Abstract
Balamuthia mandrillaris is an opportunistic pathogen that causes granulomatous amebic meningoencephalitis in animals, including humans. Based on sequence analysis of mitochondrial small-subunit-rRNA genes, we developed primers that amplify a Balamuthia-specific PCR product. These primers will be useful for retrospective analyses of fixed tissues and possible identification of Balamuthia in vivo.
Meningoencephalitis caused by the ameba Balamuthia mandrillaris is a recently recognized, and nearly always fatal, infection of the brain in mammals, including humans (3, 10, 13-15). Most infections have been identified postmortem by histological examination of an infected individual's tissue samples. While infections caused by B. mandrillaris are relatively rare, the lack of rapid identification and diagnosis of the causative agent is likely to have had serious implications regarding the outcome of disease progression (5). Balamuthia infections are further complicated by the morphological similarity of Balamuthia in tissue sections to Acanthamoeba, another amebic genus involved in encephalitis infections. Therefore, we employed a simple PCR-based method for the rapid identification of B. mandrillaris based on DNA sequence information of the B. mandrillaris mitochondrial small-subunit-rRNA gene (rns).
The use of PCR as a potential method for the rapid identification of disease organisms in a clinical setting has expanded recently (4). In the case of encephalitis, PCR is used in organism-specific procedures to identify herpes simplex encephalitis (6). PCR methods are also used to identify numerous other pathogens, including human immunodeficiency virus, hepatitis B and C viruses, various bacterial pathogens, and Acanthamoeba (1, 7, 8, 9, 11). In this study we describe a PCR-based test to specifically identify B. mandrillaris. Recently, we examined DNA sequences of nuclear small-subunit-rRNA genes (rns) from a number of B. mandrillaris isolates and found no sequence variation among them (2). However, this study also examined the mitochondrial rns from seven isolates of B. mandrillaris and found interisolate variation. The genus-specific PCR target that we have identified here is a portion of the mitochondrial rRNA gene (rns). Future application of this PCR test should facilitate examination of archived or clinical samples.
The sequence alignment of rns from seven isolates of B. mandrillaris (GenBank rns accession numbers AF477012 to AF477018) was used to identify potential PCR primers. The seven isolates were very similar across the aligned region of 1,109 sites. Our previous study of rns variation in B. mandrillaris found low levels throughout the gene but no variation in the region of our putative Balamuthia-specific primers (2). Therefore, we predicted that this primer set would work satisfactorily if B. mandrillaris DNA was present. The rns 5′ and 3′ primers chosen were designated 5′Balspec16S (5′-CGCATGTATGAAGAAGACCA-3′) and 3′Balspec16S (5′-TTACCTATATAATTGTCGATACCA-3′). These primers were predicted to yield a PCR product of 1,075 bp in Balamuthia. A GenBank search using these primers failed to identify any significant sequence similarities among other organisms. As mentioned previously, the genus Acanthamoeba has been identified as a sister genus to Balamuthia (2). Further, Acanthamoeba has also been observed in amebic encephalitis infections. Therefore, we examined the differential amplification of Balamuthia and Acanthamoeba by using the Balamuthia primers and two Acanthamoeba-specific mitochondrial 16S rRNA primers. The latter yield a PCR product of ∼950 bp from Acanthamoeba (7a).
The DNA samples used in this study were from previously obtained extractions of B. mandrillaris and Acanthamoeba sp. isolates (1, 11). Approximately 10 to 200 ng of genomic DNA was used for PCR amplifications. Putative positive Balamuthia mitochondrial 16S ribosomal DNA products were sequenced by automated fluorescent sequencing. The primer used for sequencing of the Balamuthia PCR products was mt900, previously used to confirm that PCR products were Balamuthia rns (2).
The initial screening of 5′Balspec16S and 3′Balspec16S used two B. mandrillaris isolates, CDC V039 (ATCC 50209, a female mandrill brain isolate) and CDC V188 (ATCC 50605, a male human brain isolate), and three very distinct Acanthamoeba genotypes, T4 (Acanthamoeba mauritaniensis ATCC 50253; type strain), T5 (Acanthamoeba lenticulata PD2S; ATCC 30841; type strain), and T10 (Acanthamoeba culbertsoni 409C) (12). This amplification gave products of the expected size (1,075 bp) only from B. mandrillaris. PCR amplicons from B. mandrillaris V039 then were pooled and sequenced, confirming that the fragment was the rns from this strain.
