospital, Nanjing, People's Republic of China, a 950-bed general hospital.
Susceptibility tests.
Susceptibility tests were performed by standard disk diffusion method according to National Committee for Clinical Laboratory Standards guidelines (17). Drugs tested included penicillin, oxacillin, trimethoprim-sulfamethoxazole, ciprofloxacin, chloramphenicol, clindamycin, erythromycin, gentamicin, rifampin, tetracycline, vancomycin, and teicoplanin. Spectinomycin and quinupristin-dalfopristin susceptibilities were determined as described previously (1).
Molecular typing.
Southern blot hybridization of ClaI digests with mecA and Tn554 DNA probes (5), PFGE of SmaI digests of chromosomal DNAs (5), spaA typing (26), and MLST (9) were performed as previously described. The SCCmec types were determined by a multiplex PCR strategy (19) or by PCR amplification of the ccr (cassette chromosome recombinase) gene (12) when ambiguous results were obtained with the previous methodology.
RESULTS AND DISCUSSION
All isolates studied were multiresistant (Table 1). There were, however, differences among strains from the two countries; for instance, resistance or intermediate resistance to chloramphenicol was found in each of the 14 isolates from China but only in about one-half (43%) of the 118 isolates from Taiwan, while resistance to trimethoprim-sulfamethoxazole was found in 89% of the isolates from Taiwan but in only 21% of the isolates from China. None of the strains was resistant to teicoplanin, vancomycin, and quinupristin-dalfopristin. The majority of the isolates were also susceptible to rifampin (94%) and to 500 mg of spectinomycin/liter (94%).
TABLE 1.
Phenotypic and genotypic properties of 132 MRSA isolates from Taipei, Taiwan, and Nanjing, China
| ClaI-mecA::Tn554 typea | PFGE type | No. of isolates | Clonal typeb | Antimicrobial resistance (of the majority [>50%] of the isolates)c | spaA type | MLST type | ST | SCCmec type |
|---|---|---|---|---|---|---|---|---|
| Taiwan isolates | ||||||||
| III′::B | A1 | 45 | III::B::A (n = 97; 82%) | PEN, OXA, CIP, CLI, ERY, TET, GEN, SXT | WGKAOMQ | 2-3-1-1-4-4-30 | 241 | III |
| III′::B | A7 | 4 | ||||||
| IX::B | A2 | 36 | WGKAOMQ | 2-3-1-1-4-4-30 | 241 | IIIA | ||
| IX::B | A4 | 6 | ||||||
| IX::B | A5 | 1 | ||||||
| IX::B | A9 | 1 | ||||||
| IX::B | A10 | 1 | ||||||
| XI::B | A8 | 3 | WGKAOMQ | III | ||||
| XIV::B | A1 | 3 | XIV::B::A | PEN, OXA, CIP, CLI, ERY, TET, GEN, SXT | WGKAOMQ | III | ||
| XIV::B | A3 | 2 | ||||||
| XIV::B | A6 | 1 | ||||||
| XIV::B | A9 | 1 | ||||||
| X′::B | A3 | 5 | X′::B::A | PEN, OXA, CIP, CLI, ERY, TET, GEN | WGKAOMQ | IIIA | ||
| III′::AA | A1 | 1 | III′::AA::A | PEN, OXA, CIP, CLI, ERY, TET, GEN, SXT, CHL | WGKAOMQ | III | ||
| XI::B | C | 2 | XI::B::C | PEN, OXA, CIP, CLI, ERY, TET, GEN, SXT, CHL, RIF | WGKAOMQ | 2-3-1-1-4-4-3 | 239 | IIIA |
| II::NH | B1 | 1 | II::NH::B | PEN, OXA, CLI, ERY, TET, GEN, CHL | ZDMDMOB | 19-23-15-2-19-20-15 | 59 | IV |
| II::NH | B2 | 3 | ||||||
| II::ZZ | D | 2 | II::ZZ::D | PEN, OXA, CLI, ERY, TET, GEN, CHL, RIF | NTd | 3-32-1-1-4-4-3 | 81 | II variant |
| China isolates | ||||||||
| III′::B | A11 | 7 | III::B::A (n = 10; 71%) | PEN, OXA, CIP, CLI, ERY, TET, GEN, CHL | WGKAOMQ | 2-3-1-1-4-4-3 | 239 | III |
| III′::B | A12 | 1 | ||||||
| III′::B | A16 | 2 | ||||||
| X′::B | A13 | 1 | X′::B::A | PEN, OXA, CIP, CLI, ERY, TET, GEN, CHL | WGKAOMQ | IIIA | ||
| X′::B | A14 | 2 | ||||||
| X′::B | A15 | 1 |
ClaI-mecA polymorphs and respective hybridization fragment sizes: III′, 1.9 and 4.5 kb; IX and XI, see reference 8; XIV, 1.9 and 4.3 kb; X′, 1.9 and 6 kb; II, see reference 13. Tn554 polymorphs: NH, no homology, lack of transposon; B and AA, see reference 13; ZZ, novel pattern (9.2, 7.1, 6.7, 6.1, and 4.1 kb).
