Abstract
The increasing use of influenza virus neuraminidase (NA) inhibitors (NIs) necessitates the development of reliable methods for assessing the NI susceptibility of clinical isolates. We evaluated three NA inhibition assays against a panel of five clinical isolates each of influenza virus A/H1N1, A/H3N2, and B strains and four viruses with a defined resistance genotype (R292K, H274Y, R152K, and E119V). For fluorometric enzyme assay (FA) 1 (FA-1), 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (MUNANA) at 100 μM was used as the substrate, with pretitration of the virus input. For FA-2, MUNANA at 200 μM was used as the substrate, with a fixed 1:10 dilution of input virus. For the chemiluminescence (CL) assay, the 1,2-dioxetane derivative of sialic acid at 100 μM was used as the substrate, with pretitration of the virus. Four different operators repeated the assays several times in a blinded fashion with both zanamivir and oseltamivir carboxylate (GS4071) to determine intra- and interassay variations. Mean 50% inhibitory concentration (IC50) values were lower and generally less variable with the CL assay. FA-1 displayed greater variation than the CL assay or FA-2 and the highest IC50 values with zanamivir; FA-2 showed the highest values with oseltamivir, particularly for influenza virus B, and was more variable with zanamivir than was the CL assay. All three assays detected 40-fold or greater changes in IC50 values for the resistant viruses with at least one drug. Mixing experiments, whereby increasing fractions (0, 20, 40, 60, 80, and 100%) of NA from a known NI-resistant virus were mixed with the corresponding NI-sensitive parental NA, indicated that the resolution of IC50 values was clearer with the CL assay than with FA-2 for two of the resistant variants (R152K and E119V). The FA and CL methods were reliable for the detection of NI resistance, but all assays have certain limitations. Based on reproducibility, ease of automation, time required for the assay, and greater sensitivity, the CL assay was selected for future susceptibility testing of influenza virus isolates circulating globally.
Influenza virus neuraminidase (NA) enzyme is important for the release of virions from the host cell surface and viral aggregates and may also be involved in ensuring that the virus is targeted to respiratory epithelial cells (6). Viral NA has an active site that is highly conserved across all influenza virus A subtypes and influenza virus B strains (3, 19, 20). Competitive inhibition of NA results in the inhibition of virus replication, and a new class of anti-influenza virus drugs that specifically inhibit viral NA have been developed (6, 20). Since 1999, two NA inhibitors (NIs), inhaled zanamivir and oral oseltamivir, have been licensed for use in several parts of the world. An additional drug, BCX-1812 (RWJ-270201) (1), is currently in phase III clinical trials, and further potential NIs are in preclinical development (10).
A common concern with the use of all antiviral drugs is the development of drug-resistant virus strains and the potential for the transmission of these viruses. To date, relatively few viruses with altered susceptibility to NIs have been identified, as part of either preclinical or clinical development programs (2, 5, 7, 8, 9, 12, 16). Several NA mutants with reduced susceptibility have been selected by using zanamivir in vitro (E119G/A/D and R292K), but only one virus strain (R152K) with altered susceptibility has been isolated from an immunocompromised human treated with zanamivir (5). Three virus variants with reduced susceptibility have been recovered from patients treated with oseltamivir (E119V, H274Y and, most commonly but still at a low incidence, R292K); not all of the NA mutations in these viruses were predicted from in vitro studies (9, 16). In vitro selection experiments with BCX-1812 have also produced the R292K mutation (S. Bantia, S. Ananth, L. Horn, C. Parker, U. Gulati, Y. Babu, and G. Air, Antiviral Res. 46:A60, abstr. 82, 2000).
With the potential for the worldwide use of NIs for the management of influenza, public health and regulatory concerns have been raised regarding the clinical significance and epidemiological consequences of resistant virus strains emerging in infected and treated populations. In order to respond to these concerns, an expert panel composed of scientists and clinicians representing academia and public health organizations as well as industry observers, termed the Neuraminidase Inhibitor Susceptibility Network (NISN), was established to oversee an international surveillance program (21). Generation of data on global NI resistance requires robust, reproducible assays of drug susceptibility and an understanding of the specific technical problems that limit their usefulness.
