Abstract
Inhibitors in clinical specimens can be detected by adding the target of nucleic acid amplification to the sample. Introduction of a Chlamydia trachomatis L2 434 preparation containing 12 elementary bodies (EBs) into first-void urine (FVU) from 225 nonpregnant women and 190 pregnant women before specimen processing by the assays produced false-negative rates of 0.48% (2 of 415 specimens) and 13% (44 of 338 specimens) by the APTIMA Combo 2 and the Chlamydia LCx tests, respectively. Reducing the amount of C. trachomatis added to one EB, a concentration closer to the APTIMA Combo 2 test cutoff, for a subset of 244 FVU specimens increased the number of specimens with false-negative results by the APTIMA Combo 2 assay to 7 (2.9%), suggesting that the strength of the input C. trachomatis per specimen has an influence on the number of specimens with false-negative results. Repeat testing after overnight storage and dilution decreased the APTIMA Combo 2 test false-negative rates to 0% (0 of 415 specimens) with the stronger inoculum and 0.8% (2 of 244 specimens) with the weaker inoculum; the false-negative rate of the LCx assay was reduced to 5.4% (18 of 334 specimens). When an additional 70 FVU specimens from women to which 12 EBs were added before specimen processing were tested by the LCx assay, 34 specimens had false-negative results, whereas 21 specimens had false-negative results when the C. trachomatis EBs were introduced after processing. Nine of the 21 specimens to which EBs were added after processing and all of the 34 urine specimens to which the target was added before processing remained falsely negative on repeat testing at a 1:2 dilution, suggesting that input C. trachomatis DNA was lost during processing by the LCx assay. In contrast, the APTIMA Combo 2 assay appears to have a higher sensitivity and either lost little nucleic acid during processing or demonstrated few problems with inhibitors of transcription-mediated amplification.
Chlamydia trachomatis is the most common sexually transmitted bacterium. Infections are often asymptomatic and if left untreated can cause pelvic inflammatory disease with sequelae of ectopic pregnancy, infertility, or chronic pelvic pain in women (3, 4).
Nucleic acid amplification (NAA) techniques are sensitive tools for the identification of C. trachomatis infections. While conventional tests require the invasive collection of specimens such as cervical or urethral swab specimens, NAA assays have been applied to first-void urine (FVU), vulvar, and vaginal specimens; however, some of these specimens contain substances which may contribute to false-negative results (10, 11, 14), and the results may also be influenced by pregnancy (9, 10). The false-negative results may be due to the presence of inhibitory substances in clinical specimens, the concentration of C. trachomatis added to the specimen, and/or the loss of nucleic acid from the inoculum during processing of the sample.
The APTIMA Combo 2 assay (Gen-Probe Incorporated, San Diego, Calif.) is a transcription-mediated amplification (TMA) method that uses a target capture technology during specimen processing to selectively isolate target C. trachomatis and Neisseria gonorrhoeae nucleic acids from potentially inhibitory biological substances in specimens. The test can be performed for the detection of one or both bacterial infections. In this study, we report on the prevalence of false-negative results detected when different concentrations of C. trachomatis elementary bodies (EBs) were added to FVU specimens collected from both pregnant and nonpregnant women and tested by the APTIMA Combo 2 assay and the Chlamydia LCx assay (Abbott Laboratories, Chicago, Ill.). We evaluated overnight storage at 4°C and/or dilution of the specimen as mechanisms for removing inhibitory activity. Experiments were also performed to compare the false-negative rates by the LCx assay when C. trachomatis EBs were added to FVU before processing and after processing.
MATERIALS AND METHODS
Specimens.
Specimens of FVU (defined as the first 10 to 30 ml of urine collected at any time) from 225 nonpregnant women submitted to a hospital chemistry laboratory for routine urinalysis and 190 pregnant women attending an obstetrics clinic were aliquoted and anonymously included in the study. No consent or patient information was collected. In the first series of experiments, all FVU specimens were tested with and without the addition of C. trachomatis EBs to the sample before processing by the APTIMA Combo 2 and the Chlamydia LCx assays. In the second series of experiments, 70 urine specimens from nonpregnant women were tested by adding two different concentrations of C. trachomatis EBs before and after specimen processing by the LCx test.
Determination of input C. trachomatis concentration.
