Abstract
A commercially available automated specimen preparation instrument for specific probe capture and paramagnetic separation has been developed (AmpliCap/GT-12; Roche Molecular Systems). We evaluated assay performance of the AmpliCap/GT-12 in the quantitative assay for hepatitis C virus (HCV) RNA with the AMPLICOR HCV MONITOR Test (version 2.0). Assay linearity using serial dilutions from a serum panel was observed in the range of 500 to 850,000 IU/ml, with a slightly compromised slope in the higher viral titers. The overall within-run and between-run reproducibility of the entire detection process for 3 and 5 log10 (IU/ml) of HCV RNA in samples had a standard deviation of <0.2, which was comparable to a manual method based on organic extraction and isopropanol precipitation (Roche Molecular Systems). Comparison of the test results with those obtained by the manual method showed a good correlation (R2 = 0.972, n = 86). Using heparin (3, 6.5, and 13 U/ml), dextran sulfate (0.1, 1, and 5 mM), hemoglobin (1.13, 2.25, and 4.5 g/liter), conjugated or unconjugated bilirubin (7.5, 15, and 30 mg/dl), and ATP (1.25, 2.5, and 5.0 mM) as known inhibitors, inhibition was only detected at a dextran sulfate concentration of 1 mM with the manual method but not with the AmpliCap/GT-12 extraction. In summary, the AmpliCap/GT-12 system was shown to permit a stable extraction process and accurate results for the quantitative assay of HCV RNA, successfully eliminating the inhibitory effect of dextran sulfate. This automated extraction system provides reliable and reproducible test results and saves labor; thus, it is suitable for routine diagnostic PCR.
Automated systems have been developed for amplification and detection of nucleic acid sequences for infectious agents using PCR (10). An example of this is the COBAS AMPLICOR (Roche Diagnostic Systems, Branchburg, N.J.) (6, 11, 17). In addition to offering the accuracy of automated results, this system has provided labor savings and containment of amplification and detection. However, the current manual extraction method for the COBAS AMPLICOR is time-consuming and requires meticulous technical skills to achieve reproducible results. The extraction process is a key component of nucleic acid detection, as it affects both the reliability and the reproducibility of target amplification.
Recently there has been substantial progress in automation of the extraction of nucleic acid from clinical samples. In our previous study, a prototype automated specimen preparation instrument (GT-12; Roche Molecular Systems, Pleasanton, Calif.) was developed and evaluated for specific capture of hepatitis C virus (HCV) RNA with probes and magnetic-bead-fluid (B/F) separation (14, 15). HCV RNA was isolated from serum by lysis of virus particles with a chaotropic agent, followed by hybridization of the RNA with biotinylated probes and capture of the hybridized RNA with streptavidin-coated paramagnetic particles. After washing of the hybrid-particle complexes to remove nonspecifically bound materials, the particles were resuspended in a specimen diluent and were then ready for amplification and detection using a fully automated PCR system (COBAS AMPLICOR). Based on the promising data showing that the prototype instrument of the GT-12 had an assay performance similar to that of the conventional manual method for the qualitative assay of HCV RNA, a commercially available system (AmpliCap/GT-12; Roche Diagnostics) has been developed.
Accurate quantitative assay of HCV RNA in serum is increasingly becoming important, since the pretreatment viral load in blood has been negatively correlated with the sustained response to a combination therapy with alpha interferon, and importance of monitoring of the HCV load during the treatment has been reported (3, 4, 19). One of the major advantages of nucleic acid amplification over conventional methods in diagnosis of infectious disease is the sensitive detection of agents directly from clinical specimens. However, serum may contain a variety of biological substances and therapeutic reagents that may interfere with enzymatic amplification to cause false-negative PCR results (8, 19). Such inhibition might have serious effects on quantitative values of HCV RNA. The objectives of the present study were (i) to evaluate assay performance of the AmpliCap/GT-12 in the quantitative assay for HCV RNA, including linearity, reproducibility, and comparison with a conventional manual method, and (ii) to address questions as to whether the extraction system could eliminate the inhibitory effects of a variety of known inhibitors of PCR on the quantitative assay of HCV RNA.
