Abstract
Amplified ribosomal DNA restriction analysis (rrn-ARDRA) is based on PCR amplification and restriction of a fragment of rRNA genes including 16S and 23S genes and the intergenic spacer. rrn-ARDRA was evaluated for the identification of species within the genus Streptococcus. A total of 148 type and reference strains of pyogenic, oral, and group D streptococci were examined in order to construct a database for identification of streptococci. The amplified product was a single band approximately 4,500 bp long. This amplicon was digested separately with three (HhaI, MboII, and Sau3A) restriction endonucleases. Respectively, 27, 26, and 28 major patterns were observed after HhaI, MboII, and Sau3A restrictions. Streptococcal strains belonging to different species had different patterns or different combination of patterns. An identification system based upon a combination of the three restriction patterns in a single database was then proposed. rrn-ARDRA was successfully applied to 11 clinical isolates whose identification to the species level was difficult to obtain by phenotypic analysis. Using a database of well-characterized strains, rrn-ARDRA is a powerful method for the identification of streptococcal isolates.
Streptococci are major pathogens for animals and human beings. Phenotypic studies and DNA-DNA hybridization assays have demonstrated that the genus Streptococcus may be divided in three major groups of species: pyogenic, oral, and group D streptococci (5, 6, 17, 20, 21, 27).
Identification of streptococcal species is currently based on observation of the cultural and morphological characteristics, determination of the biochemical pattern (production of enzymes and production of acid from various carbohydrate sources) (4, 5, 6, 9, 11, 17, 20), and observation of the antigenic structure according to the classification of Rebecca Lancefield (22).
Molecular tools as ribotyping (3, 8, 25, 26), oligonucleotide probing (2), PCR-based protocols (13, 23), and DNA sequence analysis (1, 19, 24) are now proposed to provide an accurate identification of streptococcal isolates. Among these, restriction fragment length polymorphism analysis was proposed as a general method for bacterial identification and typing (15). Amplified ribosomal DNA (rDNA) restriction analysis (ARDRA) was recently reported to be a rapid and efficient method of identification of bacterial isolates to the species level in mollicutes (7) and in some genera such as Acinetobacter or Mycobacterium (30, 31). The initial protocol was substantially improved by amplification of a larger fragment of the rrn operon (16S rRNA gene, 16S-23S intergenic spacer, and 23S rRNA gene) followed by the digestion with one to three endonucleases selected according to the bacterial genus studied (12, 18). The latest modification of this method uses a unique combination of three restriction endonucleases (HhaI, MboII, and Sau3A) irrespective of the bacterial species studied. This modified protocol was successfully used for identification of the genus and the species of strains belonging to the family Enterobacteriaceae (G. Giammanco, P. Nogueira, O. Bouallegue, F. Grimont, and P. A. D. Grimont, Abstr. 98th Gen. Meet. Am. Soc. Microbiol., abstr. R20, p. 482, 1998).
In the present study, we investigate the interest of the modified rrn-ARDRA protocol for identification of streptococci. One hundred and forty-eight strains were amplified and analyzed with HhaI, MboII, and Sau3A endonucleases. A database suitable for identification of streptococcal species was constructed and used to identify some clinical isolates.
MATERIALS AND METHODS
Bacterial strains.
A total of one hundred and forty-eight reference strains were included in the study (Table 1). These strains were obtained from the Australian Collection of Microorganisms (ACM), St. Lucia, Australia; the bioMérieux collection of microorganisms (API), La-Balme-les-Grottes, France; the American Type Culture Collection (ATCC), Manassas, Va.; the Culture Collection of the University of Göteborg (CCUG), Göteborg, Sweden; the Centers for Disease Control and Prevention (CDC), Atlanta, Ga.; the Collection de l'Institut Pasteur (CIP), Paris, France; le Centre National de Référence des Pneumocoques (CNRP), Créteil, France; the National Collection of Dairy Organisms (NCDO), Sheffield, United Kingdom; the National Collection of Type Cultures (NCTC), London, United Kingdom; and other depositors. Strains were cultured onto blood Columbia agar plates or brain heart infusion (BHI) broth medium (bioMérieux, La Balme-les-Grottes, France) and conserved at −80°C in BHI broth supplemented with glycerol (15%, vol/vol). For pneumococci, BHI broth was supplemented with 5% bovine ascite-serum (Bio-Rad, Marnes-la-Coquette, France), and cultures were incubated in a 5% CO2 atmosphere.