C. trachomatis L2 434 was propagated in McCoy cells for 72 h, and the EBs were purified by differential centrifugation as described previously (6). Serial 10-fold dilutions from 100 to 10−8 were made in phosphate-free saline, and the EBs were counted by direct fluorescent-antibody staining with monoclonal antibodies specific for major outer membrane proteins of C. trachomatis (Syva Behring Microtrak, San Jose, Calif.). The 10−4, 10−5, and 10−7 dilutions contained an average of 100, 12, and 1 EB(s) per 100 μl, respectively. Aliquots of the resuspended EBs in saline were prepared and frozen at −70°C.
Serial 10-fold dilutions of an EB preparation in saline were tested by both the APTIMA Combo 2 and the LCx assays in replicates of 16 to determine a concentration closest to the cutoff value of the respective tests. A volume of 100 μl of the last dilution (10−5) that yielded a positive signal by the LCx assay for all 16 replicates (approximately 12 EBs) was added to the 415 FVU specimens prior to testing by both the APTIMA Combo 2 and the LCx assays. The C. trachomatis preparation of 10−7 (approximately one EB), a concentration that is closer to the APTIMA Combo 2 test cutoff and that yielded a positive signal for all 16 replicates, was also added to a subset of 244 FVU specimens for testing only by the APTIMA Combo 2 assay. To measure the impact of adding different concentrations of C. trachomatis to the sample and the effects of the potential loss of nucleic acids during the specimen processing by the LCx assay, we tested a set of 70 FVU specimens to which 12 EBs (10−5 dilution) and 100 EBs (10−4 dilution) were added before and after the processing step of the assay.
False-negative results.
A spiked specimen was considered falsely negative if the value recorded by the test was below the assay cutoff.
Removal of inhibitory substances.
Spiked FVU specimens found to be inhibitory in the APTIMA Combo 2 and Chlamydia LCx assays were stored overnight at 4°C and retested the next day undiluted and at a dilution of 1:2 or 1:4.
Amplification assays.
The Chlamydia LCx assay was performed as outlined by the manufacturer's protocol. One milliliter of FVU was centrifuged at ≥9,000 × g for 15 min. The resulting pellet was resuspended with 1 ml of the LCx assay urine resuspension buffer, and the suspension was heated at 97°C for 15 min. After the specimen was cooled at room temperature for 15 min, 100 μl was transferred to the appropriate LCx assay amplification vial. Cycling conditions were 40 cycles of 93°C for 1 s, 59°C for 1 s, and 62°C for 1 min and 10 s. Amplification products were detected with the LCx assay analyzer.
For the APTIMA Combo 2 assay, 400 μl of the FVU specimen was added to 100 μl of Target Capture Reagent-Solution B mix, and the mixture was incubated at 62°C for 30 min, followed by a second 30-min incubation at room temperature. During this step, the released C. trachomatis rRNA was bound to coated magnetic beads. The rack of tubes containing the rRNA-bound beads was subsequently placed on a magnetic target capture system for a series of washes, during which the captured target molecules were pulled to the side of the reaction vessel and the supernatant was removed. Seventy-five microliters of the APTIMA Combo 2 assay amplification reagent and 200 μl of oil were added to the specimen tubes, and the tubes were incubated at 62°C for 10 min. The tubes were cooled to 42°C, and 25 μl of enzyme reagent was added before the mixture was incubated for another hour at 42°C. The APTIMA Combo 2 assay probe reagent (100 μl) was added, followed by an incubation of 20 min at 62°C. At this point, single-stranded chemiluminescent DNA probes labeled with acridinium ester molecules were combined with the amplicon to form stable RNA-DNA hybrids. Finally, 250 μl of selection reagent, which differentiates hybridized from unhybridized probes, was added, and the tubes were incubated at 62°C for another 10 min. After the tubes were allowed to cool to room temperature, the light emitted from the labeled RNA-DNA hybrids was measured as photon signals in a Leader HC+ luminometer. The results were interpreted with computer software provided by the manufacturer.
RESULTS
Endpoint determination for each assay.