MATERIALS AND METHODS
Clinical specimens.
Serum specimens used in this study were obtained from 90 patients referred to Tokai University Hospital for chronic liver diseases. When needed, HCV RNA was quantitatively measured by the AMPLICOR HCV MONITOR Test, version 2.0 (Roche Diagnostic Systems) (7). All samples were separated from clots within 4 h of collection, divided into aliquots, and stored at −80°C until RNAs were extracted.
RNA extraction.
HCV RNA was isolated from serum by an automated system consisting of the reagents (AmpliCap; Roche Molecular Systems) and a robotic processor (GT-12; Roche Molecular Systems) developed in cooperation with Precision System Science Co. Ltd. (Tokyo, Japan). Briefly, HCV RNA was isolated from 250 μl of serum by lysis of virus particles with 500 μl of guanidinium thiocyanate solution at 60°C for 20 min. The RNA was hybridized with biotinylated probes (KY78) that were specific to the 5′-untranslated region of the HCV genome (16) and identical to the downstream primer for amplification. The hybridized RNA was then captured with streptavidin-coated paramagnetic particles. The quantitation standard or an internal quantitative control was introduced into the specimen during the lysis reaction. After washing of the hybrid-particle complexes to remove nonspecifically bound materials, the particles were resuspended in 250 μl of a specimen diluent and were then ready for amplification and detection by PCR.
RNA was also isolated from 100 μl of serum by a manual method based on guanidinium thiocyanate lysis and isopropanol precipitation (Roche Molecular Systems). Briefly, 400 μl of a lysis solution containing guanidinium thiocyanate and the quantitation standard was added to the specimen. The nucleic acid extraction was precipitated by isopropanol and pelleted by centrifugation, and the pellet was washed with ethanol. The nucleic acid was resuspended in 1,000 μl of a specimen diluent, and 50 μl of extracted sample was used for amplification and detection by PCR.
The AmpliCap/GT-12 used a larger input volume of serum than the manual method; the former used 50 μl out of 250 μl of a specimen diluent derived from 250 μl of serum, and the latter used 50 μl out of 1,000 μl of a specimen diluent derived from 100 μl of serum.
COBAS AMPLICOR HCV Test.
The quantitative COBAS AMPLICOR HCV Tests, and AMPLICOR HCV MONITOR Test, version 2.0, have been previously described (7, 26). All samples were extracted once, and a single amplification and detection were performed on each extract except as otherwise stated. One positive and two negative controls provided with the kit were run with each batch of patient specimens.
Sample dilutions.
A serum panel from clinical specimens that was quantitated for HCV RNA levels by the AMPLICOR HCV MONITOR Test, version 2.0, was diluted serially in HCV-seronegative normal human serum. Serial dilutions of serum were prepared to provide values throughout an expected range of the assay, 500 to 850,000 IU/ml. Duplicate determinations were made on the diluted samples in the COBAS AMPLICOR HCV MONITOR Test, version 2.0.
Interference.
To assess the effect of various known inhibitors (8, 12, 21), three concentrations of heparin (Takeda Chem., Osaka, Japan), dextran sulfate (Wako Pure Chem., Osaka, Japan), hemoglobin (International Reagents, Kobe, Japan), conjugated bilirubin (International Reagents), unconjugated bilirubin (International Reagents), and ATP (Sigma Chemical Co., St. Louis, Mo.) were added to a serum sample containing 3 or 5 log10 (IU/ml) of HCV RNA. RNA was extracted from the serum by the manual method or the AmpliCap/GT-12, and was subjected to AMPLICOR HCV MONITOR Test, version 2.0.
Statistical analysis.
Before analysis, all HCV RNA results were converted to log10 (international units per milliliter). The chi-square test was used for comparison of methods.