TABLE 1.
Identification, source, and restriction patterns of streptococcal strains studied for construction of database
| Species | Strain no. | Designation in initial collection or other source | Restriction pattern
|
||
|---|---|---|---|---|---|
| HhaI | MboII | Sau3A | |||
| S. agalactiae | HDP 89543T | CCUG 4208T | H-D1b | M-B1a | S-F1b |
| HDP 96146 | CIP 104971 | H-D1a | M-B1b | S-F1a | |
| HDP 98299 | CIP 105451 | H-D1a | M-B1a | S-F1a | |
| CNRP 96/87 | Serotype Ia | H-D1a | M-B1a | S-F1a | |
| CNRP 96/126 | Serotype Ia | H-D1b | M-B1a | S-F1a | |
| CNRP 96/133 | Serotype V | H-D1c | M-B1a | S-F1a | |
| CNRP 96/138 | Serotype III | H-D1b | M-B1a | S-F1a | |
| CNRP 96/408 | Serotype V | H-D1c | M-B1a | S-F1b | |
| CNRP 96/409 | Serotype Ia | H-D1b | M-B1a | S-F1a | |
| CNRP 96/433 | Serotype V | H-D1c | M-B1a | S-F1b | |
| S. alactolyticus | HDP 90057T | CIP 103244T | H-B4 | M-H1 | S-T1 |
| S. anginosus | HDP 89578T | CIP 102921T | H-E3 | M-B5e | S-B1a |
| HDP 89551 | R. Whiley, strain UC 7895 | H-F4a | M-B5b | S-B1a | |
| HDP 92259 | R. Whiley, strain VR 348 | H-F4a | M-B5e | S-B1a | |
| HDP 93171 | R. Whiley, strain KR 455 | H-F3 | M-B5a | S-B1b | |
| HDP 93172 | R. Whiley, strain KR 687 | H-F3 | M-B5a | S-B1b | |
| HDP 93173 | CDC 1007-77 | H-C1b | M-B5a | S-B1c | |
| HDP 93174 | CDC 2236-81 | H-F4a | M-B5c | S-B1a | |
| HDP 93179 | NCTC 8037 | H-C1a | M-B5c | S-B1c | |
| HDP 93180 | ATCC 9895 | H-F4b | M-B5e | S-B1a | |
| S. bovis | HDP 89505T | CIP 102302T | H-B3b | M-E2 | S-G1c |
| S. canis | HDP 90051T | CIP 103223T | H-F1 | M-A3a | S-R1b |
| HDP 99030 | CIP 105029 | H-F1 | M-A3a | S-R1b | |
| S. caprinus | HDP 99204T | CIP 104887T | H-B3a | M-B7b | S-G2 |
| S. constellatus | HDP 89579T | NCDO 2226T | H-E2b | M-A4b | S-L1a |
| HDP 93154 | NCTC 5389 | H-E2a | M-A4a | S-L1b | |
| HDP 93155 | NCTC 11063 | H-E2a | M-A5a | S-L1b | |
| HDP 93156 | NCTC 10714 | H-E2a | M-A4c | S-L1b | |
| S. criceti | HDP 89549T | A. Coykendall, strain H56 | H-G5 | M-F2 | S-U7 |
| S. difficilis | HDP 98301T | CIP 103768T | H-D1b | M-B1 | S-F1a |
| HDP 99031 | CIP 103853 | H-D1b | M-B1 | S-F1a | |
| S. downei | HDP 90050T | CIP 103222T | H-G1 | M-F1 | S-U6 |
| S. dysgalactiae subsp. dysgalactiae | HDP 89550T | CIP 102914T | H-F2 | M-A3a | S-R1a |
| HDP 98005 | CIP 55.123 | H-F2 | M-A3a | S-R2 | |
| HDP 98110 | CIP 55.119 | H-F2 | M-A3a | S-R2 | |
| S. dysgalactiae subsp. equisimilis | HDP 98055 | CIP 105120, former type strain of S. equisimilis | H-F2 | M-A3a | S-R2 |
| HDP 97340 | ATCC 35666 | H-F2 | M-A3a | S-R1b | |
| HDP 98003 | CIP 104.339 | H-F2 | M-A3a | S-R2 | |
| HDP 98004 | CIP 55.120 | H-F2 | M-A3a | S-R2 | |
| HDP 99032 | CIP 103684 | H-F2 | M-A3a | S-R1b | |
| HDP 99033 | CIP 105034 | H-F2 | M-A3a | S-R1b | |
| S. equi subsp. equi | HDP 89501T | CIP 102910T | H-G7b | M-B6a | S-N1 |
| HDP 99034 | CIP 105022 | H-G7b | M-B6a | S-N1 | |
| HDP 99035 | CIP 105023 | H-G7b | M-B6a | S-N1 | |
| S. equi subsp. zooepidemicus | HDP 90053 | CIP 103228, former type strain of S. zooepidemicus | H-G7a | M-B6c | S-E1a |
| HDP 98092 | CIP 52.209 | H-G7d | M-B6a | S-E1b | |
| HDP 98093 | CIP A7 | H-G7d | M-B6b | S-E1b | |
| HDP 98297 | CIP 105025 | H-G7c | M-B6b | S-E1b | |
| HDP 99036 | CIP 105024 | H-G7c | M-B6a | S-E1b | |
| S. equinus | HDP 89506T | CIP 102504T | H-B3b | M-E2 | S-G1b |
| HDP 99309 | API 78.04.058, NCTC 10389 | H-B3b | M-E2 | S-G1a | |
| HDP 99313 | API 88.07.126 | H-B3b | M-E2 | S-G1b | |
| S. ferus | HDP 89545T | A. Coykendall, strain 8S1 | H-G6 | M-G5 | S-O1/PICK> |
| S. gallolyticus | HDP 98035T | ACM 36.11T | H-B3c | M-B7b | S-G1a |
| HDP 90055 | NCDO 2019 | H-B3b | M-B7a | S-G1a | |
| HDP 90299 | API 84.03.028 | H-B3b | M-B7a | S-G3 | |
| HDP 98354 | CDC 3437-70 | H-B3b | M-B7a | S-G1a | |
| S. gordonii | HDP 89552T | API 91.01.022, NCTC 7865T | H-B2a | M-G1a | S-H1a |
| HDP 84022 | API 77.09.010, NCTC 10231 | H-B2b | M-G1a | S-H1a | |
| HDP 89542 | CCUG 18374 | H-B2a | M-G1b | S-H1a | |
| HDP 99101 | CIP 103221 | H-B2a | M-G1a | S-H1a | |
| HDP 99285 | CIP 55.121 | H-B2a | M-G1a | S-H1b | |
| HDP 99287 | CIP 102337 (received as Streptococcus sp.) | H-B2a | M-G1a | S-H1c | |
| S. infantarius subsp. infantarius | HDP 90056T | NCDO 599T, CIP 103233T | H-B1 | M-E1a | S-G1b |
| HDP 90104 | Our collection, dairy product | H-B1 | M-E1c | S-G1b | |
| S. infantarius subsp. coli | HDP 90246 | NCDO 964 | H-B1 | M-E1b | S-G1b |
| HDP 90247 | NCDO 2602 | H-B1 | M-E1d | S-G1b | |
| HDP 90248 | NCDO 2620 | H-B1 | M-E1d | S-G1b | |
| S. intermedius | HDP 93161T | NCDO 2227T | H-E1b | M-B3 | S-C2 |
| HDP 99286 | CIP 105038 (received as S. gordonii) | H-E1a | M-B3 | S-C2 | |
| HDP 93162 | CDC 415-87 | H-E1a | M-B3 | S-C2 | |
| HDP 93167 | A. Coykendall, strain AC 6425 | E2c/E1a | M-A5b | S-C2 | |
