Abstract
The discrepant results available in the literature about the presence of hepatitis C virus (HCV) RNA in seminal plasma of men chronically infected by this agent are related, at least in part, to the molecular techniques used and particularly to the wide range of protocols dedicated to RNA extraction. In order to evaluate these protocols and to standardize the method of detection of HCV RNA in this fluid, a panel of coded specimens was tested blindly in 12 French laboratories; it included 14 seminal plasma specimens and four water controls spiked with HCV RNA ranging from 10 to 20,000 IU/ml and two HCV-negative seminal plasma specimens. The extraction step was performed according to methods using either silica beads (NucliSens [Organon Teknika S.A., Fresnes, France]; RNA viral kit [Qiagen, Courtaboeuf, France]) or guanidinium thiocyanate (Amplicor HCV assay; Roche Diagnostics, Meylan, France), preceded or not by a centrifugation of the seminal plasma. For the amplification step, all the laboratories performed the same reverse transcription-PCR technique (Amplicor HCV Cobas assay). The percentage of correct results ranged from 53.3 to 100, the poorest results being obtained when no centrifugation step preceded the Amplicor extraction protocol. The rate of correct results was significantly higher in laboratories using a preliminary centrifugation of the specimen (P = 0.034 by chi-square test). By contrast, the overall number of correct results was not correlated to the initial volume of sample used for the test. These results allowed us to validate standardized techniques adapted to the performance of this test on a routine basis, especially in men infected with HCV and involved in programs of medically assisted reproduction.
Hepatitis C virus (HCV) is mainly transmitted by parenteral route, and the contribution of genital secretions to the spread of the HCV outbreak is unclear. From an epidemiological point of view, discrepancies have been reported about the ability of HCV to be transmitted via sexual intercourse, with some reports supporting a possible role of this way of infection (10, 12, 13, 16), whereas other studies found no evidence of increased prevalence of HCV infection in subjects with at-risk sexual behavior (17, 22).
During the last 10 years, different studies investigated the detection of HCV RNA in seminal samples. Although the oldest ones failed to document the presence of HCV RNA in semen (8, 11, 19; S. Terada, K. Kawanishi, and K. Katayama, Letter, Ann. Intern. Med. 117:171-172, 1992), most authors using sensitive PCR techniques were able to detect HCV RNA in this fluid. Thus, and according to different parameters, including technical features and the human immunodeficiency virus (HIV) serological status of the subjects, 10 to 38% of patients showing chronic infection in blood were also positive for HCV RNA in semen (6, 14, 18; M. Leruez-Ville, J. M. Kunstmann, M. De Almeida, C. Rouzioux, and M. L. Chaix, Letter, Lancet 356:42-43, 2000).
Although no technique is available to address the infectivity of HCV RNA in seminal fluid, the possibility that this secretion has been involved in a few cases of HCV infection cannot be excluded. This opportunity for infection needs special consideration in the application of assisted reproductive techniques (ART) to couples in which the man is chronically infected with HCV. In this particular situation, it is imperative to prevent scrupulously the transmission of HCV to the female partner, to the artificially conceived embryos of the very couple or of other couples treated at the same time, and to the laboratory technicians performing ART. Recently, French legislation has introduced the systematic screening for the presence of HCV RNA in seminal samples from men who are HCV RNA positive at the blood level before initiating ART (1).
In order to evaluate the different molecular techniques used in French laboratories to detect HCV RNA in seminal plasma, a study including 12 centers performing medical virology was undertaken in May 2001 on a panel of 16 seminal specimens and four water samples spiked with different amounts of HCV RNA. The results of this investigation demonstrated the importance of the extraction step to remove inhibitors of the PCR and contributed to the definition of standardized techniques for routine screening of seminal samples in centers performing ART.
MATERIALS AND METHODS
Laboratories.
Ten public laboratories—nine from university hospitals and one from a general hospital—and two private laboratories took part in the study. All the people performing the assays were trained in nucleic acid amplification techniques on blood plasma specimens. However, four laboratories (B, E, H, and J) had no previous experience with HCV RNA detection in seminal plasma specimens.