For surveillance studies, cell culture-based assays to determine susceptibility (e.g., plaque reduction assay [PRA]) are desirable for initial screening because of their ability to detect a broad range of resistance phenotypes (15). However, the interpretation of NI susceptibility in PRA or other cell-based assays is complicated by several factors unique to NI drugs (18). Mutations in the hemagglutinin (HA) gene are selected frequently during in vitro passage in the presence of NIs and may be seen in conjunction with a mutation in the NA gene (12). Mutations in HA are generally in or near the receptor binding site and result in reduced binding to cellular receptors. Such mutations could lead to reduced virus dependence on NA function for the release of progeny virions, thereby conferring resistance to NI in cell-based assays (11, 18). However, reduced binding of HA may occur only for a specific subset of receptors. Receptors on MDCK cells, which are most often used for influenza virus growth inhibition assays, have both α2,3- and α2,6-linked terminal sialic acid residues, whereas human cells have only α2,6-linked sialic acids. Hence, a virus selected in humans and having decreased binding to α2,6-linked sugars may not exhibit reduced binding in MDCK cells and may show no reduced sensitivity to NI in a cell-based assay (5). Furthermore, viruses with HA mutations conferring apparent reduced NI susceptibility in vitro generally show full susceptibility in vivo in relevant animal models (18). Another limitation of the PRA is that NIs function by preventing the release of virus from infected cells, therefore producing smaller plaques. Although a reduction in plaque size or plaque number can be used to determine drug sensitivity, many clinical isolates do not form plaques well, so that this approach is not feasible (18).
For a global surveillance program, there is a need to determine as accurately as possible the NI susceptibility of a large number of clinical isolates both before and after the introduction of NI drugs. Enzyme inhibition assays are not subject to any of the complications of receptor specificity (12), and 96-well microtiter plate-based biochemical assays to determine enzyme inhibition have been used successfully to detect viruses with an NI drug resistance phenotype. At present, the substrate most widely used for detecting enzyme inhibition is the fluorogenic reagent 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (MUNANA), initially described by Potier et al. (14). Several published fluorometric enzyme assays (FAs) are similar in principle and vary primarily in the substrate concentration used, 30 through 1,000 μM, and whether titrated or empirical amounts of virus are used in the assay. (2, 8, 13, 17; E. Z. Baum, K. Andries, R. Willebrords, L. Ly, and K. Bush, Antiviral Res. 46:A58, abstr. 74, 2000; E. Covington, D. B. Mendel, P. Escarpe, C. Y. Tai, K. Soderbarg, and N. A. Roberts, J. Clin. Virol. 18:253-254, abstr. P-326, 2000). However, a small percentage of clinical isolates demonstrate low NA activity and cannot be evaluated with any of the available FAs (4). In order to overcome these problems, a sensitive chemiluminescence (CL) assay was developed with the 1,2-dioxetane derivative of sialic acid (NA-STAR) as the substrate (4); the CL assay with this substrate demonstrated up to 67-fold higher sensitivity for NA detection than did FAs.
Before screening large numbers of clinical isolates, it was essential to compare and validate the performance of different assays prior to choosing a single assay for the analysis of worldwide isolates. Because of the existence of both influenza type- or subtype-specific and drug-specific mutations, it was essential to select an assay that generated consistent results with each of the inhibitors. Consequently, we evaluated several candidate NI assays with two different substrates and both zanamivir and oseltamivir carboxylate (GS4071) against a panel of 15 globally representative, wild-type clinical isolates recovered during the influenza seasons of 1997 through 2000 as well as a panel of 4 NI-resistant viruses with a defined genotype. In addition, mixing experiments with viruses with a defined NI resistance genotype were also conducted to determine the ability of these assays to detect subpopulations of resistant variants.
MATERIALS AND METHODS
Viruses and cells.
A total of 15 different clinical influenza virus isolates (5 each of influenza viruses A/H1N1, A/H3N2, and B) were obtained from the repository of influenza virus strains collected by World Health Organization (WHO) laboratories during the influenza seasons of 1997 through 2000. The only criteria for selection were that the viruses were originally isolated from cell lines, since cell cultures are the major source of isolates worldwide and since egg growth can select for changes in influenza virus glycoproteins, and that they proliferated readily on Madin-Darby canine kidney (MDCK) cells. Some isolates representative of a low titer or low NA activity were included. Influenza virus clinical isolates with mutations in the NA gene and the corresponding NI-susceptible, wild-type viruses were provided by Margaret Tisdale, GlaxoSmithKline, Stevenage, United Kingdom (B/Memphis [R152K], selected during zanamivir treatment) (5), and by Noel Roberts, Roche Products Ltd., Welwyn Garden City, United Kingdom (A/H1N1 [H274Y], A/H3N2 [E119V], and A/H3N2 [R292K], selected during oseltamivir treatment) (9, 16). All viruses were expanded in cell cultures to provide stocks of sufficient volume, frozen at ≥70°C, and stored for use in two FAs (FA-1 and FA-2) as well as the CL assay. All virus stocks were identified with a code such that different operators did not know the identities.