| S. intestinalis | HDP 90052T | CIP 103224T | H-B4 | M-H1 | S-T1 |
| S. macacae | HDP 89541T | CIP 102912T | H-D4 | M-F3 | S-U8 |
| S. mitis | HDP 99284T | CIP 103335T, neo-type strain | H-A5c | M-B2c | S-A1d |
| HDP 84001 | API 78.04.067, NCTC 3168 | H-A1b | M-B2a | S-A2b | |
| HDP 84002 | CIP 103216 (received as S. sanguis biotype II) | H-A2a | M-B2b | S-A3 | |
| HDP 84009 | NCTC 10712 (received as S. sanguis biotype II) | H-A2b | M-G6 | S-A3 | |
| HDP 84010 | API 78.11.144, ATCC 10232 | H-A3a | M-F7 | S-A2a | |
| HDP 89503 | R. Facklam, strain SS-429 | H-A1b | M-B2b | S-A1d | |
| HDP 89609 | API 78.07.063 (received as S. sanguis biotype II) | H-A2a | M-E3c | S-A3 | |
| S. mutans | HDP 89502T | NCTC 10449T | H-G4 | M-G3 | S-I1a |
| HDP 97299 | ATCC 35668 | H-G4 | M-G3 | S-I1b | |
| S. oralis | HDP 89546T | CIP 102922T | H-A5a | M-E3a | S-A1a |
| HDP 89527 | NY SBE-844 | H-A2a | M-B2b | S-A1b | |
| HDP 89535 | A. Coykendall, strain AC 9811 | H-A2a | M-E3e | S-A1a | |
| HDP 89537 | A. Coykendall, strain OS51 | H-A5b | M-E3b | S-A1c | |
| S. parasanguinis | HDP 99102T | CIP 104372T | H-C1c | M-G1c | S-U5 |
| S. parauberis | HDP 98049T | CIP 103956T | H-D5a | M-D2 | S-M1 |
| HDP 90054 | NCDO 2018 | H-D5a | M-D2 | S-M1 | |
| HDP 98385 | NCDO 2056 | H-D5a | M-D2 | S-M1 | |
| HDP 98401 | NCDO 642 | H-D5a | M-D2 | S-M1 | |
| HDP 98402 | NCDO 648 | H-D5a? | M-D2 | S-M1 | |
| HDP 98403 | NCDO 649 | H-D5a? | M-D2 | S-M1 | |
| HDP 98404 | NCDO 650 | H-D5a? | M-D2 | S-M1 | |
| HDP 98405 | NCDO 689 | H-D5a? | M-D2 | S-M1 | |
| HDP 99184 | CIP 105803 | H-D5a | M-D2 | S-M1 | |
| S. pneumoniae | HDP 89540T | CIP 102911T | H-A3a | M-G2a | S-A1g |
| CNRP 70131 | P. Geslin, serotype 23F | H-A4b | M-G2c | S-U4 | |
| CNRP 70133 | P. Geslin, serotype 23F | H-A4a | M-G2c | S-U3 | |
| CNRP 70203 | P. Geslin, serotype 23F | H-A3a | M-C1c | S-V2 | |
| CNRP 70247 | P. Geslin, serotype 19F | H-A3a | M-C1b | S-A1f | |
| CNRP 70265 | P. Geslin, serotype 19F | H-A4a | M-G2b | S-A2a | |
| CNRP 70305 | P. Geslin, serotype 19F | H-A4a | M-G2b | S-A2b | |
| CNRP 70350 | P. Geslin, serotype 9V | H-A3a | M-C1a | S-A1e | |
| HDP 98040 | ATCC 6301, serotype 1 | H-A2b | M-G2a | S-A1c | |
| Strain R6 | Nontypeable strain | H-A4c | M-G2a | S-A2c | |
| S. porcinus | HDP 90049T | CIP 103218T | H-H2 | M-A1 | S-K1a |
| HDP 98090 | CIP 104385, serogroup U | H-H2 | M-A1 | S-K1b | |
| HDP 98112 | CIP 104338, serogroup P | H-H2 | M-A1 | S-K2 | |
| HDP 98089 | CIP 104384, serogroup E | H-H2 | M-A1 | S-K1b | |
| HDP 98111 | CIP 104386, serogroup V | H-H2 | M-A1 | S-K1b | |
| HDP 98149 | CIP 104389 | H-H2 | M-A1 | S-K1a | |
| HDP 98150 | CIP 104388 | H-H2 | M-A1 | S-K1a | |
| HDP 98151 | CIP 104387 | H-H2 | M-A1 | S-K1a | |
| S. pyogenes | HDP 89539T | P. Kriz, CIP 56.41T, biotype 1, serotype M1 | H-H1b | M-A2a | S-Q1a |
| HDP 93104 | P. Kriz, PHLS SF130/13, biotype 1, serotype M1 | H-H1b | M-A2a | S-Q2 | |
| HDP 93106 | P. Kriz, PZH GH1/Richard 55P serotype M3 | H-H1b | M-A2a | S-Q2 | |
| HDP 93109 | P. Kriz, RU T11/137/1, serotype M11 | H-H1b | M-A2c | S-Q1b | |
| HDP 93110 | P. Kriz, 63595-H1, serotype M12 | H-H1d | M-A2a | S-Q2 | |
| HDP 93112 | P. Kriz, NCTC 11/64, serotype M48 | H-H1d | M-A2b | S-Q2 | |
| HDP 98001 | Our collection, biotype 3 | H-H1b | M-A2b | S-Q2 | |
| HDP 98052 | Our collection, biotype 3 | H-H1c | M-A2a | S-Q3 | |
| HDP 98139 | Our collection, biotype 3 | H-H1a | M-A2a | S-Q3 | |
| HDP 98173 | Our collection, biotype 3 | H-H1b | M-A2a | S-Q3 | |
| HDP 98353 | Our collection, biotype 5 | H-H1a | M-A2a | S-Q3 | |
| HDP 98359 | Our collection, biotype 3 | H-H1a | M-A2a | S-Q3 | |
| HDP 98384 | Our collection, biotype 3 | H-H1a | M-A2a | S-Q3 | |
| S. ratti | HDP 89575T | ATCC 19645T | H-G3a | M-G4a | S-U1 |
| HDP 99416 | CIP 74.15 | H-G3b | M-G4b | S-U2 | |
| S. salivarius | HDP 89532T | CIP 102503T | H-C2 | M-E3f | S-J1 |
| S. sanguinis | HDP 84025T | API 77.09.011, NCTC 7863T | H-F5 | M-E4 | S-C1a |
| HDP 93177 | CDC 2405-81 | H-C1a | M-D3 | S-C1b | |
| HDP 99283 | CIP 103.231 | H-A1a | M-B2b | S-V1 | |
| S. sobrinus | HDP 89577T | A. Coykendall, strain SL1 | H-D3 | M-F4 | S-O1 |
| HDP 99417 | CIP 74.16 | H-G2 | M-A5b | S-U9 | |
| S. suis | HDP 90048T | CIP 103217T | H-D2 | M-D1a | S-P1 |
| S. thermophilus | HDP89530T | CIP 102303T | H-C2 | M-F5 | S-J1 |
| S. uberis | HDP 89504T | CCUG 17930T | H-E3 | M-B4 | S-D1a |
| HDP 98091 | CIP 101889 | H-E3 | M-B4 | S-D1b | |
| HDP 98406 | NCDO 640 | H-E3 | M-B4 | S-D1a | |
| HDP 98407 | NCDO 643 | H-E3 | M-B4 | S-D1b | |
| HDP 98408 | NCDO 658 | H-E3 | M-B4 | S-D1a | |
| HDP 98409 | NCDO 1513 | H-E3 | M-B4 | S-D1b | |
| HDP 98410 | NCDO 2022 | H-E3 | M-B4 | S-D1b | |
| HDP 99182 | CIP 105801 | H-E3 | M-B4 | S-D1a | |
| HDP 99183 | CIP 105802 | H-E3 | M-B4 | S-D1a | |
| S. vestibularis | HDP 90069T | CIP 103363T | H-C2 | M-E3d | S-J1 |
Preparation of DNA.