Seminal samples.
The semen samples were recovered by auto-masturbation from 16 subjects consulting for routine bacteriological analysis in one of the participating laboratories. The fraction used for the study consisted in the residual part of the specimen after the volume necessary for bacteriological testing was withdrawn. The seminal plasma was separated from the pellet by centrifugation at 800 × g for 10 min, within 2 h after ejaculation, and was kept frozen at −80°C until use. Only seminal specimens with a residual volume of at least 6 ml were selected. All the participating subjects were negative for HCV antibodies in blood.
Panel constitution.
The panel used in the multicenter study was prepared in the laboratory coordinating the study. A blood plasma specimen from a patient chronically infected with HCV (genotype 3a) was used to artificially contaminate the specimens. The HCV viral load, determined by the Monitor HCV kit (Roche Diagnostics, Meylan, France), was 5 × 105 IU/ml. The sample was mixed with RNase-free sterile distilled water to obtain four 10-fold serial dilutions ranging from 500,000 to 500 IU/ml and three 10-fold serial dilutions ranging from 25,000 to 250 IU/ml. Each of these seven concentrations was further diluted into a couple of seminal samples from two different donors. The panel consisted of 14 different seminal samples at final concentrations of 10, 20, 100, 200, 1,000, 2,000, and 20,000 IU of HCV RNA/ml (two samples per dilution) and of two uninfected seminal samples. Four additional dilutions of the HCV RNA-positive blood plasma in RNase-free sterile distilled water to give final concentrations of 20, 200, 2,000, and 20,000 IU/ml were used as controls. To check the amount of the HCV RNA added to the panel samples, additional serial dilutions were performed in RNase-free water and tested by the Cobas Monitor assay and the Cobas Amplicor assay for samples that tested negative by the former test (threshold of positivity, 600 and 50 IU/ml, respectively; Roche Diagnostics). The results are reported in Table 1.
TABLE 1.
Composition of the panel of samples artificially contaminated by different amounts of HCV RNA derived from a positive human serum specimen
| No. of seminal plasma samples in panel | No. of control samplesa | Concn of HCV RNA (IU/ml)
|
|
|---|---|---|---|
| Expected | Observedb | ||
| 2 | 0 | 0 | <600c |
| 2 | 0 | 10 | <600c |
| 2 | 1 | 20 | <600c |
| 2 | 0 | 100 | <600d |
| 2 | 1 | 200 | 110 |
| 2 | 0 | 1,000 | 1,500 |
| 2 | 1 | 2,000 | 4,400 |
| 2 | 1 | 20,000 | 30,000 |
Controls were dilutions of HCV RNA-positive blood plasma in RNase-free distilled water.
Quantified in additional serial dilution in RNase-free water by the laboratory preparing the panel before sending the specimens to the other laboratories.
Negative by qualitative Cobas Amplicor assay (threshold, 50 IU/ml).
Positive by qualitative Cobas Amplicor assay.
The 12 participating laboratories received the same panel of 20 coded frozen aliquots at a volume of 500 μl each, including 16 seminal specimens and four RNase-free water specimens. They were informed of the overall composition of the panel as described in Table 1 (number of positive or negative samples and range of viral loads). A letter code (A to L) was blindly assigned to each laboratory. In the laboratory where the panel was prepared, the person who performed the tests was different from the one who spiked and blinded the specimens.
Extraction protocols.
The RNA was extracted from an initial volume of seminal plasma ranging from 100 to 500 μl (Table 2). The techniques performed by the different laboratories are listed in Table 2; they used either silica beads (NucliSens [Organon Teknika S.A., Fresnes, France]; RNA viral kit [Qiagen, Courtaboeuf, France]) or guanidinium thiocyanate (Amplicor HCV assay; Roche Diagnostics) and were preceded in some laboratories by a high-speed centrifugation of the specimens. Whatever the technique was, the internal control (IC) of the PCR assay was added to the sample before the extraction step (after the centrifugation step when the latter was performed).