Virus stocks were expanded on MDCK cells with a maximum of three passages. MDCK cells were originally obtained from Alan Hay, Mill Hill, London, United Kingdom. All expansion cultures and experiments were carried out with phenol red-free Eagle minimum essential medium (EMEM) (Gibco, Grand Island, N.Y.) containing 10% fetal bovine serum (Summit Biotechnology, Fort Collins, Colo.), 1 mM l-glutamine, 1% HEPES, and 1% penicillin-streptomycin (all from Gibco). Infection media consisted of EMEM supplemented with 0.14% bovine serum albumin fraction V and 2.5 μg of tosylsulfonyl phenylalanyl chloromethy ketone-trypsin/ml (both obtained from Worthington Biochemical Co., Lakewood, N.J.).
Compounds, reagents, and materials.
Zanamivir was provided by Margaret Tisdale, and oseltamivir carboxylate (GS4071) was provided by Noel Roberts. Oseltamivir phosphate (GS4104) is the ethyl ester prodrug that is converted to the biologically active oseltamivir carboxylate in vivo (13). MUNANA, 2-(N-morpholino)ethanesulfonic acid (MES), CaCl2, dimethyl sulfoxide, NaOH, and sodium acetate were all purchased from Sigma Chemical Co., St. Louis, Mo. NA-STAR CL substrate and Light Emission Accelerator II (Sapphire II enhancer in 0.1 M diethanol amine [pH 10]) were obtained from Applied Biosystems/Tropix (Foster City, Calif.). Tissue culture flasks, 96-well microtiter plates, and microplate press-on adhesive sealing film were purchased from Costar, and black 96-well Optiplates, white 96-well Optiplates, and TopSeal-a 96-well microplate press-on adhesive sealing film were acquired from Packard (Meriden, Conn.).
NA assays.
Three different NA enzyme inhibition assays were chosen for evaluation. The principle features of these methods are summarized in Table 1.
TABLE 1.
Summary of three influenza virus NI assaysa
| Assay format and condition | Characteristic of:
|
||
|---|---|---|---|
| CL assay | FA-1 | FA-2 | |
| Virus titration | Yes | Yes | No |
| Volume of virus and buffer (μl) | 50 | 20 | 50 |
| Substrate | 5 μl of NA-STAR (100 μM) | 30 μl of MUNANA (100 μM) | NA |
| Titration conditions | 15 min, 37°C, shaking | 1 h, 37°C | NA |
| Inhibition assay | |||
| Signal/noise ratio | 40:1 | 2:1 | NA |
| Volume of virus (μl) | 40 | 10 | 50 μl (1/10 dilution) |
| Volume of inhibitor (μl) | 10 | 10 | 50 |
| Inhibitor concn (nM) | 0.028-550 | 0.0038-1,000 | 0.01-10,000 |
| Inhibitor preincubation | 30 min, RT | 30 min, 37°C | 45 min, RT |
| Substrate | 5 μl of NA-STAR (100 μM) | 30 μl of MUNANA (100 μM) | 50 μl of MUNANA (200 μM) |
| Incubation | 15 min, 37°C shaking | 1 h, 37°C | 2 h, 37°C |
| Stop solution | 55 μl of Sapphire II enhancer | 150 μl of 0.14 M NaOH in 83% ethanol | 100 μl of 0.14 M NaoH in 83% ethanol |
| Substrate half-life | 5 min | Hours | Hours |
| Assay duration | 1 h | 2 h 30 min | 2 h 45 min |
| Instrumentation | NORTHSTAR Luminometer | FluoroCount fluorometer | FluoroCount fluorometer |
All assays were done with 96-well microtiter plates. RT, room temperature; NA, not applicable.
FA-1.
The unmodified assay described by Barnett et al. (2) (FA-1) requires the titration of input virus by making serial twofold dilutions and then graphically determining the virus and enzyme concentrations which fall in the linear part of the curve for the inhibition assay. A signal-to-noise ratio of ≥2 is considered optimal for use in the inhibition assay. Equal volumes of drug and the appropriate virus dilution were mixed and incubated for 30 min on black Optiplates (Table 1). The final drug concentrations in the assay ranged from 0.0038 to 1,000 nM in serial 1:4 dilutions. The reaction was initiated by the addition of buffer (32.5 mM MES [pH 6.5], 4 mM CaCl2) and substrate (100 μM MUNANA). After 1 h of incubation (37°C with shaking), the reaction was stopped by the addition of 150 μl of freshly prepared 0.14 M NaOH in 83% ethanol.
FA-2.
Previously described modifications of the above assay (8; Covington et al., J. Clin. Virol. 18:253-254, abstr. P-326, 2000) were incorporated into FA-2. This assay uses a twofold higher substrate concentration and eliminates the initial titration of input virus required in FA-1 by use of a standardized 1:10 dilution of the original virus isolate cell culture supernatant in EMEM. The drug concentrations in this assay ranged from 0.01 to 10,000 nM in serial 10-fold dilutions. The reaction was initiated by the addition of buffer (32.5 mM MES [pH 6.5], 4 mM CaCl2) and substrate (200 μM MUNANA). After 2 h of incubation (37°C with shaking, although shaking is not necessary), the reaction was stopped by the addition of 100 μl of freshly prepared 0.14 M NaOH in 83% ethanol. Once the reaction is initiated, this method requires a total incubation time of 2 h 45 min at 37°C.