Bacteria (10 ml of a mid-log-phase culture) were harvested by centrifugation and washed twice in TE buffer (10 mM Tris, 1.0 mM Na2EDTA [pH 8.0]). Pellets were suspended in 500 μl of TES buffer (10 mM Tris, 1.0 mM Na2EDTA, 1 M sucrose [pH 8.0]) for lysis. Mutanolysin (5 U/ml; Sigma, St. Louis, Mo.) and lysozyme (10 mg/ml; Boehringer, Mannheim, Germany) were added, and the mixture was incubated overnight at 37°C. Bacterial membrane disruption was then done with proteinase K (0.4 mg/ml) and sodium dodecyl sulfate (1%, vol/vol). High-molecular-weight bacterial genomic DNA was purified from lysates by two sequential phenol-chloroform extraction and ethanol precipitation steps. DNA was dissolved in a buffer containing 10 mM Tris, 1 mM EDTA, and RNase (20 mg/ml). Its concentration and its integrity were determined visually after agarose (Bioprobe) gel electrophoresis (0.8%, wt/vol; 100 V; 1 h). Gels were stained by immersion in a 50-μg/ml ethidium bromide solution. DNA samples were diluted to a concentration of 1 μg/μl and microdialyzed (pore size, 0.22 μm; Millipore) before amplification.
Amplification.
Oligonucleotide primers Ad (5′-AGAGTTTGATCMTGGCTCAG) and O24/3 (5′-CGACATCGAGGTGCCAAA) were derived from conserved regions present at the 5′ end of the 16S rRNA gene and the 3′ end of the 23S rRNA gene, respectively (Giammanco et al., Abstr. 98th Gen. Meet. Am. Soc. Microbiol.). Purified DNA (1 μg) was added to a 74-μl aliquot of PCR mixture containing 0.4 pmol (each) of primers Ad and O24/3 (Genset, Paris, France), 250 μmol (each) of deoxynucleoside triphosphate (Pharmacia Biotech, Orsay, France), and 0.05 U of Taq polymerase (Gibco BRL, Cergy Pontoise, France) in 1× reaction buffer. Magnesium chloride (1.1 mM; Gibco BRL), 0.05% (vol/vol) wetting agent (Gibco BRL), and 0.08% (wt/vol) bovine serum albumin (Sigma) were added to the mixture. Amplification was done in a Perkin-Elmer 2400 system as follows: initial denaturation at 95°C for 5 min, 30 cycles of denaturation at 94°C for 45 s and annealing at 60°C for 60 s, and extension at 72°C for 3 min. Finally, a 7-min extension period at 72°C was carried out. Positive (DNA from S. agalactiae CNRP 96/409) and negative controls were included in each run of amplification. The presence of a specific PCR product was controlled by agarose gel electrophoresis (0.8%, wt/vol; 100 V; 1 h). The molecular weight of the amplification product was calculated using a 1-kb marker from Sigma.
Restriction endonuclease digestion and analysis.
Restriction was carried out for a 4-h period at 37°C in a 20-μl mixture containing 2 U of restriction enzymes HhaI, MboII, and Sau3AI (Amersham), 2 μl of the supplied specific incubation buffer, and 10 to 16 μl of PCR product. The volume of PCR product used in the restriction mixture was adjusted on the basis of the fluorescence intensity of the observed fragment in the control gel. Restriction was stopped by addition of 5 μl of 1× TE buffer (glycerol [50%, vol/vol], bromophenol blue 0.07%, wt/vol]). Before electrophoresis, restriction endonuclease digestions were heat inactivated as recommended by Giammenco et al. (14). Restriction fragment patterns were analyzed by gel electrophoresis at 120 V for 5 to 6 h in a 1.5% (wt/vol) Metaphor agarose (FMC BioProduct, Rockport, Maine) and 1.5% (wt/vol) standard agarose (Bioprobe) gel in 0.5× TAE buffer. DNA ladder 50-2000 (Bio-Rad) was loaded in four lanes of each gel for an accurate determination of the molecular weights of the restriction fragments. After migration, gels were stained by immersion in a 50-μg/ml BET solution for 1 h. Gels were then photographed and video acquired using Visiomic software (Genomic, Grenoble, France). The produced TIFF-formatted file was then reopened with the RestrictoScan program from the Taxotron package (Institut Pasteur, Paris, France). In each lane, restriction fragments were detected and their positions were consecutively noted. Calculation of the molecular size of each fragment was done using the Schaffer and Sederoff algorithm implemented in RestrictoTyper program. The sizes of the fragments were then entered in a database. Clustering of restriction patterns based on the single-linkage, average-linkage, and unweighted pair-group method using arithmetic averages (UPGMA) algorithms were done using the Adanson and Dendrograph programs from the Taxotron package.
In silico analysis.
In order to test the reliability of our method, virtual restriction analyses were performed on the nucleotide sequence of the rrn operon from S. pyogenes ATCC 700294 (serotype M1). The sequences were retrieved from the S. pyogenes sequence data published by the University of Oklahoma Health Sciences Center (28). In silico analysis was performed using the Geneman program from the Lasergene software (DNAStar, Madison, Wis.). Results were compared to those provided by the experimental restriction patterns of the amplified rrn operon of S. pyogenes HDP 89539T (serotype M1), HDP 93106 (serotype M3), and HDP 93112 (serotype M48).
Identification of atypical streptococcal isolates.
Eleven strains recently isolated from clinical specimens were studied. These isolates were identified as streptococci with conventional methods based upon morphological, cultural, and biochemical characteristics but exhibited either unexpected serological markers by the agglutination assay (Bio-Rad) or atypical biochemical traits when the rapid ID32STREP galleries (bioMérieux) were used. The three restriction patterns were first compared to their respective data base but, in an effort to allow a definitive identification, the three data bases were combined using the average-linkage algorithm. Restriction patterns of unknown isolates were then simultaneously compared to the complete data base, and a bacterial identification was then proposed. When analysis of the restriction patterns allowed identification of species represented in the library by a small number of strains, we confirmed the rrn-ARDRA identification by 16S rDNA sequence analysis as previously described (26).
RESULTS
Amplification and restriction of rrn operon.