ClaI-mecA polymorphs III′, IX, and XI present minor variations compared to pattern III and were found to be genetically related (21). Therefore strains belonging to these mecA polymorphs were grouped under type III for the definition of clonal types.
Abbreviations: PEN, penicillin; OXA, oxacillin; CIP, ciprofloxacin; CLI, clindamycin; ERY, erythromycin; TET, tetracycline; GEN, gentamicin; SXT, trimethoprim-sulfamethoxazole; CHL, chloramphenicol; RIF, rifampin.
NT, there was no amplification with the set of primers used.
While the 132 MRSA isolates from Taiwan and China could be separated into six ClaI-mecA polymorphs and three ClaI-Tn554 polymorphs (Table 1), the great majority of isolates were characterized as ClaI-mecA polymorph III (13) or the genetically related polymorphs IX, XI, and III′ (21) (n = 110) and Tn554 polymorph B (13) (n = 125).
The PFGE analysis grouped the 132 MRSA strains into four types (Table 1), of which pattern A was assigned to a surprisingly large proportion of the isolates (Taiwan, n = 110, 93%; China, n = 14, 100%). This pattern showed 10 different subtypes in Taiwan and 6 in China, but by far the most frequent were A1 in Taiwan (n = 49, 42%) and A11 in China (n = 7, 50%). There are no common PFGE subtypes shared by the strains from the hospitals in the two countries.
The combination of the three molecular typing methods (ClaI-mecA and ClaI-Tn554 hybridization and PFGE) distributed the 118 MRSA isolates from Taiwan and the 14 isolates from China into seven and two clonal types, respectively (Table 1). However, the overwhelming majority of the strains from Taipei, Taiwan, and Nanjing, China (82 and 71%, respectively), belonged to the same clone, III::B::A. Interestingly, this clone was apparently already present in 1994 in a hospital in another city of Taiwan (15). In addition, the MRSA strain responsible for a hospital-acquired outbreak initiated by a surgeon carrier in the beginning of 1997 in a hospital in northern Taiwan showed the same PFGE pattern, A (29). According to a recent study involving 22 hospitals distributed throughout Taiwan, the high prevalence of MRSA in the country was, at least in past, due to the spreading of a single predominant strain (30) having a PFGE pattern very similar, if not identical, to PFGE pattern A, shown by the majority of MRSA isolates that we studied (Fig. 1). It is probable that clone III::B::A was spread in different cities in Taiwan and had at least a 5-year existence in the country.
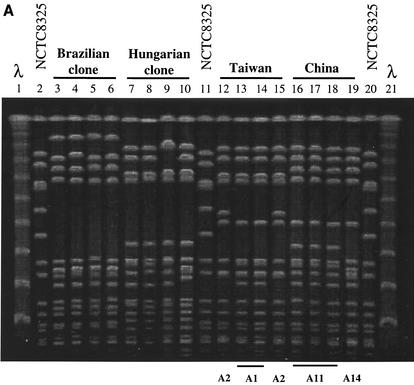
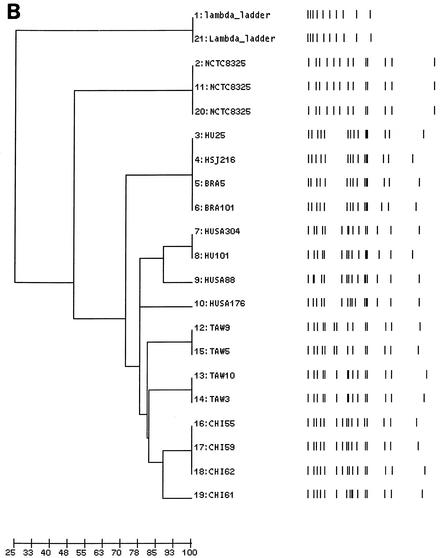
FIG. 1.