Fluorometric determinations by both assays were quantified immediately with a Packard FluoroCount fluorometer. The excitation wavelength was 365 nm, and the emission wavelength was 450 nm.
CL assay.
The method of Buxton et al. (4) requires the growth of cells and virus in phenol red-free media, since residual phenol red can interfere with the assay. The virus was initially titrated in twofold dilutions in 32.5 mM MES (pH 6.0)-4 mM CaCl2, and the dilution giving a signal-to-noise ratio of 40 was used. Final drug concentrations ranged from 0.028 to 550 nM in serial 1:3 dilutions. The appropriate viral NA dilution (40 μl) was preincubated with 10 μl of drug for 30 min at room temperature on white Optiplates. The reaction was started by the addition of 5 μl of a 1:9 dilution of NA-STAR prepared in 32.5 mM MES (pH 6.0)-4 mM CaCl2. The final concentration of substrate used in the assay was 100 μM. The reaction mixture was incubated at 37°C for 15 min with shaking. CL light emission was triggered by the addition of 55 μl of Light Emission Accelerator II to each well. The half-life of the mixture is 5 min, so that rapid automation is required to optimize sensitivity. All assay results were immediately read with an Applied Biosystems (Foster City, Calif.) NORTHSTAR Luminometer.
Assessment of interassay variation.
Four different operators performed these experiments. Each isolate was assayed by each operator at least three times and at different positions in the batch (i.e., beginning, middle, and end); each assay involved duplicate wells for each drug dilution. Repetitions within isolate and drug took place over two consecutive days.
Determination of lower limits of detection of resistant subpopulations.
To determine the lower limits of detection of the CL assay and FA-2, known NI-resistant mutants and corresponding parental wild-type virus were combined in various proportions (0, 20, 40, 60, 80, and 100% resistant viruses) based on NA activity determined prior to assay of the mixtures. All virus mixtures were identified with a code to “blind” the operators from the known virus combinations, and the unknown stocks were stored at ≤−70°C until assayed. Each mixture was assayed with each of the three methods and evaluated with duplicate wells by a single operator three times.
Statistical analysis.
Fifty percent inhibitory concentration (IC50) values were calculated by using the Robosage Microsoft Excel add-in for curve fitting and calculation of IC50 values. The equation used in the Robosage program for curve fitting and calculation of IC50 values for all viruses tested was y = Vmax × {1 − [x/(K + x)]}. The description of this enzyme kinetics model is as follows. This equation describes a simple hyperbolic inhibition curve with a zero baseline. x is the inhibitor concentration, y is the response being inhibited (for example, y could be the velocity of an enzymatic reaction), and Vmax is the limiting response as x approaches zero. As x increases without bound, y tends toward its lower limit, zero. K is the IC50 for the inhibition curve, that is, y is 50% Vmax when x = K.
Descriptive measures, such as the mean, standard deviation (SD), minimum, maximum, fold difference [(maximum − minimum)/minimum], and coefficient of variation (CV), were used to summarize the data. Graphic presentations of the raw data were used to identify possible sources of variability and to characterize any patterns in the data. No formal hypothesis testing was performed.
RESULTS
Assay performance.
The observed IC50 values for each drug were affected by assay type, influenza virus type or subtype, and drug tested. In general, no pattern was apparent with operator or date of assay for either NI (data not shown). Tables 2 and 3 summarize the descriptive statistics for repetitions performed with clinical isolates and the drugs zanamivir and oseltamivir carboxylate (GS4071), respectively, for the three different assays.
TABLE 2.