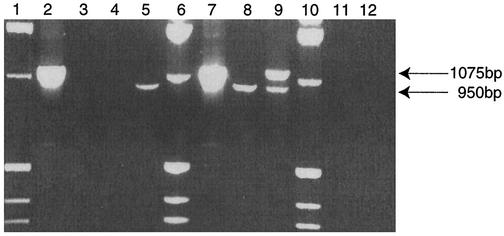
Next we performed a multiplex PCR that contained a mixture of ameba DNA and different primer combinations (Fig. 1). Multiplex PCR used DNA from A. mauritaniensis (genotype T4) and B. mandrillaris isolate V039. Lanes 1, 6, and 10 contain a 1-kb DNA ladder, which includes a fragment of 1,018 bp. This fragment aids in rapid identification of PCR products of the two genera because it migrates on gels between the 1,075-bp Balamuthia amplicon and the 950-bp amplicon of Acanthamoeba. Lane 2 contains B. mandrillaris DNA, incubated with the specific Balamuthia rns primers, which produced an amplimer of the expected size for this genus. Lane 3 contained B. mandrillaris DNA incubated with the Acanthamoeba-specific primers. No amplification was observed in this lane. Lane 4 contained A. mauritaniensis DNA incubated with the B. mandrillaris-specific primers, and as expected, no amplification was observed. Next, in lane 5, Acanthamoeba DNA incubated together with the Acanthamoeba-specific primers gave the expected Acanthamoeba product. Lanes 7 through 9 show multiplexing with multiple DNAs (A. mauritaniensis and B. mandrillaris) and multiple primer sets. Lane 7 used the two DNAs with the B. mandrillaris-specific primer set, and only the expected amplimer (1,075 bp) was observed. Lane 8 had DNA from the two species incubated with the Acanthamoeba-specific primer set, and only the PCR product of the predicted size of 950 bp was observed. Most significantly, lane 9 combined genomic DNA from B. mandrillaris and A. mauritaniensis with both primer sets. In this multiplexed experiment, the respective bands of 950 bp for Acanthamoeba and 1,075 bp for B. mandrillaris were observed. Lane 11 has human DNA treated with both primer sets, and no amplification was observed. Finally, no amplification was observed in the negative control (lane 12), which consisted of both primer sets and no DNA.
FIG. 1.
B. mandrillaris multiplex rns PCR. Amplification was performed in 50-μl reaction mixtures with 2.5 mM MgCl2; cycle conditions were 40 cycles of 1 min at 94°C, 2 min at 48°C, and 3 min at 72°C followed by a 15-min final extension at 72°C. Lanes 1, 6, and 10, 1-kb marker; lane 2, Balamuthia DNA with Balamuthia-specific primers; lane 3, Balamuthia DNA with Acanthamoeba-specific primers; lane 4, Acanthamoeba DNA with Balamuthia-specific primers; lane 5, Acanthamoeba DNA with Acanthamoeba-specific primers; lanes 7 to 9, Acanthamoeba and Balamuthia DNA with Balamuthia-specific primers (lane 7), with an Acanthamoeba-specific primer set (lane 8), and with both genus-specific primers sets (lane 9); lane 11, human DNA with both specific primer sets, lane 12, negative control. Amplicons produced by these mitochondrial 16S genus-specific primer sets are indicated on the right.
In this study we demonstrate the ability of primers based on the sequence of the mitochondrial rns to specifically amplify a product from whole-cell DNA of B. mandrillaris, even in the presence of DNA from Acanthamoeba. Also, this primer set does not amplify products from human DNA, which would be a potential source of contaminating DNA in a clinical setting. Future application of this very sensitive diagnostic test will be in the retrospective analysis of archived encephalitis samples and in the screening of clinical specimens in an effort to detect these organisms in early stages of infection, when therapy may be possible.
Acknowledgments
The work of G.C.B., J.R.C., T.J.B., and P.A.F. was funded by Public Health Service grant EY09073 awarded to P.A.F. by the National Eye Institute.