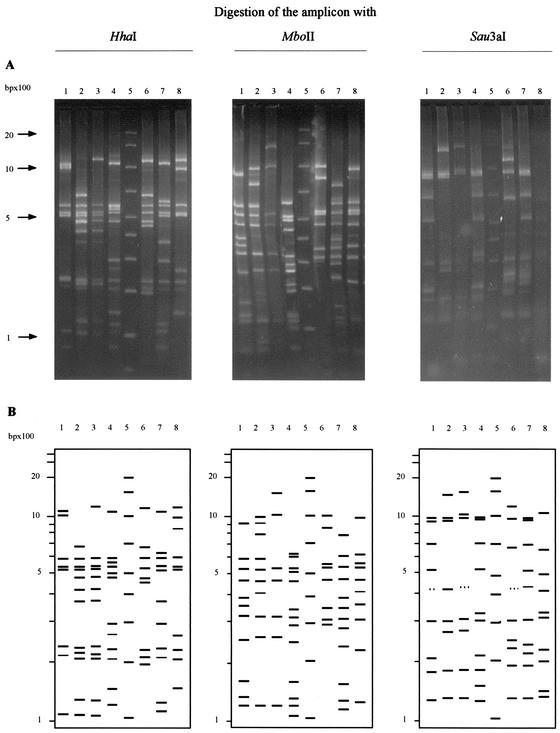
We have evaluated a total of 148 strains representative of 38 streptococcal species or subspecies mostly isolated from humans or animals. The PCR-based protocol allowed production of a single amplification product with an estimated size between 4,400 and 4,500 bp. This size was consistent with the size predicted after analysis of the published sequence of S. pyogenes ATCC 700294 (28). Electrophoretic analysis of the amplified product digested with HhaI, MboII, and Sau3A separately disclosed 7 to 14 bands ranging from 30 up to 1,500 bp (Fig. 1). BET staining and densitometric detection of smaller bands (i.e., bands under 100 bp) were not sufficiently reliable between runs, and these bands were not recorded for further analysis.
FIG. 1.
Restriction analysis of amplifed operon with HhaI, MboII, and Sau3A restriction endonucleases. 1, S. pyogenes HDP 89539T; 2, S. infantarius HDP 90056T; 3, S. equinus HDP 89506T; 4, S. porcinus HDP 90049T; 5, molecular size marker; 6, S. pneumoniae HDP 89540T; 7, S. pyogenes HDP 93110; 8, S. constellatus HDP 89579T. (A) Visualization of the bands obtained after migration and BET staining. (B) Schematic representation of agarose gel. Only bands ranging from 100 to 2,000 bp are used for identification and are represented here.
In silico restriction analysis.
Since the published sequence of the entire chromosome of S. pyogenes ATCC 700294 includes six amplified fragments of the rrn operon, we considered different sequences corresponding to positions 268.758 to 273.233 (4,476 bp), 515.696 to 520.172 (4,477 bp), 1.587.525 to 1.583.047 (4,479 bp), 1.772.590 to 1.768.155 (4,476 bp), 1.828.809 to 1.824.334 (4,475 bp), and 1.834.809 to 1.830.335 (4,475 bp). Each fragment was virtually digested with the three endonucleases. The six restriction patterns were then combined together and compared with the experimental patterns obtained with S. pyogenes HDP 89539T, HDP 93106, and HDP 93112. The real patterns displayed all the predicted bands and also few extra bands after HhaI and Sau3A digestion. However, these additional bands are very faint and may correspond to strain-specific alterations of the DNA sequence of only one or a few copies of the operon.
Restriction length polymorphism analysis.
Visual observation of restriction patterns demonstrated a high interspecies variability and a low intraspecies variability (Fig. 1 and 2). In some species, the patterns successively obtained with the three restriction endonucleases were very homogeneous even though a large set of strains was studied (S. agalactiae, S. equi, S. gordonii, S. parauberis, S. uberis, and S. pyogenes strains). Thus, the construction of a similarity matrix and the statistical clustering of patterns improved the detection of small alterations in patterns obtained for strains of the same species. A computerized database was then built with restriction patterns using the average-linkage algorithm. The fragment size tolerance (experimental error) was set to 5%, which is superior to the usual interrun variations. Results of clustering analysis were identical when carried out by using UPGMA, single-linkage, or average-linkage analysis (data not shown). Initially the clustering analysis was done with each endonuclease separately, and a designation was assigned to each pattern, corresponding to a capital letter for major subdivisions of the dendrogram (distance between patterns > 0.25), an Arabic number for the different cluster of patterns inside these subdivisions (distance between 0.25 and 0.05), and a lowercase letter for minor variations of pattern (distance between patterns < 0.05) (Table 1).
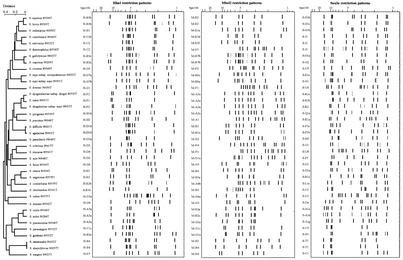
FIG. 2.
Diagrammatic representation showing restriction patterns database for rrn-ARDRA obtained with type strains of major streptococcal species. Each restriction pattern was named separately in each restriction endonuclease database. The designation was assign to each digestion pattern corresponding to a capital letter for major subdivisions of the dendrogram (similarity index over 0.75), an arabic number for the different cluster of patterns inside of these subdivisions (similarity index between 0.75 and 0.95) and a lowercase letter for minor variations of pattern (similarity of patterns comprised between 0.95 and 0.99). Dendrogram was calculated using the UPGMA algorithm, for a database limited to the restriction pattern of the type strains of the species.
HhaI analysis alone allowed the delineation of most of the species, except those within the “mitis” group, including S. mitis, S. oralis, and S. pneumoniae (cluster designated as H-A with large strain-specific variations). Patterns obtained for S. bovis and S. gallolyticus isolates are also combined together (H-B3). Subspecies of S. dysgalactiae and S. equi were not delineated after digestion with HhaI endonuclease (patterns H-F2). Contrarily, restriction patterns of strains belonging to S. anginosus were assigned to different clusters (H-C1, H-E3, H-F3, and H-F4).
In the MboII database, all S. anginosus strains were grouped together (M-B5 patterns). Restriction patterns obtained with S. dysgalactiae subsp. equisimilis, S. dysgalactiae subsp. dysgalactiae, and S. canis were also merged (M-A3). At the same time, subspecies of S. equi were not distinguishable (patterns M-B6).
Patterns obtained with Sau3A displayed a large variability among S. pyogenes or S. porcinus strains, which were divided in two or three clusters (S-Q1, -Q2, and -Q3; S-K1 and -K2). Within S. dysgalactiae, two clusters of patterns were observed (S-R1 and -R2) irrespective of the subspecies delineation according to reference identification. In contrast to HhaI and MboII, the patterns obtained with Sau3A for the two subspecies S. equi subsp. equi and S. equi subsp. zooepidemicus were separated in two different clusters (S-N1 and S-E1, respectively). Restriction patterns obtained for oral streptococci belonging to the mitis group (S. mitis, S. oralis, and S. pneumoniae) were not grouped in the same branch of the dendrogram as was observed with HhaI or MboII endonuclease.