RESULTS
Linearity study.
A serum panel diluted serially in HCV seronegative normal human serum was isolated by the AmpliCap/GT-12 assay, and then HCV RNA was measured in duplicate by the COBAS AMPLICOR HCV MONITOR Test, version 2.0. The dilutions were evaluated in a plot of the mean of duplicate observed values versus the expected values. These data showed that the assay was almost linear over the range of 500 to 850,000 IU/ml of HCV RNA. The linearity becomes slightly compromised at higher titers of HCV RNA, i.e., >500,000 IU/ml (Fig. 1).
FIG. 1.
Assay linearity in serial dilutions of a serum sample for HCV RNA levels as determined by the AMPLICOR HCV MONITOR Test using RNA extracted by the AmpliCap/GT-12 system. For linearity study, serial dilutions of a serum panel were subjected to HCV RNA extraction by the AmpliCap/GT-12, and measured for HCV RNA in the COBAS AMPLICOR HCV MONITOR Test, version 2.0. Data are means of duplicate assays.
Within-run and between-run reproducibility.
Within-run and between-run reproducibilities were evaluated by using 3 and 5 log10 (IU/ml) of HCV RNA in samples. For within-run reproducibility, 10-fold determinations were made on three lots of reagents of AMPLICOR HCV MONITOR Test, version 2.0. The standard deviations (SDs) of the log10 (units per milliliter) were calculated and plotted against the average log10 (units per milliliter) for each sample. Serum HCV RNA levels, tested 10 times in the same round for within-run reproducibility, showed that the SDs were 0.12 to 0.17 and 0.06 to 0.11 for 3 and 5 log10 (IU/ml) of HCV RNA in samples, respectively, and the coefficients of variation were 3.3 to 4.9% and 1.4 to 2.3%, respectively, which were comparable to those obtained in the manual method (Table 1). Because a separate sample aliquot was processed for each test performed, these variations represent the total variability of the assay.
TABLE 1.
Within- and between-run precision for the AmpliCap/GT-12 system and AMPLICOR MONITOR Test, version 2.0a
| Precision group and extraction method | Lot of reagents | Result with indicated HCV RNA titer (IU/ml)
|
|||||
|---|---|---|---|---|---|---|---|
| 3 log10
|
5 log10
|
||||||
| Mean | SD | CV (%) | Mean | SD | CV (%) | ||
| Within run | |||||||
| Manual | 1 | 3.51 | 0.14 | 4.1 | 4.99 | 0.06 | 1.1 |
| 2 | 3.64 | 0.15 | 4.2 | 4.80 | 0.05 | 1.1 | |
| 3 | 3.42 | 0.18 | 5.4 | 4.85 | 0.10 | 2.0 | |
| AmpliCap/GT-12 | 1 | 3.59 | 0.15 | 4.1 | 4.69 | 0.11 | 2.3 |
| 2 | 3.45 | 0.17 | 4.9 | 4.73 | 0.10 | 2.1 | |
| 3 | 3.58 | 0.12 | 3.3 | 4.78 | 0.06 | 1.4 | |
| Between run | |||||||
| Manual | 3.26 | 0.12 | 3.7 | 4.58 | 0.08 | 1.8 | |
| AmpliCap/GT-12 | 3.43 | 0.11 | 3.1 | 4.90 | 0.16 | 3.2 | |
RNA was extracted by either the manual method or the AmpliCap/GT-12 system and tested 10 times in the same round for within-run reproducibility on three lots of reagents of the AMPLICOR HCV MONITOR Test, version 2.0. RNA was also tested over 12 days for between-run reproducibility on one lot of reagents of the AMPLICOR HCV MONITOR Tests, version 2.0. The SDs of the log10 (units per milliliter) were calculated and plotted against the average log10 (units per milliliter) for each sample. CV, coefficient of variation.