Zanamivir IC50 values for each clinical isolate in three NI assays
| Influenza virus type | Isolate | IC50 (nM) in the following assay:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CL assay (n = 12)
|
FA-1 (n = 12)
|
FA-2 (n = 6)
|
||||||||
| Mean (SD) | CV | Rangea (fold change) | Mean (SD) | CV | Rangea (fold change) | Mean (SD) | CV | Rangea (fold change) | ||
| B | 97012351 | 1.00 (0.14) | 13.90 | 0.76-1.26 (0.7) | 6.77 (10.14) | 149.89 | 0.51-26.40 (50.8) | 4.66 (3.14) | 67.37 | 0.69-7.67 (10.1) |
| 97012942 | 1.07 (0.17) | 15.88 | 0.81-1.35 (0.7) | 5.28 (8.84) | 167.42 | 0.65-27.70 (41.6) | 5.19 (3.65) | 70.26 | 0.53-8.22 (14.5) | |
| 97013118 | 1.40 (0.37) | 26.58 | 1.04-2.51 (1.4) | 4.49 (8.34) | 185.89 | 0.76-29.80 (38.2) | 5.35 (3.70) | 69.23 | 0.70-9.21 (12.1) | |
| 97013406 | 1.03 (0.19) | 18.48 | 0.73-1.29 (0.8) | 5.44 (8.40) | 154.55 | 0.81-23.70 (28.3) | 2.89 (1.75) | 60.55 | 0.39-5.25 (12.5) | |
| 98010075 | 1.11 (0.13) | 12.11 | 0.91-1.34 (0.5) | 7.72 (10.54) | 136.45 | 0.56-30.00 (52.6) | 4.83 (3.22) | 66.73 | 0.67-7.59 (10.3) | |
| A/H1N1 | 98010152 | 0.89 (1.36) | 151.87 | 0.27-4.94 (17.3) | 1.86 (0.86) | 45.98 | 0.76-3.20 (3.2) | 1.21 (0.63) | 51.67 | 0.21-1.92 (8.1) |
| 98013212 | 0.27 (0.04) | 15.24 | 0.22-0.34 (0.5) | 1.32 (0.74) | 56.01 | 0.39-2.49 (5.4) | 0.50 (0.27) | 53.46 | 0.16-0.74 (3.6) | |
| 98014203 | 0.26 (0.03) | 12.40 | 0.19-0.30 (0.6) | 1.91 (1.19) | 62.32 | 0.48-3.84 (7.0) | 1.40 (1.11) | 79.26 | 0.19-2.86 (14.1) | |
| 98026506 | 0.26 (0.07) | 25.40 | 0.17-0.41 (1.6) | 2.53 (1.64) | 64.94 | 0.60-5.74 (8.6) | 0.89 (0.48) | 54.45 | 0.13-1.35 (9.4) | |
| 98027803 | 0.45 (0.68) | 150.39 | 0.22-2.61 (10.9) | 5.15 (5.82) | 112.98 | 0.42-18.40 (42.8) | 0.81 (0.53) | 65.52 | 0.11-1.32 (11.0) | |
| A/H3N2 | 97011746 | 1.63 (0.68) | 41.41 | 0.22-2.24 (9.0) | 3.96 (4.77) | 120.47 | 1.21-14.40 (10.9) | 2.96 (1.88) | 63.44 | 0.50-4.29 (7.4) |
| 97011803 | 1.70 (0.72) | 42.40 | 0.26-2.36 (8.1) | 2.88 (1.72) | 59.67 | 1.03-6.56 (5.4) | 2.55 (1.67) | 65.65 | 0.37-4.03 (9.9) | |
| 97012203 | 1.82 (0.43) | 23.53 | 1.21-2.90 (1.4) | 5.26 (6.47) | 123.08 | 1.06-18.80 (16.7) | 2.93 (2.06) | 70.19 | 0.35-5.41 (60.2) | |
| 98010134 | 1.63 (0.27) | 16.64 | 1.29-2.07 (0.6) | 6.08 (7.75) | 127.54 | 0.85-21.40 (24.2) | 1.86 (1.23) | 66.22 | 0.32-3.15 (8.8) | |
| 98026522 | 2.07 (0.92) | 44.25 | 1.54-4.94 (2.2) | 13.11 (14.02) | 106.98 | 1.77-43.80 (23.7) | 6.22 (4.40) | 70.69 | 0.89-11.00 (11.4) | |
Ranges are given as minimum to maximum.
TABLE 3.