TABLE 2.
Qualitative RT-PCR results obtained by 12 laboratories for detection of HCV RNA in 16 seminal samples according to protocol of RNA extraction
| Laboratory | RNA extraction protocol
|
No. of results
|
% Correctb | |||||
|---|---|---|---|---|---|---|---|---|
| Vol (μl) | Dilution of sample used | Concn of RNA by centrifugation at 4°C | Assay | Positive (n = 13) | Indeterminatea (n = 5) | Negative (n = 2) | ||
| A | 200 | Undiluted | None | NucliSens | 13 | 3 | 2 | 100 |
| B | 500 | Undiluted | None | NucliSens | 11 | 4 | 1 | 80 |
| C | 100 | Undiluted | None | Nuclisens | 9 | 0 | 2 | 73.3 |
| D | 250 | 1:1 | 21,000 × g for 1 h | Qiagen RNA | 13 | 1 | 2 | 100 |
| E | 200 | Undiluted | None | Amplicor | 7 | 1 | 1 | 53.3 |
| F | 140 | Undiluted | None | Qiagen RNA | 11 | 1 | 2 | 86.7 |
| G | 200 | Undiluted | 23,000 × g for 1 h | Amplicor | 12 | 1 | 2 | 93.3 |
| H | 200 | Undiluted | None | Amplicor | 9 | 1 | 0 | 60 |
| I | 200 | 1:2 | 28,100 × g for 1 h | Amplicor | 12 | 3 | 2 | 93.3 |
| J | 250 | 1:1 | 20,000 × g for 1 h | Qiagen RNA | 12 | 3 | 2 | 93.3 |
| K | 200 | 1:2 | 24,000 × g for 1 h | Amplicor | 12 | 0 | 2 | 93.3 |
| L | 200 | 1:1 | None | NucliSens | 10 | 0 | 2 | 80 |
Corresponds to the five samples listed in Table 1 with a viral load below the threshold of sensitivity of 50 IU/ml.
Taking into consideration the 13 positive and the 2 negative samples.
Reverse transcription (RT)-PCR assay.
In all the laboratories, the extracted RNA was reverse-transcribed and amplified using the Amplicor HCV Cobas assay (Roche Diagnostics). According to the manufacturer's recommendations, the results were validated when the optical density (OD) of the IC was ≥0.25 and interpreted as negative, indeterminate, or positive; when the OD of HCV RNA was <0.15; or when the OD of HCV RNA was between 0.15 and 1, or ≥1, respectively. The combination of OD <0.15 for HCV RNA and <0.25 for IC was considered as a noninterpretable result.
When a center performed several determinations for the same sample, the result transmitted to the statistician responsible for the methodology of the study (S. Laporte) was taken into account for the data synthesis.
Statistical analysis.
The chi-square test was used to compare qualitative variables. The link between quantitative variables was examined using Spearman's rank correlation test. The threshold for significance was 0.05.