(A) PFGE of SmaI macrorestriction fragments of MRSA clinical isolates belonging to the China/Taiwan, Hungarian (7, 18), and Brazilian clones (2, 3, 27). Lanes 1 and 21, lambda molecular weight marker; lanes 2, 11, and 20, reference strain NCTC 8325; lanes 3 to 6, HU25, HSJ216, BRA5, and BRA101, respectively; lanes 7 to 10, HUSA304, HU101, HUSA88, and HUSA176, respectively; lanes 12 to 15, TAW9 (A2), TAW10 (A1), TAW3 (A1), and TAW5 (A2), respectively; lanes 16 to 19, CHI55 (A11), CHI59 (A11), CHI62 (A11), and CHI61 (A14), respectively. (B) Computer-generated dendrogram based on Jaccard matching of pattern similarity of the PFGE of Fig. 1A. The scale at the bottom of the dendrogram represents similarity.
Representatives of PFGE pattern A were compared to strains belonging to previously characterized clones (Fig. 1A), namely, the Brazilian and the Hungarian MRSA clones, which share some molecular properties (identical mecA and Tn554 polymorphisms). It was found that PFGE type A showed a high degree of similarity with both clones, in particular with the Hungarian MRSA (Fig. 1A and B).
Several selected isolates from Taiwan and China representing the major clonal lineage (PFGE type A) were also examined by spaA typing, MLST, and SCCmec typing (Table 1). All shared the same spaA repeat motif, WGKAOMQ, which was very similar to the one described for the Hungarian and Brazilian clones (18, 20). MLST analysis showed a single-locus variant on the basis of a comparison of the ST of isolates from Taiwan (ST241) and China (ST239): ST239 possesses allele 3 at yqiL, which is found in several other distantly related lineages, whereas ST241 possesses allele 30. According to information found in the MLST database, allele 30 is only found in ST241 and differs from allele 3 at a single nucleotide site, suggesting that allele 30 arose from allele 3 by a point mutation. Strains from China showed an ST identical to the one described for the Hungarian and Brazilian clones (ST239) (20). All isolates were assigned to SCCmec type III or to variant IIIA, two SCCmec types that differ mainly by the presence or absence of plasmid pT181 and that are characteristic of the Hungarian and Brazilian clones, respectively (19). ST241 and ST239 have also been detected in Thailand (10), which may indicate a possible spread of the Nanjing/Taipei clone to or from other Asiatic countries.
The application of spaA typing, MLST, and SCCmec assignment to isolates belonging to the minor clones XIV::B::A, X′::B::A, III′::AA::A, and XI::B::C confirmed that they were related to the major clonal lineage III::B::A. Representatives of two other minor groups of isolates were also characterized: four strains were clonal type II::NH::B, and two strains were clonal type II::ZZ::D. The strain representing clonal type II::NH::B had a rare spaA type and belonged to ST59, which was previously detected only in a few MSSA isolates and in a single MRSA isolate from the United States and which is considered one of the most divergent MRSA ST (10). The strain representing clonal type II::ZZ::D was nontypeable by spaA and belonged to a new multilocus ST (ST81), which is a double-locus variant of ST239.
Our study documents the dominance of variants of a single multiresistant clone (III::B::A or ST239- and ST241-III and -IIIA) among MRSA isolates (126 of 132 or 95%) from the hospitals in Taipei, Taiwan, and in Nanjing, People's Republic of China. This clone shows high degree of similarity in MLST type, spaA type, PFGE pattern, and the structural type of the SCCmec element to the Brazilian and Hungarian epidemic MRSA (22). MRSA with an identical genetic background (ST239) has already been recovered in 1982 in Australia and in 1987 in the United States (22), as well as in several countries of Europe, South America, and Asia since the 1990s (2, 10, 16). The results of our study document the dissemination of this highly epidemic MRSA lineage to Taiwan and China.