Serum HCV RNA levels, tested on one lot of reagents over 12 days for between-run reproducibility, showed that the SD of the log10 of HCV RNA levels for 3 and 5 log10 (IU/ml) of HCV RNA in samples was 0.11 and 0.16, respectively, and the coefficients of variation were 3.1 and 3.2%, respectively, which were comparable to those obtained in the manual method (Table 1).
Comparison with the manual method.
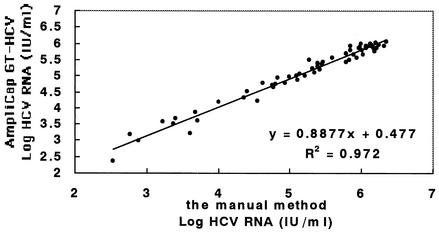
A total of 90 clinical samples were subjected to RNA extraction by either the manual method or the automated method, and measured in singlet using the version 2.0 AMPLICOR HCV MONITOR tests. Of the samples, 86 had levels of HCV RNA that quantified both assay. The overall correlation of the two assays was good (y = 0.8877x + 0.477; r2 = 0.972) (Fig. 2).
FIG. 2.
HCV RNA levels for 86 clinical specimens as determined by the AMPLICOR HCV MONITOR Test using RNA extracted by the manual method or the AmpliCap/GT-12 system. A total of 90 clinical samples were subjected to RNA extraction by either the manual method or AmpliCap/GT-12, and measured in singlet using the version 2.0 AMPLICOR HCV MONITOR Test. Of the samples, 86 had levels of HCV RNA that were quantifiable by both assays.
Interference study.
Using heparin (3, 6.5, and 13 U/ml), dextran sulfate (0.1, 1, and 5 mM), hemoglobin (1.13, 2.25, and 4.5 g/liter), conjugated or unconjugated bilirubin (7.5, 15, and 30 mg/dl), and ATP (1.25, 2.5, and 5.0 mM) as known inhibitors, inhibition was only detected with dextran sulfate at a concentration of 1 mM by the manual method but not by the AmpliCap/GT-12 extraction (Table 2).
TABLE 2.
Study of interference of heparin and dextran sulfate on the AmpliCap/GT-12 system and AMPLICOR MONITOR Test, version 2.0a
| Reagent concn | HCV RNA titer (IU/ml) obtained by extraction method
|
|||
|---|---|---|---|---|
| Manual
|
AmpliCap/GT-12
|
|||
| 3 log10 | 5 log10 | 3 log10 | 5 log10 | |
| Heparin (U/ml) | ||||
| Blank | 1.00 × 103 | 1.78 × 105 | 1.17 × 103 | 1.36 × 105 |
| 3 | 8.22 × 102 | 1.40 × 105 | 1.86 × 103 | 8.38 × 104 |
| 6.5 | 8.82 × 102 | 2.00 × 105 | 1.76 × 103 | 6.70 × 104 |
| 13 | 9.38 × 102 | 1.65 × 105 | 1.05 × 103 | 5.98 × 104 |
| Dextran sulfate (mM) | ||||
| Blank | 8.45 × 102 | 1.18 × 105 | 1.11 × 103 | 7.90 × 104 |
| 0.1 | 1.29 × 103 | 1.04 × 105 | 8.17 × 102 | 9.53 × 104 |
| 1.0 | Inhibition | Inhibition | 2.07 × 103 | 9.00 × 104 |
| 5 | Inhibition | Inhibition | 1.23 × 103 | 6.97 × 104 |
Heparin or dextran sulfate was added to a serum specimen containing 3 or 5 log10 IU of HCV/ml. RNA was extracted by either the conventional manual method or the AmpliCap/GT-12 system, and the HCV RNA was amplified by the AMPLICOR MONITOR Test, version 2.0. Data are means of duplicate assays. For blanks, normal saline was added to serum.