Oseltamivir carboxylate (GS4071) IC50 values for each clinical isolate in three NI assays
| Influenza virus type | Isolate | IC50 (nM) in the following assay:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CL assay (n = 12)
|
FA-1 (n = 12)
|
FA-2 (n = 6)
|
||||||||
| Mean (SD) | CV | Rangea (fold change) | Mean (SD) | CV | Rangea (fold change) | Mean (SD) | CV | Rangea (fold change) | ||
| B | 97012351 | 3.99 (0.85) | 21.34 | 2.89-5.80 (1.0) | 13.29 (4.52) | 33.99 | 7.85-26.00 (2.4) | 67.88 (22.74) | 33.49 | 43.60-107.00 (1.5) |
| 97012942 | 3.63 (0.73) | 20.07 | 2.86-5.57 (0.9) | 13.05 (1.85) | 14.20 | 9.81-15.70 (0.6) | 65.75 (19.01) | 28.92 | 44.40-96.80 (1.2) | |
| 97013118 | 4.81 (1.75) | 36.45 | 2.99-9.35 (2.1) | 12.81 (3.34) | 26.11 | 7.24-19.70 (1.7) | 71.05 (23.02) | 32.40 | 45.90-114.00 (1.5) | |
| 97013406 | 5.32 (2.46) | 46.26 | 0.54-8.75 (15.2) | 23.23 (9.74) | 41.92 | 0.37-36.20 (96.8) | 88.87 (12.48) | 14.04 | 68.10-104.00 (0.5) | |
| 98010075 | 3.58 (0.59) | 16.49 | 2.55-4.53 (0.8) | 13.55 (4.20) | 30.98 | 8.49-22.40 (1.6) | 64.32 (19.98) | 31.07 | 41.00-94.90 (1.3) | |
| A/H1N1 | 98010152 | 0.48 (0.17) | 35.06 | 0.36-0.98 (1.7) | 2.16 (1.33) | 61.76 | 0.66-4.26 (5.4) | 3.56 (1.06) | 29.75 | 2.04-4.71 (1.3) |
| 98013212 | 0.47 (0.13) | 28.46 | 0.37-0.82 (1.2) | 2.82 (4.14) | 146.85 | 0.45-14.60 (31.4) | 3.42 (0.81) | 23.74 | 2.19-4.17 (0.9) | |
| 98014203 | 0.47 (0.09) | 19.35 | 0.36-0.64 (0.8) | 2.19 (1.02) | 46.67 | 1.11-4.41 (3.0) | 6.78 (5.79) | 85.47 | 1.96-16.90 (7.6) | |
| 98026506 | 0.43 (0.10) | 22.81 | 0.29-0.65 (1.2) | 4.68 (6.77) | 144.60 | 0.93-24.50 (25.3) | 3.70 (1.44) | 38.80 | 1.38-5.27 (2.8) | |
| 98027803 | 0.40 (0.29) | 73.91 | 0.01-1.05 (104) | 5.31 (6.65) | 125.21 | 1.73-25.30 (13.6) | 2.79 (1.43) | 51.24 | 0.81-5.21 (5.4) | |
| A/H3N2 | 97011746 | 0.54 (0.16) | 29.37 | 0.32-0.90 (1.8) | 0.84 (0.54) | 63.88 | 0.33-1.90 (4.8) | 1.77 (0.30) | 17.02 | 1.41-2.13 (0.5) |
| 97011803 | 0.52 (0.14) | 26.64 | 0.41-0.94 (1.3) | 0.78 (0.30) | 38.29 | 0.37-1.19 (2.2) | 1.60 (0.25) | 15.90 | 1.25-1.85 (0.5) | |
| 97012203 | 0.37 (0.07) | 18.21 | 0.25-0.53 (1.1) | 0.92 (1.04) | 112.21 | 0.20-3.13 (14.7) | 0.96 (0.13) | 13.89 | 0.75-1.11 (0.5) | |
| 98010134 | 0.41 (0.03) | 7.55 | 0.37-0.48 (0.3) | 0.44 (0.18) | 40.86 | 0.07-0.72 (9.3) | 1.05 (0.20) | 19.02 | 0.77-1.25 (0.6) | |
| 98026522 | 0.30 (0.10) | 32.96 | 0.22-0.59 (1.7) | 0.23 (0.24) | 103.75 | 0.07-0.94 (12.4) | 0.75 (0.16) | 21.81 | 0.59-0.99 (0.7) | |
Ranges are given as minimum to maximum.
CL assay.
For the CL assay, the ranges of IC50 values across all repetitions and clinical isolates were 0.17 to 4.94 nM for zanamivir and 0.01 to 9.35 nM for oseltamivir carboxylate. Across repetitions for a particular isolate, the IC50 values were consistent, and the SDs demonstrated low variability for each drug. With zanamivir, the fold differences between the minimum and maximum IC50 values ranged from 0.5 to 1.4, 0.5 to 17.3, and 0.6 to 9.0 for individual influenza virus B, A/H1N1, and A/H3N2 isolates, respectively. With oseltamivir carboxylate, the fold differences between the minimum and maximum IC50 values were 0.8 to 15.2, 0.8 to 104, and 0.3 to 1.8, respectively. The single high fold difference was due to a very low value (0.01 nM) observed for one influenza virus A/H1N1 isolate (Table 3). Similarly, the CVs for the isolates tested with zanamivir ranged from 12 to 152, and only two CVs, for A/H1N1 isolates 98010152 and 98027803, were greater than 100. The CVs for oseltamivir carboxylate ranged from 8 to 74, so that all of the CVs for the isolates tested fell below 100.
In addition, the IC50 values for each influenza type and subtype fell within a relatively narrow range in the CL assay. With zanamivir, the ranges of mean IC50 values were 1.0 to 1.4, 0.3 to 0.9, and 1.6 to 2.1 nM for influenza virus B, A/H1N1, and A/H3N2 isolates, respectively (Table 2). The corresponding values for oseltamivir carboxylate were 3.6 to 5.3, 0.4 to 0.5, and 0.3 to 0.5 nM, respectively (Table 3).
FAs.