Acknowledgments
This work was partially supported by project POCTI/1999/ESP/34872 from Fundação para a Ciência e Tecnologia, Portugal, and by project SDH.IC.I.01.13 from Fundação Calouste Gulbenkian, Portugal, awarded to H. de Lencastre. M. Aires de Sousa was supported by grant BD/13731/97 from PRAXIS XXI, and M. I. Crisóstomo was supported by grant BD/5205/01 from Fundação para a Ciência e Tecnologia, Portugal. The strains from Taipei, Taiwan, and Nanjing, People's Republic of China, were obtained through Project Resist with a grant from Rhône-Poulenc Rorer, S. A., awarded to Alexander Tomasz and Hermínia de Lencastre. The Multilocus Sequence Typing website (http://www.mlst.net), which is hosted at Imperial College London and at the University of Bath, was used for the studies described in this work.
REFERENCES
- 1.Aires de Sousa, M., H. de Lencastre, I. Santos Sanches, K. Kikuchi, K. Totsuka, and A. Tomasz. 2000. Similarity of antibiotic resistance patterns and molecular typing properties of methicillin-resistant Staphylococcus aureus isolates widely spread in hospitals in New York City and in a hospital in Tokyo, Japan. Microb. Drug Resist. 6:253-258. [DOI] [PubMed] [Google Scholar]
- 2.Aires de Sousa, M., M. Miragaia, I. Santos Sanches, S. Ávila, I. Adamson, S. T. Casagrande, M. C. C. Brandileone, R. Palacio, L. Dell'Acqua, M. Hortal, T. Camou, A. Rossi, M. E. Velazquez-Meza, G. Echaniz-Aviles, F. Solorzano-Santos, I. Heitmann, and H. de Lencastre. 2001. Three-year assessment of methicillin-resistant Staphylococcus aureus clones in Latin America, 1996 to 1998. J. Clin. Microbiol. 39:2197-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aires de Sousa, M., I. Santos Sanches, M. L. Ferro, M. J. Vaz, Z. Saraiva, T. Tendeiro, J. Serra, and H. de Lencastre. 1998. Intercontinental spread of a multidrug-resistant methicillin-resistant Staphylococcus aureus clone. J. Clin. Microbiol. 36:2590-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, M. L., S. C. Chang, H. J. Pan, P. R. Hsueh, L. S. Yang, S. W. Ho, and K. T. Luh. 1999. Longitudinal analysis of methicillin-resistant Staphylococcus aureus isolates at a teaching hospital in Taiwan. J. Formos. Med. Assoc. 98:426-432. [PubMed] [Google Scholar]
- 5.de Lencastre, H., I. Couto, I. Santos, J. Melo-Cristino, A. Torres-Pereira, and A. Tomasz. 1994. Methicillin-resistant Staphylococcus aureus disease in a Portuguese hospital: characterization of clonal types by a combination of DNA typing methods. Eur. J. Clin. Microbiol. Infect. Dis. 13:64-73. [DOI] [PubMed] [Google Scholar]
- 6.de Lencastre, H., I. Santos Sanches, and A. Tomasz. 2000. CEM/NET: clinical microbiology and molecular biology in alliance, p. 451-456. In A. Tomasz (ed.), Streptococcus pneumoniae—molecular biology and mechanisms of disease. Mary Ann Liebert Inc., Larchmont, N.Y.
- 7.de Lencastre, H., E. P. Severina, H. Milch, M. K. Thege, and A. Tomasz. 1997. Wide geographic distribution of a unique methicillin-resistant Staphylococcus aureus clone in Hungarian hospitals. Clin. Microbiol. Infect. 3:289-296. [DOI] [PubMed] [Google Scholar]
- 8.Dominguez, M. A., H. de Lencastre, J. Liñares, and A. Tomasz. 1994. Spread and maintenance of a dominant methicillin-resistant Staphylococcus aureus (MRSA) clone during an outbreak of MRSA disease in a Spanish hospital. J. Clin. Microbiol. 32:2081-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiramatsu, K., L. Cui, M. Kuroda, and T. Ito. 2001. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 9:486-493. [DOI] [PubMed] [Google Scholar]
- 12.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kreiswirth, B., J. Kornblum, R. D. Arbeit, W. Eisner, J. Maslow, A. McGeer, D. E. Low, and R. Novick. 1993. Evidence for a clonal origin of methicillin resistance in Staphylococcus aureus. Science 259:227-230. [DOI] [PubMed] [Google Scholar]
- 14.Li, J., A. J. Weinstein, and M. Yang. 2001. Surveillance of bacterial resistance in China (1998-1999). Zhonghua Yi Xue Za Zhi 81:8-16. [PubMed] [Google Scholar]
- 15.Liu, P. Y., Z. Y. Shi, Y. J. Lau, B. S. Hu, J. M. Shyr, W. S. Tsai, Y. H. Lin, and C. Y. Tseng. 1996. Use of restriction endonuclease analysis of plasmids and pulsed-field gel electrophoresis to investigate outbreaks of methicillin-resistant Staphylococcus aureus infection. Clin. Infect. Dis. 22:86-90. [DOI] [PubMed] [Google Scholar]
- 16.Melter, O., I. Santos Sanches, J. Schindler, M. Aires de Sousa, R. Mato, V. Kovarova, H. Zemlickova, and H. de Lencastre. 1999. Methicillin-resistant Staphylococcus aureus clonal types in the Czech Republic. J. Clin. Microbiol. 37:2798-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Committee for Clinical Laboratory Standards. 1995. Performance standards for antimicrobial disk susceptibility test. National Committee for Clinical Laboratory Standards, Villanova, Pa.