DISCUSSION
In this study, we evaluated the performance of the AmpliCap/GT-12 system for the quantitative assay of HCV RNA by the COBAS AMPLICOR HCV MONITOR Test, version 2.0. Comparison of the test results with those obtained by the manual method showed that the overall correlation of the two assays was good (r2 = 0.972). Assay linearity was observed in the 3-log dynamic range over HCV RNA titers of 500 to 850,000 IU/ml but with a slightly compromised slope at higher titers (i.e., >500,000 IU/ml) in comparison with that obtained by the manual method based on organic extraction and precipitation (7, 26). The underestimation of high-titer sera by the Roche Amplicor HCV Monitor assay has been indicated previously (5). This is more evident in the quantitative assay of HCV RNA using the AmpliCap/GT-12, because this assay uses a larger input volume of serum than the manual method.
False-negative results because of amplification inhibitors present in clinical specimens are problematic in PCR assays (8, 18). Therapeutic reagents for infectious agents may have inhibitory effects (5, 24), and novel reagents will be constantly developed and introduced into the treatment of infectious diseases (13, 22). It is increasingly important to accurately measure viral loads in serum, which may contain various therapeutic reagents. A potential use of dextran sulfate had once been explored for the treatment of human immunodeficiency virus (1), where the prevalence of HCV infection is high (2). The inclusion of the internal quantitative control allows one to assure amplification of the target. However, the use of an internal control, when added during the initial lysis step, does not by itself distinguish cases of failure of extraction from the presence of an interfering substance. In such cases the internal control may not amplify. An ideal robust extraction system should efficiently extract both the internal control and the target and effectively eliminate inhibitors. The AmpliCap/GT-12 system can efficiently extract both the internal control and the target and eliminate effects of potent inhibitors such as dextran sulfate as well as other substances that may be present in serum (1, 13). This enhanced the reliability and reproducibility of the extraction process and thus the entire nucleic acid detection. The extraction method used in the AmpliCap/GT-12 system is theoretically suitable for eliminating any inhibitors in the serum, because it is based on specific capture with probes and magnetic B/F separation. On the other hand, conventional manual methods such as that used in this study are not able to eliminate dextran sulfate, which is thought to bind nucleic acids (5).
The inhibitory effect of heparin on PCR has been problematic in hemodialysis patients (23; E. Beutler, T. Gelbart, and W. Kuhl, Letter, BioTechniques 9:166, 1990), who have a high prevalence of HCV infection (21). In contrast to the qualitative assay, there was a lack of inhibition in the quantitative assay of HCV RNA, when RNA was extracted by the manual method from sera in the presence of heparin. This can be explained by the impact of different preparation processes for nucleic acid amplification and detection used in the qualitative and quantitative assays of HCV RNA. In the latter, nucleic acid extracted from 100 μl of serum is diluted in 1,000 μl of a specimen diluent by 10-fold, and thus the final concentration of residual heparin could be diluted to the extent that the inhibitory effect was negligible. This is supported by our previous report that 10-fold dilution of the serum sample was enough to eliminate the inhibitory effect in the former (15). There was also a lack of inhibition when RNA was extracted by the AmpliCap/GT-12 system from sera in the presence of heparin, in spite of using a larger input volume of serum than the manual method.
One of the major advantages of automating the extraction is the ability to provide a stable extraction process among samples. Using 3 and 5 log10 (IU/ml) for within-run and between-run precision of the combination of the AmpliCap/GT-12 system and the COBAS AMPLICOR HCV MONITOR Test, the SDs were <0.2. Considering that these data are comparable to the reproducibility of the COBAS AMPLICOR HCV MONITOR Test (SD < 0.2 [7, 26]), the AmpliCap/GT-12 system has permitted a stable extraction process among samples. The system is characterized by automated separation of nucleic acid for amplification by target-specific probe capture, specimen preparation with high throughput (96 reactions in 2.5 h), and minimal hands-on reagent preparation and machine setup. The ability to automate the extraction in addition to the measurement improved the assay performance in terms of not only stability of the extraction process but also labor efficiency. The reliability of the GT-12 system has been recognized, as shown by its use in major reference laboratories and in blood screening for the Red Cross Center in Japan (9).