RESULTS
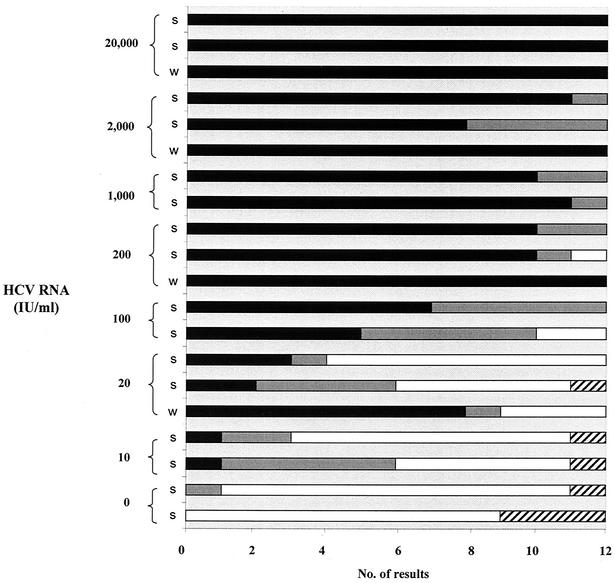
Figure 1 illustrates the overall distribution of the results obtained by the 12 laboratories according to the amount of HCV RNA contained in the 20 samples. All the specimens containing 20,000 IU of HCV RNA/ml were found to be positive. The deleterious effects of inhibitors of the RT-PCR in seminal plasma are illustrated by the discrepant results obtained for the two different specimens contaminated with the same amount of HCV RNA, i.e., 2,000 IU/ml: one of them was positive in 11 of 12 laboratories, and the other was positive in only 8 of them. This finding is also supported by the number of positive results, which was significantly higher in water-diluted specimens than in seminal samples, especially for samples with low amounts of HCV RNA (P = 0.09 by chi-square test) (Table 3).
FIG. 1.
Cumulative results from the 12 laboratories of the 20 specimens of the panel (four RNase-free water samples [w] and 16 seminal specimens [s]) according to RNA HCV concentration. Bars illustrate the number of positive results in each category: positive (black columns), indeterminate (gray columns), negative (white columns), noninterpretable (hatched columns).
TABLE 3.
Comparative detection of HCV RNA in specimens diluted in RNase-free water and in seminal plasma
| HCV RNA load (IU/ml) | No. (%) of specimens detected positive for HCV RNA
|
Pa | |
|---|---|---|---|
| RNase-free water | Seminal plasma | ||
| 20,000 | 12/12 (100) | 24/24 (100) | |
| 2,000 | 12/12 (100) | 19/24 (79.2) | NS |
| 200 | 12/12 (100) | 20/24 (83.3) | NS |
| 20 | 8/12 (66.7) | 5/24 (20.8) | 0.01 |
| Total | 44/48 (91.7) | 68/96 (70.8) | |
Determined by chi-square test (NS, not significant).
Among the 12 laboratories, the number of positive results varied from 8 to 15 and the percentage of correct results ranged from 53.3 to 100 (Table 2). The lowest scores were obtained by laboratories E and H, which used the Amplicor extraction assay with the protocol described for the detection of HCV RNA in blood plasma. In laboratories G, I, and K, which performed a centrifugation step before using the same test, the percentage of correct results was 93.3 (Table 2). Four laboratories (A, B, C, and L) used the NucliSens RNA extraction procedure, with percentages of good results varying from 73.3 to 100. In centers D, F, and J, which used the viral RNA kit from Qiagen, this percentage ranged from 86.7 to 100, again with better results for the laboratories using an initial centrifugation of the specimens (Table 2).
As a whole, the number of positive results was 69 of 90 (76.7%) in the five laboratories that used a protocol including an initial centrifugation step of the specimens and 80 of 126 (63.5%) in the seven laboratories that omitted this step (P < 0.05 by chi-square test). The number of correct results obtained by the different laboratories was not correlated to the initial volume of sample used for the test (rS = 0.002, P = 0.85).
DISCUSSION
The present multicenter quality control study was undertaken to validate the use of molecular techniques originally designed for the detection of HCV RNA in blood for the search of this marker in seminal fluids. As documented previously (3), these techniques are particularly needed in the context of ART to control the absence of viral RNA in semen from men chronically infected with HCV. In France, a recent legislation has dictated rules that require the systematic testing of seminal plasma for the presence of HCV RNA in this population (1). The low concentrations of HCV RNA that were chosen in this quality control study correspond to the viral loads previously reported in this kind of sample (3, 14; Leruez-Ville et al., letter).
Before sending the samples to the collaborative laboratories, it was important to check that the specimens of the panel had been spiked with correct amounts of HCV RNA compared to the expected values after serial dilutions in water of the human blood plasma used to contaminate the seminal fluids (Table 1).