REFERENCES
- 1.Booton, G. C., D. J. Kelly, Y.-W. Chu, D. V. Seal, E. Houang, D. S. C. Lam, T. J. Byers, and P. A. Fuerst. 2002. 18S ribosomal DNA typing and tracking of Acanthamoeba species isolates from corneal scrape specimens, contact lenses, lens cases, and home water supplies of Acanthamoeba keratitis patients in Hong Kong. J. Clin. Microbiol. 40:1621-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Booton, G. C., J. R. Carmichael, G. S. Visvesvara, T. J. Byers, and P. A. Fuerst. Genotyping of Balamuthia mandrillaris based on nuclear 18S and mitochondrial 16S rRNA genes. Am. J. Trop. Med. Hyg., in press. [PubMed]
- 3.Canfield, P. J., L. Vogelnest, M. L. Cunningham, and G. S. Visvesvara. 1997. Amoebic meningoencephalitis caused by Balamuthia mandrillaris in an orangutan. Aust. Vet. J. 75:97-100. [DOI] [PubMed] [Google Scholar]
- 4.Fredricks, D. N., and D. A. Relman. 1999. Application of polymerase chain reaction to the diagnosis of infectious diseases. Clin. Infect. Dis. 29:475-488. [DOI] [PubMed] [Google Scholar]
- 5.Gordon, S. M., J. P. Steinberg, M. H. DuPuis, P. E. Kozarsky, J. F. Nickerson, and G. S. Visvesvara. 1992. Culture isolation of Acanthamoeba species and leptomyxid amebas from patients with amebic meningoencephalitis, including two patients with AIDS. Clin. Infect. Dis. 15:1024-1030. [DOI] [PubMed] [Google Scholar]
- 6.Lakeman, F. D., R. J. Whitley, et al. 1995. Diagnosis of herpes simplex encephalitis: application of polymerase chain reaction to cerebrospinal fluid from brain-biopsied patients and correlated with disease. J. Infect. Dis. 171:857-863. [DOI] [PubMed] [Google Scholar]
- 7.Ledee, D. R., J. Hay, T. J. Byers, D. V. Seal, and C. M. Kirkness. 1996. Acanthamoeba griffini. Molecular characterization of a new corneal pathogen. Investig. Ophthalmol. Vis. Sci. 37:544-550. [PubMed] [Google Scholar]
- 7a.Ledee, D. R., G. C. Booton, M. H. Awwad, S. Sharma, R. K. Aggarwal, I. A. Niszl, M. B. Markus, P. A. Fuerst, and T. J. Byers. Advantages of using mitochondrial 16S rDNA sequences to classify clinical isolates of Acanthamoeba Investig. Ophthalmol. Vis. Sci., in press. [DOI] [PubMed]
- 8.Quentin, R., R. Ruimy, A. Rosenau, J. M. Musser, and R. Christen. 1996. Genetic identification of cryptic genospecies of Haemophilus causing urogenital and neonatal infections by PCR using specific primers targeting genes coding for 16S rRNA. J. Clin. Microbiol. 34:1380-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Razman, N. N., E. J. Loftus, L. J. Burgart, M. Rooney, K. P. Batts, R. H. Wiesner, D. N. Fredricks, D. A. Relman, and D. H. Persing. 1997. Diagnosis and monitoring of Whipple disease by polymerase chain reaction. Ann. Intern. Med. 126:520-527. [DOI] [PubMed] [Google Scholar]
- 10.Rideout, B. A., C. H. Gardiner, I. H. Stalis, J. R. Zuba, T. Hadfield, and G. S. Visvesvara. 1997. Fatal infections with Balamuthia mandrillaris (a free-living amoeba) in gorillas and other old world primates. Vet. Pathol. 34:15-22. [DOI] [PubMed] [Google Scholar]
- 11.Schroeder, J. M., G. C. Booton, J. Hay, I. A. Niszl, D. V. Seal, M. B. Markus, P. A. Fuerst, and T. J. Byers. 2001. Use of subgenic 18S rDNA PCR and sequencing for generic and genotypic identification of Acanthamoeba from human cases of keratitis and from sewage sludge. J. Clin. Microbiol. 39:1903-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stothard, D. R., J. M. Schroeder-Diedrich, M. H. Awwad, R. J. Gast, D. R. Ledee, S. Rodriguez-Zaragoza, C. L. Dean, P. A. Fuerst, and T. J. Byers. 1998. The evolutionary history of the genus Acanthamoeba and the identification of eight new 18s rRNA gene sequence types. J. Eukaryot. Microbiol. 45:45-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uribe-Uribe, N. O., M. Becerra-Lomeli, I. Alvarado-Cabrero, P. M. Sashida, P. O. Nevares, S. Sosa, L. G. Chavez Macias, J. Ceballos, A. J. Martinez, G. S. Visvesvara, and J. E. Olivera-Rabiela. 2001. Granulomatous amebic encephalitis by Balamuthia mandrillaris. Patologia 39:141-148. [Google Scholar]
- 14.Visvesvara, G. S., A. J. Martinez, F. L. Schuster, G. J. Leitch, S. V. Wallace, T. K. Sawyer, and M. Anderson. 1990. Leptomyxid ameba, a new agent of amebic meningoencephalitis in humans and animals. J. Clin. Microbiol. 28:2750-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Visvesvara, G. S., F. L. Shuster, and A. J. Martinez. 1993. Balamuthia mandrillaris, N. G., N. Sp., agent of amebic meningoencephalitis in humans and other animals. J. Eukaryot. Microbiol. 40:504-514. [DOI] [PubMed] [Google Scholar]