The simultaneous observation of restriction patterns obtained with the three endonucleases was then proposed to improve the differentiation of strains corresponding to the different species and subspecies (Fig. 1 and 2). Minor alterations observed in one of the restriction patterns (band shift or addition or suppression of a single band) have a limited effect upon clustering when the two patterns obtained with the other restriction endonucleases are similar. At the same time, strains belonging to different subspecies and distinguished only by patterns obtained with one endonuclease were separated in the cumulative database. All the strains belonging to the 36 species studied, including type strains, reference strains, and clinical isolates, were included in separated clusters. The phylogenic tree obtained for cumulative analysis of the three sets of data after UPGMA analysis of type strains is presented in Fig. 2. A distance index of <0.05 was found for four pairs of related streptococcal species: S. equinus and S. bovis, S. gallolyticus and S. caprinus, S. agalactiae and S. difficilis, and S. alactolyticus and S. intestinalis, respectively. These results suggest that each of these couples corresponds to a single species. S. dysgalactiae subsp. dysgalactiae, S. dysgalactiae subsp. equisimilis, and S. canis appeared close to each other in cumulative analysis. In contrast, restriction patterns obtained for S. equi subsp. equi and S. equi subsp. zooepidemicus were clearly separated.
Identification of clinical isolates.
A total of 11 streptococcal strains identified with atypical characteristics were subjected to rrn-ARDRA analysis. Biochemical identification was confirmed in three cases and changed to another species in seven cases (Table 2). In four cases, the rrn-ARDRA identification was confirmed by the 16S rDNA analysis, including one strain which was initially unidentified according to its biochemical patterns and assigned to S. anginosus.
TABLE 2.
Identification of atypical or phenotypically unidentified streptococcal strains by rrn-ARDRA
| Strain no. | Conventional identification | Atypical characteristic(s)a | Restriction pattern
|
rrn-ARDRA identification | ||
|---|---|---|---|---|---|---|
| HhaI | MboII | Sau3a | ||||
| HDP 99045 | S. porcinus | Lancefield group B | H-H2 | M-A1 | S-K1a | S. porcinus |
| HDP 99520 | S. dysgalactiae | Lancefield group A | H-F2 | M-A3a | S-R2 | S. dysgalactiae |
| HDP 99296 | S. suis | β-GLU (−), MβDG (−), mannitol (+) | H-D2 | M-D1a | S-P1 | S. suis |
| HDP 91257 | S. bovis | Trehalose (−), raffinose (+), pullulan (+) | H-B1 | M-E1d | S-G1b | S. infantarius |
| HDP 99201 | S. sanguinis | β-GAR (−), trehalose (+), tagatose (−), dextran (+) | H-C1 | M-G1c | S-U5 | S. macacae |
| HDP 99294 | S. suis | ADH (−), β-GUR (−), β-MAN (+), raffinose (+) | H-B3b | M-E2 | S-G1c | S. equinus |
| HDP 99374 | S. suis | β-GAL (+), glycogen (−) | H-E1a | M-B3 | S-C2 | S. intermedius |
| HDP 99401 | S. parasanguinis | ADH (−), dextran (−) | H-A4a | M-E3e | S-A1a | S. oralis |
| HDP 99419 | S. suis | Lactose (−), β-GAL (+), β-NAG+, tagatose (+) | H-G2 | M-A5a | S-P1 | S. sobrinus |
| HDP 2000/341 | S. alactolyticus | α-GAL (−), mannitol (−), pullulan (+) | H-C2 | M-E3f | S-J1 | S. salivarius |
| HDP 2000/051 | Unidentified coccus from suppuration | ADH (−), β-GLU (−), VP (−), Lancefield group G | H-F4a | M-B5e | S-B1b | S. anginosus |
Atypical characteristics include atypical results for abilities to grow on the indicated substrates or to produce, the indicated compounds. Abbreviations: β-NAG, β-N-acetyl glycosylase; α-GAL, α-galactosidase; β-GAL, β-galactosidase; β-GUR, β-glucuronidase; β-GLU, β-glucosidase; β-MAN, β-mannosidase; β-NAG, N-acetyl glycosidase; VP, production of acetoin; ADH, arginine dihydrolase; MβDG, methyl-β-d-glucopyranoside.
DISCUSSION
Among molecular tools of bacterial identification, the determination of rRNA gene restriction patterns (ribotyping) has been extensively used (15). However, this methodology requires several steps, including transfer onto membranes and hybridization steps. Therefore, some alternative techniques have been proposed in an effort to obtain faster results. PCR amplification of DNA followed by restriction analysis of amplification products has attracted a growing interest. ARDRA (i.e., amplification and polymorphism of restriction analysis of 16S rRNA genes) has been proposed for the identification of species within some genera, such as Acinetobacter, Mycobacterium, and Mycoplasma (7, 30, 31). However, analysis restricted to the 16S rRNA gene appeared to be of limited value in some other genera (18). It was then suggested that inclusion of the 23S rRNA gene and 16-23S spacer in the analysis may improved the differentiation between species in some genera (18; Giammanco et al., Abstr. 98th Gen. Meet. Am. Soc. Microbiol.) as it allows the study of a larger amount of genetic information. Indeed, due to evolutionary constraints, there is a minimal variation into the 16S or the 23S rRNA sequences, whereas the 16S-23S intergenic sequence demonstrated an enhanced variability. The similarity of the sequences of 16S rRNA gene between different streptococcal species was determined to be between 91 and 99% (1). In contrast, the 16S-23S rRNA internal spacer is subjected to a minimal selective pressure compared to rRNA genes. Insertion, deletion, and single-base changes are frequent in this DNA fragment, and sequence identity between the intergenic spacers of different streptococcal species is limited to between 50 and 80% (10). In our study, a single band corresponding to the amplification of the rrn operon of streptococci was observed on the electropherogram. This indicates that the potential difference of lengths between the rrn operons of distinct species or between the six different rrn operons of the streptococcal strains (28) remains within the resolution of the agarose gel we used.
To determine the advantage and the reliability of the improved rrn-ARDRA protocol of identification, we have investigated 148 type and reference strains representative of the 39 major species and subspecies isolated from humans or animals. All these strains have been phenotypically characterized by their initial depositor, and most of them have also been identified by DNA-DNA hybridizations. A large interspecific variability of restriction profiles was detected among the species studied. When available, three to seven reference strains of each species were included in the study. The results of these investigations suggest that restriction patterns were conserved within species. So, restriction patterns obtained with strains of S. pyogenes were homogeneous and similar to the profiles predicted by DNA sequence analysis. In fact, controversial data were published about the presence or the absence of variable restriction sites in different copies of the 16S rRNA gene within the bacterial chromosome. Some studies reported the absence of intraspecies variations of 16S rRNA genes (18, 30), but a large variability of restriction patterns of 16S RNA was observed within Abiotrophia adiacens in the study of a large collection of Abiotrophia strains (23). Gurtler et al. (16) have also reported highly variable restriction patterns in a recent analysis of the clostridial species. Alterations of one or a few copies of the rrn operon may have resulted in faint bands on the electropherogram, but these additional bands, like the bands due to minor or incomplete digestion, did not appear essential for the differentiation between the restriction patterns of different species (31). Some other minor variations of restriction patterns appeared unpredictable according to the species studied or to the restriction endonuclease used. These variations were assumed to be strain specific.