One of the main goals of this comparative study was to define the best protocols adapted to the extraction of HCV RNA from seminal fluids; indeed, this step was shown to be critical for the detection of viral genomes in semen by molecular techniques (5, 7, 20). Our results clearly demonstrate that different extraction protocols can be applied with close sensitivity scores. However, when the Amplicor extraction assay was used, a preliminary centrifugation step was shown to be essential to the quality of the test (Table 2). As a whole, laboratories performing the centrifugation step were shown to have the best results (P < 0.05; Table 2). The beneficial effect of this centrifugation step can be related to its ability to remove most of the inhibitors of PCR, which may reach high concentrations in some seminal samples: lactoferrin, peroxides, and mostly zinc residues are thought to be the main substances that interfere with the action of Taq polymerases (4, 15). For laboratories using an extraction technique different from NucliSens, similar results were obtained in terms of sensitivity with the Qiagen assay and the modified Amplicor assay. As the latter was faster and cheaper to perform, it was preferred for the consensus technique (see below). Figure 1 clearly shows that, even for samples spiked with high concentrations of HCV RNA (1,000 or 2,000 IU/ml), the number of false-negative or weakly positive results is higher in semen samples than in water controls. The presence of these inhibitors has been widely documented in previous studies dealing with the detection of HIV (4, 19) or HCV RNA (14, 19) in seminal fluids, especially when the standard Amplicor extraction assay was used. The efficacy of the extraction techniques based on the NucliSens extraction technique appeared to be less influenced by inhibitors, as previously shown for the detection of HIV RNA (2, 9, 21).
Although all laboratories had a wide experience of molecular techniques, four of them were not trained in the testing of seminal samples and two of the latter exhibited the poorest results (Table 2). Rather than to a lack of experience, these findings are probably due to the omission of the centrifugation step prior to the use of the Amplicor extraction assay. Similarly, the volume of seminal sample used for performing the reaction is not critical, as illustrated by laboratories A and B that performed the same extraction method on 200 and 500 μl of seminal fluid, respectively, with close results (Table 2). By contrast with blood, the volume of seminal specimens available for virological testing may be critical (as illustrated by the difficulty to organize quality control studies on a multicenter basis with this kind of specimen); a sample volume of at least 200 μl appears to be sufficient for this assay.
All the laboratories included in this study spontaneously used the Cobas Amplicor assay to perform the amplification of HCV RNA. Actually, this technique is easy to perform, widely automated, and licensed in many countries for diagnostic use with blood samples. The inclusion of an IC of amplification is critical to detect samples with high amounts of PCR inhibitors not removed by the treatments discussed above. A high sensitivity of the technique (<50 IU/ml when combined with an efficient extraction step in this study) is also required, since low amounts of HCV RNA have been reported in seminal samples (Leruez-Ville et al., letter). It must be noted that it was not possible to define precisely the actual sensitivity of the assay for seminal specimens since the number of replicates was limited by the volume of samples and the number of measures performed in each laboratory. No conclusions can be drawn from the results obtained from the samples with low viral loads (listed as “intermediate” in Table 2) since they were under the threshold of detection of the standard Amplicor protocol. As well as sensitivity variations in the assays used, the differences concerning the detection of these samples could result in random distribution of spiked HCV RNA between tubes.
On the basis of the results of this multicenter study, simple consensus techniques for the detection of HCV RNA in seminal plasma can be proposed as follows: (i) the volume of specimen suggested for the test is 200 μl of seminal plasma, (ii) a predilution of the sample in the same volume of RNA-free water (1:1) is recommended, (iii) the extraction step is based either on the NucliSens test as recommended by the manufacturer or on the Amplicor HCV extraction assay necessarily preceded by a centrifugation step of the specimen (at least 20,000 × g for 1 h), (iv) the amplification step is performed by RT-PCR using the Cobas Amplicor assay according to the manufacturer's instructions. This consensus technique is intended to be tested by a wider panel of French laboratories.