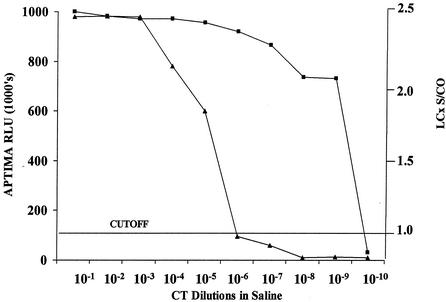
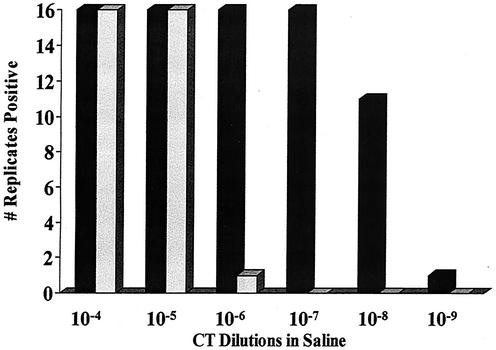
The effects of serial 10-fold dilution of C. trachomatis L2 434 EBs in saline tested by the APTIMA Combo 2 and LCx assays are shown in Fig. 1. Endpoints of positivity were seen with 12 EBs (10−5 dilution) and 0.01 EBs (10−9 dilution) in the LCx and APTIMA Combo 2 assays, respectively. By the APTIMA Combo 2 test, there was a steep decline in readings, from a positive reading of 731,000 relative light units (RLU) at a dilution of 10−9 to a negative reading of 10,000 RLU at 10−10. A similar pattern was observed with the LCx assay signal/cutoff (s/co) ratio between endpoint dilutions of 10−5 to 10−7. While the LCx assay with 12 EBs (10−5) tested positive in 16 replicates, the last dilution to produce a positive signal in all 16 replicates of the APTIMA Combo 2 assay contained 1 EB (10−7) (Fig. 2).
FIG. 1.
Effect of serial 10-fold dilutions of C. trachomatis L2 434 (CT) in relation to the cutoff in the APTIMA Combo 2 and LCx assays. ▴, APTIMA Combo 2 assay; ▪, LCx assay.
FIG. 2.
Determination of endpoints showing all replicates of C. trachomatis L2 434 (CT) positive by the APTIMA Combo 2 and LCx assays. ▪, APTIMA Combo 2 assay; ░⃞, LCx assay.
False-negative results for urine specimens spiked before processing.
A total of 415 FVU specimens collected from 190 pregnant women and 225 nonpregnant women were tested by the APTIMA Combo 2 and LCx assays with and without 12 C. trachomatis EBs. The data for one LCx assay run were eliminated from the data analysis due to invalid controls, thereby reducing the total number of FVU specimens tested by the LCx assay to 338 (159 pregnant women, 179 nonpregnant women). With an input C. trachomatis concentration of 12 EBs, which were introduced into the urine before processing, the APTIMA Combo 2 and LCx assays yielded negative results for 0.48% (2 of 415) and 13% (44 of 338) of the spiked specimens, respectively (Table 1). There was no significant difference in the number of spiked FVU specimens from pregnant and nonpregnant women with false-negative results by either assay (0 versus 0.89% for the APTIMA Combo 2 assay [P = 0.50], 11.9 versus 13.9% for the LCx assay [P = 0.58]).
TABLE 1.
Effect of adding C. trachomatis EBs to urine specimens from pregnant and nonpregnant women before processing by the APTIMA Combo 2 and LCx assays
| Population | No. of negative urine specimens/total no. of C. trachomatis EB-spiked urine specimens (%) |
||
|---|---|---|---|
| APTIMA Combo 2 assay |
LCx assay with 12 EBs | ||
| 12 EBs | 1 EB | ||
| Pregnant women | 0/190 (0) | 3/119 (2.5) | 19/159 (11.9) |
| Nonpregnant women | 2/225 (0.89) | 4/125 (3.2) | 25/179 (13.9) |
| All women | 2/415 (0.48) | 7/244 (2.9) | 44/338 (13.0) |
When 244 of the FVU specimens were tested by the APTIMA Combo 2 assay after the addition of one EB before specimen processing, the proportions of samples with false-negative results increased to 2.5% (3 of 119 specimens) for pregnant women, 3.2% (4 of 125 specimens) for nonpregnant women, and 2.9% (7 of 244 specimens) overall. In these experiments, only 5 of 42 (11.9%) of the specimens with false-negative results by the APTIMA Combo 2 or LCx assay were falsely negative in both assays. None of the 42 specimens with false-negative results were from C. trachomatis-infected patients. Indigenous C. trachomatis nucleic acids were detected in 6 of 298 (2.0%) urine specimens with sufficient volume for testing by the APTIMA Combo 2 test and 2 of 223 (0.9%) urine specimens with sufficient volume for testing by the LCx assay.