In FA-1, the ranges of IC50 values across all repetitions and clinical isolates were 0.39 to 43.80 nM for zanamivir and 0.07 to 36.20 nM for oseltamivir carboxylate. The results of this assay demonstrated considerable variability. With zanamivir, the fold differences between the minimum and maximum IC50 values ranged from 28.3 to 52.6, 3.2 to 42.8, and 5.4 to 24.2 for individual influenza virus B, A/H1N1, and A/H3N2 isolates, respectively. With oseltamivir carboxylate, the corresponding fold differences ranged from 0.6 to 96.8, 3.0 to 31.4, and 2.2 to 14.7, respectively. Compared to the results of the CL assay, in which 2 of 15 viruses showed >10-fold differences between the minimum and maximum values on repeated testing with each drug (total of 4), 10 of 15 isolates tested with zanamivir and 6 of 15 isolates tested with oseltamivir carboxylate (total of 16) showed >10-fold differences (P value, 0.0022; two-sided Fisher's exact test). Similarly, the CVs for isolates tested with zanamivir ranged from 46 to 186, and 10 of 15 were greater than 100. With oseltamivir carboxylate, the CVs ranged from 14 to 147, and 5 of 15 were greater than 100.
In FA-2, the ranges of IC50 values across all repetitions and isolates were 0.11 to 11.00 for zanamivir and 0.59 to 114.00 for oseltamivir carboxylate. The wide range observed with oseltamivir carboxylate was bimodal in distribution, such that the IC50 values for influenza virus A/H1N1 and H3N2 isolates fell within 0.59 to 16.90 nM and those for the influenza virus B isolates were much higher, ranging from 41.00 to 114.00 nM. This assay showed variability that was intermediate between those of the CL and FA-1 methods. With zanamivir, the fold differences between the minimum and maximum IC50 values ranged from 10.1 to 14.5, 3.6 to 14.1, and 7.4 to 60.2 for individual influenza virus B, A/H1N1, and A/H3N2 isolates, respectively. With oseltamivir carboxylate, the corresponding fold differences ranged from 0.5 to 1.5, 0.9 to 7.6, and 0.5 to 0.7, respectively. Nine of 15 isolates tested with zanamivir (P value, 0.021 [versus the CL assay]) and none of 15 isolates tested with oseltamivir carboxylate (P values, 0.209 [versus the CL assay] and 0.115 [versus FA-1]) showed >10-fold differences (total of nine). The CVs for isolates tested with zanamivir ranged from 52 to 79, and those for isolates tested with oseltamivir carboxylate ranged from 14 to 85, so that all CVs were below 100.
Effects of viruses and drugs.
Wide variations on repeated testing (defined as >10-fold differences between the minimum and maximum IC50 values) tended to be limited to particular virus, drug, and assay combinations. One influenza virus A/H1N1 isolate (98027803) showed increased variability across all three assays with zanamivir and two of three assays with oseltamivir carboxylate. With zanamivir, both FA-1 and FA-2 showed increased variability (>10-fold differences between the minimum and maximum values) for all influenza virus B isolates and the majority of influenza virus A/H3N2 isolates. With oseltamivir carboxylate, FA-1 and particularly FA-2 showed higher IC50 values for influenza virus B isolates than did the CL assay. The ranges of mean oseltamivir carboxylate IC50 values for the five influenza virus B isolates were 3.6 to 5.3 nM for the CL assay, 12.8 to 23.2 nM for FA-1, and 64.3 to 88.9 nM for FA-2 (Table 3).
Testing of prototype resistant variants.
All three assays were able to detect clinical isolates with defined NA resistance mutations (Table 4), although the magnitude of the changes in observed IC50 values varied with the assay, the drug, and the particular NA. Each assay showed at least a 40-fold change in IC50 values for at least one drug against each of the mutant NAs compared to the susceptible, parental NA. All three assays showed over 350-fold changes in IC50 values for the A/Texas/36/91(H1N1) H274Y mutant, over 8,000-fold changes for the A/H3N2 R292K mutant, and over 40-fold changes for the A/H3N2 E119V mutant tested with oseltamivir carboxylate. In general, zanamivir retained inhibitory activity for each of these mutant NAs, although the two FAs showed more changes than did the CL assay for the A/H3N2 R292K mutant. In contrast, the influenza virus B isolates displayed the greatest variation in the pattern of inhibition, and the corresponding changes in IC50 values varied substantially (Table 4). The greatest discrimination between parental and mutant NAs in IC50 values was observed with zanamivir in FA-1 (148-fold) and, to a lesser extent, in FA-2 (44-fold) and with oseltamivir carboxylate (76-fold) in the CL assay.
TABLE 4.