As previously shown for HIV (7), the interest of quality control assays performed on a multicenter basis is essential in order to standardize the detection of viral genomes in seminal fluids, especially in with regard to minimizing the risk of virus transmission by techniques of medically assisted reproduction.
Acknowledgments
This work was supported by grant number 2001/069 from the French Agence Nationale de Recherche sur le SIDA and was performed in the name of the AC-11 group of this agency.
REFERENCES
- 1.Anonymous. 2001. Arrêté du 10 Mai modifiant l'arrêté du 12 Janvier 1999 relatif aux règles de bonnes pratiques cliniques et biologiques en assistance médicale à la procréation. J. Officiel République Française 112:7735-7736. [Google Scholar]
- 2.Ball, J., R. Curran, W. L. Irving, and A. Dearden. 1991. HIV-1 in semen: determination of proviral and viral titres compared to blood, and quantification of semen leukocyte populations. J. Med. Virol. 59:356-363. [DOI] [PubMed] [Google Scholar]
- 3.Bourlet, T., R. Levy, A. Maertens, J. C. Tardy, F. Grattard, H. Cordonier, J. L. Laurent, J. F. Guerin, and B. Pozzetto. 2002. Detection and characterization of hepatitis C virus RNA in seminal plasma and spermatozoon fractions of semen from patients attempting medically assisted conception. J. Clin. Microbiol. 40:3252-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen, M. S., I. F. Hoffman, R. A. Royce, P. Kasembe, J. R. Dyer, C. C. Daly, D. Zimba, P. L. Vernazza, M. Maida, S. A. Fiscus, J. J. Eron, and the AIDSCAP Malawi Research Group. 1997. Reduction of concentration of HIV-1 in semen after treatment of urethritis: implications for prevention of sexual transmission of HIV-1. Lancet 349:1868-1873. [DOI] [PubMed] [Google Scholar]
- 5.Coombs, R., C. Speck, J. Hughes, W. Lee., R. Sampoleo., S. O. Ross, J. Dragavon, G. Peterson, T. M. Hooton, A. C. Collier, L. Corey, L. Koutsky, and J. N. Krieger. 1998. Association between culturable human immunodeficiency virus type 1 (HIV-1) in semen and HIV-1 RNA levels in semen and blood: evidence for compartmentalization of HIV-1 between semen and blood. J. Infect. Dis. 177:320-330. [DOI] [PubMed] [Google Scholar]
- 6.Fiore, R. J., D. Potenza, L. Monno, A. Appice, M. DiStefano, A. Gianelli, L. LaGrasta, C. Romanelli, C. DiBari, and G. Pastore. 1995. Detection of HCV RNA in serum and seminal fluid from HIV-1 co-infected intravenous drug addicts. J. Med. Virol. 46:364-367. [DOI] [PubMed] [Google Scholar]
- 7.Fiscus, S. A., D. Brambilla, R. W. Coombs, B. Yen-Lieberman, J. Bremer, A. Kovacs, S. Rasheed, M. Vahey, T. Schutzbank, P. Reichelderfer, and other members of the AIDS clinical group genital secretions working group. 2000. Multicenter evaluation of methods to quantitate human immunodeficiency virus type 1 RNA in seminal plasma. J. Clin. Microbiol. 38:2348-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fried, M. W., M. Shindo, T. L. Fong, P. C. Fox, J. H. Hoofnagle, and A. M. Di Bisceglie. 1992. Absence of hepatitis C viral RNA from saliva and semen of patients with chronic hepatitis C. Gastroenterology 102:1306-1308. [PubMed] [Google Scholar]