Table 2 summarizes the results of repeat testing after overnight storage of the series of 415 urine specimens with false-negative results. Two urine specimens which yielded false-negative results by the APTIMA Combo 2 assay after the addition of 12 EBs and after storage at 4°C overnight gave a positive signal when they were tested the next day undiluted and diluted 1:4. Only two of seven urine specimens with false-negative results with a C. trachomatis inoculum containing one EB remained negative. There was sufficient volume to retest only 40 of the 44 urine specimens that were false negative by the LCx assay. Of these 40 specimens, 18 remained negative after overnight storage at 4°C and retesting at a 1:4 dilution. Thus, with storage and retesting after dilution, the false-negative rates were reduced to 0.8% for the APTIMA Combo 2 assay and 5.4% for the LCx assay.
TABLE 2.
Effect of storage and retesting of urine specimens to which C. trachomatis was introduced before processing by the APTIMA Combo 2 and LCx assaysa
| Dilution | No. of negative urine specimens/no. of false-negative specimens after addition of C. trachomatis EBs (%) |
||
|---|---|---|---|
| APTIMA Combo 2 assay |
LCx assayb with 12 EBs | ||
| 12 EBs | 1 EB | ||
| Neat | 0/2 (0) | 2/7 (28.6) | 19/40 (47.5) |
| 1:4 | 0/2 (0) | 2/7 (28.6) | 18/40 (45.0) |
| False-negative result reduction | 0/415 (0) | 2/244 (0.8) | 18/334 (5.4) |
The specimens were kept at 4°C overnight.
There was sufficient volume to retest only 40 of 44 false-negative urine specimens by the LCx assay.
Contribution of processing or inhibitory substances to false-negative results for spiked specimens.
Table 3 summarizes the effect of adding 100 and 12 EBs of C. trachomatis before and after processing of 70 FVU specimens by the LCx assay. Whether C. trachomatis was introduced into the specimens before or after the processing step of the test protocol, fewer false-negative results were detected when 100 EBs were added, as only 2 of 70 urine specimens to which the EBs were added before and after processing tested falsely negative. When only 12 EBs were added, 30% (21 of 70) of the FVU specimens tested negative when C. trachomatis was added after processing, whereas 48.6% (34 of 70) of the FVU specimens tested negative when the target nucleic acid was added before processing. Repeat testing after overnight storage at 4°C and dilution of the sample 1:2 had the largest impact on the results for the specimens when the EBs were introduced after processing. Of the two samples that generated negative results after the addition of 100 EBs before processing, one retested positive the next day. None of the 34 samples for which false-negative results were detected when 12 EBs were added before specimen processing became positive on repeat testing. The two samples that were negative upon the addition of 100 EBs after processing retested positive, and 9 of the 21 FVU specimens that were false negative when 12 EBs were introduced after processing remained negative upon repeat testing the next day.
TABLE 3.
Effect of introducing two different concentrations of C. trachomatis EBs to urine before and after processing on the number of samples whose results fall below the test cutoff (false negative) by the LCx assay
| No. of C. trachomatis EBs added | No. of negative urine specimens/total no. of C. trachomatis EB-spiked urine specimens (%) |
|||
|---|---|---|---|---|
| Before processing |
After processing |
|||
| Original test | Repeat test with 1:2 dilution | Original test | Repeat test with 1:2 dilution | |
| 100 | 2/70 (2.9) | 1/2 (50) | 2/70 (2.9) | 0/2 (0) |
| 12 | 34/70 (48.6) | 34/34 (100) | 21/70 (30) | 9/21 (42.8) |
DISCUSSION
The APTIMA Combo 2 assay was 100-fold more sensitive than the LCx test for the detection of serial dilutions of C. trachomatis L2 434 in saline. The last dilution to consistently produce a positive signal in 16 replicates of the APTIMA Combo 2 assay was 10−7 (1 EB), whereas for the LCx assay the last dilution was 10−5 (12 EBs). Clinical evaluations are needed to determine whether this greater analytical sensitivity of the APTIMA Combo 2 test can be translated to the analysis of patient specimens. In a study by Martin et al. (D. H. Martin, C. Cammarata, L. Ivor, K. Smith, and C. Knott, Abstr. 100th Gen. Meet. Am. Soc. Microbiol., abstr. C-387, 2000), in which an infected patient was defined as positive if the patient specimen was positive by any two NAA tests, the sensitivities for detection of C. trachomatis in swab and urine specimens were reported to be 86.7 and 86.7%, respectively, for the APTIMA Combo 2 assay and 85.0 and 60.0%, respectively, for the LCx assay. In another study (M. L. Theodore, N. Dalesio, B. J. Wood, T. C. Quinn, and C. Gaydos, Abstr. 39th Annu. Meet. Infect. Dis. Soc. Am., abstr. 926, 2001), 327 urine samples from both symptomatic and asymptomatic men and women were tested by the Abbott LCx assay, the Becton Dickinson ProbeTec ET assay, and the Gen-Probe APTIMA Combo 2 assay. With a “gold standard” for a positive result defined as positive results by any two NAA tests, the sensitivities for C. trachomatis detection were found to be 94.7% for the LCx assay and 100% for the APTIMA Combo 2 assay.