- 18.Oliveira, D. C., I. Crisóstomo, I. Santos Sanches, P. Major, C. R. Alves, M. Aires de Sousa, M. K. Thege, and H. de Lencastre. 2001. Comparison of DNA sequencing of the protein A gene polymorphic region with other molecular typing techniques for typing two epidemiologically diverse collections of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 39:574-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2001. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 7:349-361. [DOI] [PubMed] [Google Scholar]
- 21.Oliveira, D. C., S. W. Wu, and H. de Lencastre. 2000. Genetic organization of the downstream region of the mecA element in methicillin-resistant Staphylococcus aureus isolates carrying different polymorphs of the antibiotic resistance gene. Antimicrob. Agents Chemother. 44:1906-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of methicillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2:180-189. [DOI] [PubMed] [Google Scholar]
- 23.Roberts, R. B., A. de Lencastre, W. Eisner, E. P. Severina, B. Shopsin, B. N. Kreiswirth, A. Tomasz, and the MRSA Collaborative Study Group. 1998. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in 12 New York hospitals. J. Infect. Dis. 178:164-171. [DOI] [PubMed] [Google Scholar]
- 24.Sá-Leão, R., I. Santos Sanches, D. Dias, I. Peres, R. M. Barros, and H. de Lencastre. 1999. Detection of an archaic clone of Staphylococcus aureus with low-level resistance to methicillin in a pediatric hospital in Portugal and in international samples: relics of a formerly widely disseminated strain? J. Clin. Microbiol. 37:1913-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanches, I. S., M. Ramirez, H. Troni, M. Abecassis, M. Pádua, A. Tomasz, and H. de Lencastre. 1995. Evidence for the geographic spread of a methicillin-resistant Staphylococcus aureus (MRSA) clone between Portugal and Spain. J. Clin. Microbiol. 33:1243-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teixeira, L., C. A. Resende, L. R. Ormonde, R. Rosenbaum, A. M. S. Figueiredo, H. de Lencastre, and A. Tomasz. 1995. Geographic spread of epidemic multiresistant Staphylococcus aureus clone in Brazil. J. Clin. Microbiol. 33:2400-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, F. D., Y. Y. Chen, and C. Y. Liu. 1998. Prevalence of nosocomial respiratory tract infections in the surgical intensive care units of a medical center. Zhonghua Yi Xue Za Zhi 61:589-595. [PubMed] [Google Scholar]
- 29.Wang, J. T., S. C. Chang, W. J. Ko, Y. Y. Chang, M. L. Chen, H. J. Pan, and K. T. Luh. 2001. A hospital-acquired outbreak of methicillin-resistant Staphylococcus aureus infection initiated by a surgeon carrier. J. Hosp. Infect. 47:104-109. [DOI] [PubMed] [Google Scholar]
- 30.Wang, J. T., Y. C. Chen, T. L. Yang, and S. C. Chang. 2002. Molecular epidemiology and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus in Taiwan. Diagn. Microbiol. Infect. Dis. 42:199-203. [DOI] [PubMed] [Google Scholar]
- 31.Wu, K. C., H. H. Chiu, J. H. Wang, N. S. Lee, H. C Lin, C. C. Hsieh, F. J. Tsai, C. T. Peng, and Y. C. Yseng. 2002. Characteristics of community-acquired methicillin-resistant Staphylococcus aureus in infants and children without known risk factors. J. Microbiol. Immunol. Infect. 35:53-56. [PubMed] [Google Scholar]