In summary, the AmpliCap/GT-12 system, based on the specific capture with probes and magnetic B/F separation, was shown to permit a stable extraction process and accurate results for a quantitative assay of HCV RNA, successfully eliminating the inhibitory effect of dextran sulfate from serum. This system provides reliable and reproducible test results and saves labor; thus, it is suitable for routine diagnostic PCR.
Acknowledgments
We thank Nicholas Smith, Roche Diagnostics K.K., for reviewing the manuscript.
REFERENCES
- 1.Abrams, D. I., S. Kuno, R. Wong, K. Jeffords, M. Nash, J. B. Molaghan, et al. 1989. Oral dextran sulfate (UA001) in the treatment of the acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. Ann. Intern. Med. 110:183-188. [DOI] [PubMed] [Google Scholar]
- 2.Bruno, R., P. Sacchi, M. Puoti, V. Soriano, and G. Filice. 2002. HCV chronic hepatitis in patients with HIV. Clinical management issues. Am. J. Gastroenterol. 97:1598-1606. [DOI] [PubMed] [Google Scholar]
- 3.Carreno, V. 2002. Present treatment expectations and risks of chronic hepatitis C. Clin. Microbiol. Infect. 8:74-79. [DOI] [PubMed] [Google Scholar]
- 4.Castro, F. J., J. I. Seteban, A. Juarez, S. Sauleda, L. Viladomiu, M. Martell, F. Moreno, H. Allende, R. Esteban, and J. Guardia. 2002. Early detection of nonresponse to interferon plus ribavirin combination treatment of chronic hepatitis C. J. Viral Hepat. 9:202-207. [DOI] [PubMed] [Google Scholar]
- 5.Demeke, T., and R. P. Adams. 1992. The effects of plant polysaccharides and buffer additives on PCR. BioTechniques 12:322-324. [PubMed] [Google Scholar]
- 6.DiDomenico, N., H. Link, R. Knobel, T. Caratsch, W. Weschler, Z. G. Loewy, and M. Rosenstraus. 1996. COBAS AMPLICOR: fully automated RNA and DNA amplification and detection system for routine diagnostic PCR. Clin. Chem. 42:1915-1923. [PubMed] [Google Scholar]
- 7.Eralis, M., E. R. Ashwood, and D. R. Hillyard. 2000. Performance characteristics of the COBAS AMPLICOR hepatitis C virus MONITOR test, version 2.0. Am. J. Clin. Pathol. 114:180-187. [DOI] [PubMed] [Google Scholar]
- 8.Greenfield, L., and T. J. White. 1993. Sample preparation methods, p. 122-137. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology. Principles and applications. American Society for Microbiology, Washington, D.C.