The construction of a combined database using the three restriction patterns obtained separately for each strain was proposed to overtake the strain variability and achieve a better delineation of species (Giammanco et al., Abstr. 98th Gen. Meet. Am. Soc. Microbiol.). Using the triple combination of HhaI, MboII, and Sau3A restriction endonucleases, strains belonging to the same species are found to exclusively cluster in the same restriction patterns. When 3 to 10 strains were studied, none of the strains were assigned to an inadequate cluster with the noticeable exceptions of S. dysgalactiae and S. canis, as well as S. pneumoniae, S. mitis, and S. oralis. We observed that enlargement of the available database of patterns usually induces a better clustering of the restriction patterns around the type strain of the species. The dendrogram that we obtained using average-linkage or UPGMA clustering methods confirms the major subdivisions of the classification of streptococci and the results based on rRNA (1) or superoxide dismutase (19, 24) sequence analysis
The classification of S. anginosus, S. constellatus, and S. intermedius, which has now been clarified (33), is clearly delineated using rrn-ARDRA. However, the similarity of their HhaI and Sau3A patterns confirms the phylogenic proximity of these three species (33). In the case of S. uberis and S. parauberis, where biochemical or serological differentiation is not possible (2, 34), rrn-ARDRA clearly distinguishes between the two species. We also noticed the inclusion of strains belonging to the species S. bovis, S. caprinus, S. difficilis, or S. intestinalis into the respective clusters of S. equinus, S. gallolyticus, S. agalactiae, and S. alactolyticus. Our data appear consistent with the results of DNA-DNA reassociation experiments which have recently demonstrated the synonymy of these species.
The strains of S. porcinus, S. pyogenes, or S. pneumoniae cluster together within their respective species, including whatever Lancefield's serogroups or serotypes they belong, allowing a rapid species identification. Since similar conserved patterns were observed for different strains within the same species, rrn-ARDRA may have a reduced epidemiological (typing) interest. rrn-ARDRA could not differentiate between the two subspecies of S. dysgalactiae and S. canis. These species are part of a large and heterogeneous group of C, G, or L streptococci associated with human and animal infections. HhaI, MboII, and Sau3A patterns are quite similar for these three species as well, although DNA homology between species is between 45 and 75% (32) and 16S rRNA similitude value is limited to 96 to 97% (1). However, the classification of this group remained controversial as no test was clearly capable of differentiating these species (9, 29, 32).
Similarly, identification of strains belonging to S. pneumoniae, S. oralis, and S. mitis to the species level is also difficult with rrn-ARDRA. These three species constitute a single cluster of restriction patterns and the distinct differences seen between the three type strains are not observed with other species. Similar results were found using analysis of superoxide dismutase (19, 24) or the 16S rRNA gene sequence (1). S. pneumoniae, S. oralis, and S. mitis were demonstrated to form a bacterial complex in which both biochemical or genotypical differentiation may be difficult (K. Poulsen and M. Kilian, ASM Conf. Streptococcal Genet., abstr. 2D-05, 1998). In contrast, species belonging to the streptococcal group of S. sanguinis, S. parasanguinis, and S. gordonii displayed restriction patterns clearly separated from the S. pneumoniae, S. oralis, and S. mitis complex.
The data presented here show that PCR amplification followed by restriction analysis of the amplified products is a reliable and easy-to-use method of identification of streptococci. It has been demonstrated on a limited scale to be capable of identifying clinical isolates that were phenotypically mis- or not identified and appears to be a promising method for identification of pathogenic streptococci.
As observed for identification of Enterobacteriaceae or Acinetobacter, the performances of rrn-ARDRA were found excellent for identification of gram-positive, catalase negative cocci belonging to the genus Streptococcus. In order to improve these performances, determination of restriction patterns of large set of bacteria have to be conducted and databases of patterns have to be extended. Using our modified rrn-ARDRA protocol, a single database which combine the three restriction patterns obtained for each strain, may allow a rapid identification irrespective of the Gram staining or the morphological characteristics of the isolate. We propose to use this method in reference centers for identification of phenotypically unidentified isolates after construction and evaluation of specific databases of interest.
REFERENCES
- 1.Bentley, R. W., J. A. Leigh, and M. D. Collins. 1991. Intrageneric structure of Streptococcus based on comparative analysis of small-subunit rRNA sequences. Int. J. Syst. Bacteriol. 41:487-494. [DOI] [PubMed] [Google Scholar]
- 2.Bentley, R. W., J. A. Leigh, and M. D. Collins. 1993. Development and use of species-specific oligonucleotide probes for differentiation of Streptococcus uberis and Streptococcus parauberis. J. Clin. Microbiol. 31:57-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouvet, A., F. Grimont, and P. A. D. Grimont. 1991. Intraspecies variations in nutritionally variant streptococci: rRNA gene restriction patterns of Streptococcus defectivus and Streptococcus adjacens. Int. J. Syst. Bacteriol. 41:483-486. [DOI] [PubMed] [Google Scholar]
- 4.Bouvet, A., F. Grimont, M. D. Collins, F. Benaoudia, C. Devine, B. Regnault, and P. A. D. Grimont. 1997. Streptococcus infantarius sp. nov. related to S. bovis and S. equinus. Adv. Exp. Med. Biol. 418:393-395. [DOI] [PubMed] [Google Scholar]
- 5.Bridge, P. D., and P. H. A. Sneath. 1983. Numerical taxonomy of Streptococcus. J. Gen. Microbiol. 129:565-597. [DOI] [PubMed] [Google Scholar]
- 6.Coykendall, A. L. 1989. Classification and identification of the viridans streptococci. Clin. Microbiol. Rev. 2:315-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng, S., C. Hiruki, J. A. Robertson, and G. W. Stemke. 1992. Detection by PCR and differentiation by restriction length polymorphism of Acholeplasma. Spiroplasma, Mycoplasma, and Ureaplasma, based upon 16S rRNA genes. PCR Methods Appl. 1:202-204. [DOI] [PubMed] [Google Scholar]
- 8.Doit, C., F. Grimont, R. A. Whiley, B. Regnault, P. A. D. Grimont, J. M. Hardie, and A. Bouvet. 1994. Ribotypes of the ‘Streptococcus milleri'-group allow discrimination between strains of Streptococcus constellatus, Streptococcus intermedius and Streptococcus anginosus, p. 531-532. In A. Totolian (ed.), Pathogenic streptococci, present and future. Lancer. St. Petersburg, Publications, Russia.