Previous studies defining inhibition rates for NAA assays have tested specimens to which C. trachomatis inoculum near the cutoff value for each test was added; results below the test cutoff for positivity were presumed to be due to the presence of inhibitors in the specimen (10, 14). Those studies introduced C. trachomatis after processing. In the APTIMA Combo 2 assay, a raw EB preparation cannot be added after processing because it is only at the processing step when the cells are lysed and the bacterial rRNA is extracted.
Addition of 12 C. trachomatis EBs to each specimen before processing showed a false-negative rate of 0.48% (2 of 415 specimens) by the APTIMA Combo 2 assay. Seven of 244 (2.9%) FVU specimens to which one EB of C. trachomatis was added tested negative. This observation indicates the importance of testing specimens with C. trachomatis at a concentration near the cutoff value of an assay to provide a true indication of the test's susceptibility to amplification inhibitors. Inhibition may not be detected with an EB preparation that is too strong. On the other hand, if the inoculum is too weak, it may be lost during specimen processing, thereby showing an inflated inhibitor prevalence. The latter scenario may have occurred in the LCx assay. When the EBs were added before processing, 13% (44 of 338) of the spiked samples tested negative. In a previous study, we added C. trachomatis to the specimens after processing and found a lower inhibition rate of 3.9% (10), with all of the initial negative samples retesting positive after overnight storage and dilution. Other investigators have found similar inhibition rates. Berg et al. (2) detected an inhibition rate of 2.6% with urine specimens collected from men attending a sexually transmitted disease clinic. Gaydos et al. (8) reported no LCR inhibitors in urine specimens from 148 pregnant women. The urine specimens from the last two studies were either frozen at −70°C or stored at 4°C for days prior to testing with a C. trachomatis inoculum. While this may account for the lower rates because some inhibitors of the LCx assay or PCR are labile with storage (10, 14), in those studies C. trachomatis was also introduced into the specimens after specimen processing.
False-negative results may be observed in NAA tests when the number of targets in a sample is low because of sampling variability. When inhibitors are responsible for false-negative NAA test results, they may interfere with cell lysis, nucleic acid extraction, or target capture, causing nucleic acid degradation. Alternatively, they may interfere with the enzyme activity required for amplification (15). The larger number of false-negative results observed in the LCx assay in the present study may have been created by the step in the testing protocol at which the C. trachomatis EB inoculum was added to the specimen. Experiments with 70 urine specimens to which C. trachomatis was added before processing and after processing (Table 3) showed that fewer false-negative results were found when the EB preparation was added after specimen processing than before specimen processing (21 versus 34). Examination of the LCx assay s/co ratios (data not shown) for the 34 samples to which EBs were added before the specimen processing step of the test protocol revealed that the s/co ratio for none of them was near the LCx assay cutoff of 1.0 (s/co ratio range, 0.02 to 0.32). On the other hand, of the 21 specimens with false-negative results that were identified when EBs were introduced to the samples after processing, 12 retested positive at a 1:2 dilution. Eight of the nine samples that retested negative were on the borderline of positivity (s/co ratio range, 0.67 to 0.98; mean, 0.85). Although the number of specimens containing 100 C. trachomatis EBs with false-negative results was small, the two samples that were identified when the inoculum was added after processing retested positive, whereas only one of two false-negative specimens became positive when EBs were added before processing became positive upon repeat testing. These important observations indicate that a loss of positivity may be due to the presence of inhibitors in the sample and/or the loss of nucleic acids during specimen processing when C. trachomatis is added to the urine before initiation of the test protocol.