- 9.Gupta, P., J. Mellors, L. Kingsley, S. Riddler, M. K. Singh, S. Schreiber, M. Cronin, and C. R. Rinaldo. 1997. High viral load in semen of human immunodeficiency virus type 1-infected men at all stages of disease and its reduction by therapy with protease and nonnucleoside reverse transcriptase inhibitors. J. Virol. 71:6271-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halfon, P., H. Riflet, C. Renou, Y. Quentin, and P. Cacoub. 2001. Molecular evidence of male-to-female sexual transmission of hepatitis C virus after vaginal and anal intercourse. J. Clin. Microbiol. 39:1204-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu, H. H., T. Wright, D. Luba, M. Martin, S. M. Feinstone, G. Garcia, and H. B. Greenberg. 1991. Failure to detect hepatitis C virus genome in human secretions with the polymerase chain reaction. Hepatology 14:763-767. [DOI] [PubMed] [Google Scholar]
- 12.Kao, J. H., Y. T. Hwang, P. J. Chen, P. M. Yang, M. Y. Lai, T. H. Wang, and D. S. Chen. 1996. Transmission of hepatitis C virus between spouses: the important role of exposure duration. Am. J. Gastroenterol. 91:2087-2090. [PubMed] [Google Scholar]
- 13.Karmochkine, M., F. Carrat, A. J. Valleron, and G. Raguin. 1998. Transmission modes of hepatitis C virus. Presse Med. 27:871-876. [PubMed] [Google Scholar]
- 14.Levy, R., J. C. Tardy, T. Bourlet, H. Cordonier, F. Mion, J. Lornage, and J. F. Guerin. 2000. Transmission risk of hepatitis C virus in assisted reproductive techniques. Hum. Reprod. 15:1083-1085. [DOI] [PubMed] [Google Scholar]
- 15.Mayer, K. H., and D. J. Anderson. 1995. Heterosexual HIV transmission. Infect. Agents Dis. 4:273-284. [PubMed] [Google Scholar]
- 16.Mesquita, P. E., C. F. Granato, and A. Castelo. 1997. Risk factors associated with hepatitis C virus (HCV) infection among prostitutes and their clients in the city of Santos, Sao Paulo State, Brazil. J. Med. Virol. 51:338-343. [PubMed] [Google Scholar]
- 17.Osella, A. R., M. A. Massa, S. Joekes, N. Blanch, M. R. Yacci, S. Centonze, and S. Sileoni. 1998. Hepatitis B and C virus sexual transmission among homosexual men. Am. J. Gastroenterol. 93:49-52. [DOI] [PubMed] [Google Scholar]
- 18.Pasquier, C., M. Daudin, L. Righi, L. Berges, L. Thauvin, A. Berrebi, P. Massip, J. Puel, L. Bujan, and J. Izopet. 2000. Sperm washing and virus nucleic acid detection to reduce HIV and hepatitis C virus transmission in serodiscordant couples wishing to have children. AIDS 14:2093-2099. [DOI] [PubMed] [Google Scholar]
- 19.Semprini, A. E., T. Persico, V. Thiers, M. Oneta, R. Tuveri, P. Serafini, A. Boschini, S. Giuntelli, G. Pardi, and C. Brechot. 1998. Absence of hepatitis C virus and detection of hepatitis G virus/GB virus C RNA sequences in the semen of infected men. J. Infect. Dis. 177:848-854. [DOI] [PubMed] [Google Scholar]
- 20.Shepard, R. N., J. Schock, K. Robertson, D. C. Shugars, J. Dyer, P. Vernazza, C. Hall, M. S. Cohen, and S. A. Fiscus. 2000. Quantitation of human immunodeficiency virus type 1 RNA in different biological compartments. J. Clin. Microbiol. 38:1414-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vernazza, P. L., B. L. Gillian, J. Dyer, A. C. Frank, S. A. Fiscus, M. S. Cohen, and J. J. Eron. 1997. Quantification of HIV in semen: correlation with antiviral treatment and immune status. AIDS 11:987-993. [PubMed] [Google Scholar]
- 22.Wyld, R., J. R. Robertson, R. P. Brettle, J. Mellor, L. Prescott, and P. Simmonds. 1997. Absence of hepatitis C virus transmission but frequent transmission of HIV-1 from sexual contact with doubly-infected individuals. J. Infect. 35:163-166. [DOI] [PubMed] [Google Scholar]