- 9.Efstratiou, A., G. Colman, G. Hahn, J. F. Timoney, J. M. Bouefgras, and D. Monget. 1994. Biochemical differences among human and animal streptococci of Lancefield group C or group G. J. Med. Microbiol. 41:145-148. [DOI] [PubMed] [Google Scholar]
- 10.Forsman, P., A. Tilsala-Timisjarvi, and T. Alatossava. 1997. Identification of staphylococcal and streptococcal causes of bovine mastitis using 16S-23S rRNA spacer regions. Microbiology 143:3491-3500. [DOI] [PubMed] [Google Scholar]
- 11.Freney, J., S. Bland, J. Etienne, M. Desmonceaux, J. M. Boeufgras, and J. Fleurette. 1992. Description and evaluation of the semiautomated 4-hour rapid ID32Strep method for identification of streptococci and members of related genera. J. Clin. Microbiol. 30:2657-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcià-Arata, M. I., P. Gerner-Smidt, F. Baquero, and A. Ibrahim. 1997. PCR-amplified 16S and 23S rDNA restriction analysis for the identification of Acinetobacter strains at the DNA group level. Res. Microbiol. 148:777-784. [DOI] [PubMed] [Google Scholar]
- 13.Garnier, F., G. Gerbaud, P. Courvalin, and M. Galimand. 1997. Identification of clinical relevant viridans group streptococci to the species level by PCR. J. Clin. Microbiol. 35:2337-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giammanco, G. M., F. Grimont, and P. A. D. Grimont. 1999. MboII endonuclease heat inactivation before agarose gel electrophoresis to prevent artifactual bands in restriction patterns. BioTechniques 27:886-887. [DOI] [PubMed] [Google Scholar]
- 15.Grimont, F., and P. A. D. Grimont. 1986. Ribosomal ribonucleic acid gene restriction patterns as potential taxonomic tools. Ann. Inst. Pasteur Microbiol. B 137:165-175. [DOI] [PubMed] [Google Scholar]
- 16.Gurtler, V., V. A. Wilson, and B. C. Mayall. 1991. Classification of medically important clostridia using restriction endonuclease site difference of PCR-amplified 16S rDNA. J. Gen. Microbiol. 137:2673-2679. [DOI] [PubMed] [Google Scholar]
- 17.Hardie, J. M. 1986. Genus Streptococcus, p. 1043-1071. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. The Williams & Wilkins Co., Baltimore, Md.
- 18.Ibrahim, A., P. Gerner-Smidt, and S. Sjöstedt. 1996. Amplification and restriction endonuclease digestion of a large fragment of genes coding for rRNA as a rapid method for discrimination of closely related pathogenic bacteria. J. Clin. Microbiol. 34:2894-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawamura, Y., R. A. Whiley, S. E. Shu, T. Ezaki, and J. Hardie. 1999. Genetic approaches to the identification of the mitis group within the genus Streptococcus. Microbiology 145:2605-2613. [DOI] [PubMed] [Google Scholar]
- 20.Kilian, M., L. Mikkelsen, and J. Henrichsen. 1989. Taxonomic study of viridans streptococci: description of Streptococcus gordonii sp. nov. and emended description of Streptococcus sanguis (White and Niven 1946). Streptococcus oralis (Bridge and Sneath 1982) and Streptococcus mitis (Andrewes and Horder 1906). Int. J. Syst. Bacteriol. 39:417-484. [Google Scholar]
- 21.Kilpper-Bälz, R., and K. H. Schleifer. 1984. Nucleic acid hybridization and cell wall composition studies of pyogenic streptococci. FEMS Microbiol. Lett. 24:355-364. [Google Scholar]
- 22.Lancefield, R. C. 1933. A serological differentiation of human and other groups of hemolytic streptococci. J. Exp. Med. 59:571-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohara-Nemoto, Y., S. Tajika, M. Sasaki, and M. Kaneko. 1997. Identification of Abiotrophia adiacens and Abiotrophia defectiva by 16S rRNA gene PCR and restriction fragment length polymorphism analysis. J. Clin. Microbiol. 35:2458-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poyard, C., G. Quesne, S. Coulon, P. Berche, and P. Trieu-Cuot. 1998. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependant superoxide dismutase. J. Clin. Microbiol. 36:41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudney, J. D., and C. J. Larson. 1993. Species identification of oral viridans streptococci by restriction fragment polymorphism analysis of rRNA genes. J. Clin. Microbiol. 31:2467-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schlegel, L., F. Grimont, M. D. Collins, B. Régnault, P. A. D. Grimont, and A. Bouvet. 2000. Streptococcus infantarius sp. nov., Streptococcus infantarius subsp. infantarius subsp. nov., and Streptococcus infantarius subsp coli subsp. nov., isolated from humans and food. Int. J. Syst. E vol. Microbiol. 50:1425-1434. [DOI] [PubMed] [Google Scholar]
- 27.Schleifer, K. H., and R. Klipper-Bälz. 1987. Molecular and chemotaxonomic approches to the classification of streptococci, enterococci and lactococci: a review. Syst. Appl. Microbiol. 10:1-19. [Google Scholar]
- 28.Suvorov, A. N., and J. J. Ferretti. 1996. Physical and genetic chromosomal map of an M-type 1 strain of Streptococcus pyogenes. J. Bacteriol. 178:5546-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vandamme, P., B. Pot, E. Falsen, K. Kersters, and L. A. Devriese. 1996. Taxonomic studies of Lancefield streptococcal group C, G, and L (Streptococcus dysgalactiae) and proposal of S. dysgalactiae subsp. equisimilis subsp. nov. Int. J. Syst. Bacteriol. 46:774-781. [DOI] [PubMed] [Google Scholar]
- 30.Vaneechoutte, M., H. De Beenhouwer, C. Claeys, G. Verschraegen, A. De Rouck, N. Paepe, A. Elaichouni, and F. Portaels. 1993. Identification of Mycobacterium species with amplified rDNA restriction analysis. J. Clin. Microbiol. 31:2061-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaneechoutte, M., L. Dijkshoorn, I. Tjernberg, A. Elaichouni, P. De Vos, C. Claeys, and G. Verschraegen. 1995. Identification of Acinetobacter genomic species by amplified ribosomal DNA restriction analysis. J. Clin. Microbiol. 33:11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vieira, V. V., L. M. Teixera, V. Zahner, H. Momen, R. R. Facklam, A. G. Steigerwalt, D. J. Brenner, and A. C. D. Castro. 1998. Genetic relationships among the different phenotypes of Streptococcus dysgalactiae strains. Int. J. Syst. Bacteriol. 48:1231-1243. [DOI] [PubMed] [Google Scholar]
- 33.Whiley, R. A., and D. Beighton. 1991. Emended description and recognition of Streptococcus constellatus, Streptococcus intermedius and Streptococcus anginosus as distinct species. Int. J. Syst. Bacteriol. 41:1-5. [DOI] [PubMed] [Google Scholar]
- 34.Williams, A., and M. D. Collins. 1990. Molecular taxonomic studies on Streptococcus uberis types I and II. Description of Streptococcus parauberis sp. nov. J. Appl. Bacteriol. 68:485-490. [DOI] [PubMed] [Google Scholar]