There were no differences in the false-negative rate of either the LCx assay or the APTIMA Combo 2 test for urine specimens from pregnant and nonpregnant women. In a previous study (10), the inhibitor prevalence rates for the LCx assay were 1 and 3.1%, respectively, for urine specimens from 101 pregnant women and 287 nonpregnant women. Pregnancy also caused no difference in the inhibitor prevalence rate for the Amplified CT TMA test assessed in that study. Only PCR was inhibited by urine from pregnant women more often than it was inhibited by urine from nonpregnant women (9.9 versus 3.1% [P = 0.011]) (10). Jensen et al. (9) reported that the inhibitor prevalence rate for urine samples in the LCx assay was greatly affected by the pregnancy status of 1,136 women because 15 cervical swabs were positive for C. trachomatis by direct fluorescent-antibody staining-confirmed enzyme immunoassay, but the corresponding urine samples were negative by the LCx assay. As the specimens were not tested with a C. trachomatis inoculum, the differences may be accounted for by inconsistent sampling or loss of nucleic acids during processing of the urine specimens. To definitively test for amplification inhibitors, C. trachomatis needs to be added to the clinical specimen after specimen processing.
Of the C. trachomatis-spiked urine specimens, only 5 of 42 tested false negative by both the APTIMA Combo 2 and LCx assays, suggesting that if all specimens truly contained inhibitors, the substances and/or the mechanisms causing amplification inhibition are different. Urinalysis and microscopic examination of both inhibitory and noninhibitory urine specimens in a previous study (10) showed a strong association between the presence of certain urinary substances and inhibition of various amplification assays. Logistic regression analysis of the data revealed that beta-human chorionic gonadotropin and crystals are associated with PCR inhibition, that nitrites are associated with LCR inhibition, and that hemoglobin, nitrites, and crystals are associated with TMA method inhibition. PCR has also been shown to be inhibited by bile salts and complex polysaccharides in feces, heme in blood, and urea in urine (1, 15), while the LCx assay has been inhibited by phosphate (11).
When 12 EBs were added to the samples, after overnight storage at 4°C, none of the urine specimens with false-negative results by the APTIMA Combo 2 assay remained negative. Only two of the seven (28.6%) urine specimens found to be negative with the addition of one EB remained negative after retesting. The false-negative rate for the LCx assay was lowered from 13 to 5.7% after storage. This finding is similar to those reported in other studies, in which amplification inhibitors were removed by dilution, prolonged storage of the specimen prior to amplification, or freezing and thawing (2, 5, 7, 8, 10, 12, 13).
Several important principles have been demonstrated in these studies. The sensitivities of NAA tests for the detection of nucleic acids may fall short of 100% because of the presence of inhibitors in clinical specimens and/or the loss of low levels of nucleic acids during specimen processing. Inhibitor prevalence rates measured after the addition of C. trachomatis to the specimens after sample processing are a truer estimate because the loss of nucleic acids during processing is not a factor. The addition to a specimen of a C. trachomatis concentration that is too close to the test's cutoff runs the risk of loss of nucleic acids and the creation of artificially negative results which may be assumed to be the result of the presence of inhibitors, whereas use of an EB preparation with a concentration that is too strong may overpower any inhibitors in the sample.
We have found the prevalence of NAA test inhibitors in urine specimens tested by the Gen-Probe APTIMA Combo 2 assay to be very low (range, 0.48 to 2.9%). The rate was dependent upon the amount of C. trachomatis EBs added to the FVU specimens prior to the processing step of the assay. None of the inhibitory urine specimens detected were from infected patients, and there were no differences in rates of inhibitory activity on the basis of pregnancy. Most of the inhibitory activity detected in the APTIMA Combo 2 assay was removed by overnight storage at 4°C. With this low susceptibility to amplification inhibitors and a high sensitivity level, the APTIMA Combo 2 assay has the potential to detect additional cases of C. trachomatis in urine specimens.
Acknowledgments
We thank the nurses of the St. Joseph's Healthcare Centre Obstetrics and Gynecology Department, Michael St. Pierre, and the technologists of the Chemistry Department of St. Joseph's Healthcare Centre for collecting urine specimens for this study.