- 9.Japanese Red Cross NAT Screening Research Group. 2000. Nationwide nucleic acid amplification testing of hepatitis B virus, hepatitis C virus and human immunodeficiency virus type 1 for blood transfusion and follow-up study of nucleic acid amplification positive donors. Jpn. J. Infect. Dis. 53: 116-123. [PubMed] [Google Scholar]
- 10.Jungkind, D. 2001. Molecular testing for infectious disease. Science 294:1553-1555. [DOI] [PubMed] [Google Scholar]
- 11.Jungkind, D., S. DiRenzo, K. G. Beavis, and N. S. Silverman. 1996. Evaluation of automated COBAS AMPLICOR PCR system for detection of several infectious agents and its impact on laboratory management. J. Clin. Microbiol. 34:2778-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masukawa, A., H. Miyachi, T. Ohshima, and Y. Ando. 1998. Inhibitory effects of therapeutic reagents on the PCR detection of hepatitis C virus in serum and their elimination by RNA extraction method. Jpn. J. Clin. Pathol. 47: 949-955. (In Japanese with English abstract.) [PubMed]
- 13.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M.-H. Ling, S. Cort, and J. K. Albrecht. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 14.Miyachi, H., A. Masukawa, T. Ohshima, H. Fusegawa, T. Hirose, C. Impraim, and Y. Ando. 1998. Monitoring of inhibitors of enzymatic amplification in polymerase chain reaction and evaluation of efficacy of RNA extraction for the detection of hepatitis C virus using the internal control. Clin. Chem. Lab. Med. 36:571-575. [DOI] [PubMed] [Google Scholar]
- 15.Miyachi, H., A., Masukawa, T. Ohshima, T. Hirose, C. Impraim, and Y. Ando. 2000. Automated specific capture of hepatitis C virus RNA with probes and paramagnetic particle separation. J. Clin. Microbiol. 38:18-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nolte, F. S., C. Thurmond, and M. W. Fried. 1995. Preclinical evaluation of AMPLICOR hepatitis C virus test for detection of hepatitis C virus RNA. J. Clin. Microbiol. 33:1775-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poljak, M., K. Seme, and S. Koren. 1997. Evaluation of the automated COBAS AMPLICOR hepatitis C virus detection system. J. Clin. Microbiol. 35:2983-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rolfs, A., I. Schuller, U. Finckh, and I. Weber-Rolfs. 1992. PCR: clinical diagnostics and research, p. 51-67. Springer-Verlag, Berlin, Germany.
- 19.Rosen, H. R., R. R. Ribeiro, L. Weinberger, S. Wolf, M. Chung, D. R. Gretch, D. R., and A. S. Perelson. 2002. Early hepatitis C viral kinetics correlate with long-term outcome in patients receiving high dose induction followed by combination interferon and ribavirin therapy. J. Hepatol. 37:124-130. [DOI] [PubMed] [Google Scholar]
- 20.Roth, W. K., J.-H. Lee, B. Ruster, and S. Zeuzem. 1996. Comparison of two quantitative hepatitis C virus reverse transcriptase PCR assays. J. Clin. Microbiol. 34:261-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silini, E., F. Bono, A. Cerino, V. Piazza, E. Solcia, and M. U. Mondelli. 1993. Virological features of hepatitis C virus infection in hemodialysis patients. J. Clin. Microbiol. 31:2913-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Squires, K. E. 2001. An introduction to nucleoside and nucleotide analogues. Antivir. Ther. 6(Suppl. 3):1-14. [PubMed] [Google Scholar]
- 23.Willems, M., H. Moshage, F. Nevens, J. Fevery, and S. H. Yap. 1993. Plasma collected from heparinized blood is not suitable for HCV-RNA detection by conventional RT-PCR assay. J. Virol. Methods 42:127-130. [DOI] [PubMed] [Google Scholar]
- 24.Yedidag, E. N., A. J. Kofferon, K. H. Mueller, B. Kaplan, D. B. Kaufman, J. P. Fryer, F. P. Stuart, and M. Abecassis. 1996. Acyclovir triphosphate inhibits the diagnostic polymerase chain reaction for cytomegalovirus. Transplantation 62:238-242. [DOI] [PubMed] [Google Scholar]
- 25.Yu, M.-L., W.-L. Chuang, S.-C. Chen, Z.-Y. Lin, M.-Y. Hsieh, L.-Y. Wang, and W.-Y. Chang. 1999. Clinical application of the Quantiplex HCV RNA 2.0 and Amplicor HCV Monitor assays for quantifying serum hepatitis C virus RNA. J. Clin. Pathol. 52:807-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu, M.-L., W.-L. Chuang, C.-Y. Dai, S.-C. Chen, Z.-Y. Lin, M.-Y. Hsieh, et al. 2000. Clinical evaluation of the automated COBAS AMPLICOR HCV MONITOR Test version 2.0 for quantifying serum hepatitis C virus RNA and comparison to the Quantiplex HCV version 2.0 test. J. Clin. Microbiol. 38:2933-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]