REFERENCES
- 1.Abu Al-Soud, W., and P. Radstrom. 1998. Capacity of nine thermostable DNA polymerases to mediate DNA amplification in the presence of PCR-inhibiting samples. Appl. Environ. Microbiol. 64:3748-3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg, E. S., G. Anestad, H. Moi, G. Storvold, and K. Staug. 1997. False-negative results of a ligase chain reaction assay to detect Chlamydia trachomatis due to inhibitors in urine. Eur. J. Clin. Microbiol. Infect. Dis. 16:727-731. [DOI] [PubMed] [Google Scholar]
- 3.Cates, W., and J. N. Wasserheit. 1991. Genital chlamydia infections: epidemiology and reproductive sequelae. Am J. Obstet. Gynecol. 164:1771-1781. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 1993. Recommendation for prevention and management of Chlamydia trachomatis infections. Morb. Mortal. Wkly. Rep. 42:1-39. [Google Scholar]
- 5.Chernesky, M., S. Chong, D. Jang, K. Luinstra, M. Faught, and J. Mahony. 1998. Inhibition of amplification of Chlamydia trachomatis plasmid DNA by the ligase chain reaction associated with female urines. Clin. Microbiol. Infect. 4:397-400. [DOI] [PubMed] [Google Scholar]
- 6.Chernesky, M. A., D. Jang, J. Sellors, K. Luinstra, S. Chong, S. Castriciano, and J. B. Mahony. 1997. Urinary inhibitors of polymerase chain reaction and testing of multiple specimens may contribute to lower assay sensitivities for diagnosing Chlamydia trachomatis infected women. Mol. Cell. Probes 11:243-249. [DOI] [PubMed] [Google Scholar]
- 7.de Barbeyrac, B., M. Geniaux, C. Hocke, M. Dupon, and C. Bebear. 2000. Detection of Chlamydia trachomatis in symptomatic and asymptomatic populations with urogenital specimens by AMP CT (Gen-Probe Incorporated) compared to others commercially available amplification assays. Diagn. Microbiol. Infect. Dis. 37:181-185. [DOI] [PubMed] [Google Scholar]
- 8.Gaydos, C. A., M. R. Howell, T. C. Quinn, J. C. Gaydos, and K. T. McKee, Jr. 1998. Use of ligase chain reaction with urine versus cervical culture for detection of Chlamydia trachomatis in an asymptomatic military population of pregnant and nonpregnant females attending Papanicolaou smear clinics. J. Clin. Microbiol. 36:1300-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jensen, I. P., P. Thorsen, and B. R. Moller. 1997. Sensitivity of ligase chain reaction assay of urine from pregnant women for Chlamydia trachomatis. Lancet 349:329-330. [DOI] [PubMed] [Google Scholar]
- 10.Mahony, J. B., S. Chong, D. Jang, K. Luinstra, M. Faught, D. Dalby, J. Sellors, and M. A. Chernesky. 1998. Urine specimens from pregnant and nonpregnant women inhibitory to amplification of Chlamydia trachomatis nucleic acid by PCR, ligase chain reaction, and transcription-mediated amplification: identification of urinary substances associated with inhibition and removal of inhibitory activity. J. Clin. Microbiol. 36:3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Notomi, T., Y. Ikeda, A. Okadome, and A. Nagayama. 1998. The inhibitory effect of phosphate on the ligase chain reaction used for detecting Chlamydia trachomatis. J. Clin. Pathol. 51:306-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pasternack, R., P. Vuorinen, and A. Miettinen. 1997. Evaluation of the Gen-Probe Chlamydia trachomatis transcription-mediated amplification assay with urine specimens from women. J. Clin. Microbiol. 35:676-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stary, A., S. Tomazic-Allen, B. Choueiri, J. Burczak, K. Steyrer, and H. Lee. 1996. Comparison of DNA amplification methods for the detection of Chlamydia trachomatis in first-void urine from asymptomatic military recruits. Sex Transm. Dis. 23:97-102. [DOI] [PubMed] [Google Scholar]
- 14.Verkooyen, R. P., A. Luijendijk, W. M. Huisman, W. H. F. Goessens, J. A. J. W. Kluytmans, J. H. van Rijsoort-Vos, and H. A. Verbrugh. 1996. Detection of PCR inhibitors in cervical specimens by using the AMPLICOR Chlamydia trachomatis assay. J. Clin. Microbiol. 34:3072-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson, I. G. 1997. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 63:3741-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]