Inhibitory concentrations of zanamivir and oseltamivir for parental and mutant NAs of influenza viruses recovered in clinical trials
| Isolate | NA | IC50 (nM) in the following assay with the indicated druga:
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CL assay
|
FA-1
|
FA-2
|
|||||||||||||||||
| Zanamivir
|
Oseltamivir
|
Zanamivir
|
Oseltamivir
|
Zanamivir
|
Oseltamivir
|
||||||||||||||
| Mean | SD | Fold change | Mean | SD | Fold change | Mean | SD | Fold change | Mean | SD | Fold change | Mean | SD | Fold change | Mean | SD | Fold change | ||
| B/Memphis/20/96 | 152R | 3.5 | 0.8 | 6.7 | 0.3 | 3.2 | 0.2 | 13.3 | 2.1 | 5.8 | 0.7 | 92.2 | 4.4 | ||||||
| 152K | 33.7 | 5.8 | 9.6 | 509.2 | 154.0 | 76 | 473.1 | 163.9 | 147.8 | 412.7 | 71.0 | 31 | 255.5 | 42.7 | 44.1 | 1,154 | 290 | 12.5 | |
| A/Texas/36/91 (H1N1) | 274H | 0.5 | 0.05 | 0.4 | 0.04 | 0.5 | 0.1 | 0.8 | 0.04 | 0.8 | 0.0 | 1.5 | 0.1 | ||||||
| 274Y | 0.7 | 0.05 | 1.4 | 253.9 | 20.2 | 634.8 | 0.8 | 0.2 | 1.6 | 282.8 | 12.3 | 353.5 | 1.0 | 0.1 | 1.3 | 680.0 | 61.5 | 453.3 | |
| A/Sydney/5/97 (H3N2) | 292R | 1.8 | 0.1 | 0.4 | 0.04 | 1.2 | 0.1 | 0.4 | 0.01 | 3.5 | 0.2 | 0.9 | 0.1 | ||||||
| 292K | 6.7 | 0.4 | 3.7 | 3,877 | 517 | 9,693 | 29.4 | 2.8 | 24.5 | 4,576 | 1,633 | 11,440 | 47.0 | 4.3 | 13.4 | 7,271 | 1,195 | 8,079 | |
| A/Wuhan/359/95- like (H3N2) | 119E | 0.7 | 0.05 | 0.3 | 0.01 | 1.6 | 0.5 | 1 | 0.7 | 6.1 | 1.7 | 0.5 | 0.1 | ||||||
| 119V | 1.3 | 0.1 | 1.9 | 15.6 | 2.8 | 52 | 1.4 | 0.2 | 0.9 | 106.4 | 11.1 | 106.4 | 6.1 | 0.4 | 1 | 167.7 | 30.0 | 335.4 | |
IC50 values are the mean of six independent assays. Fold change is the fold change in IC50 values for the mutant virus compared to the wild-type virus.
Detection of resistant subpopulations.
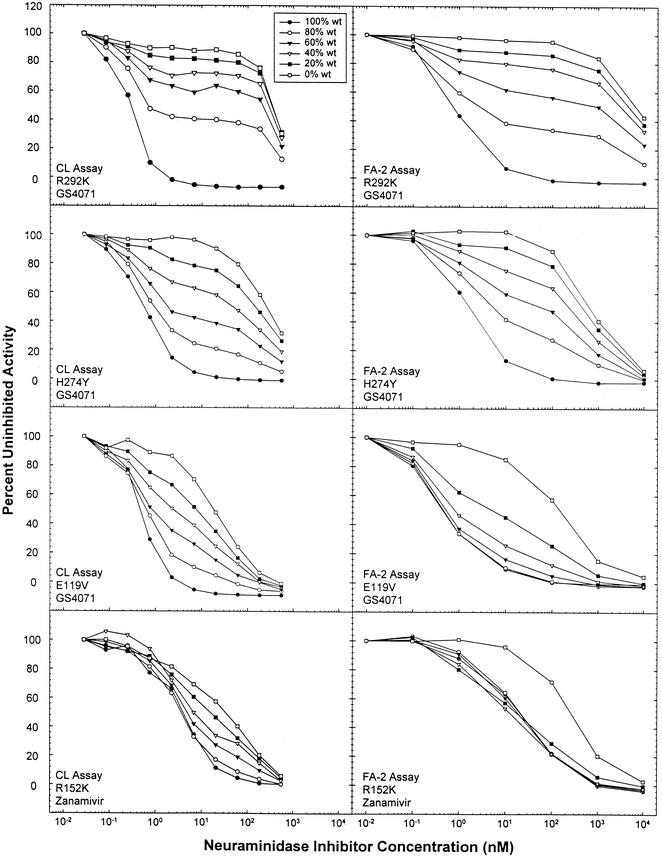
The mixing experiments were performed on the basis of relative NA activity and not infectious virus, so that the fractions of mixtures represented measurable NA signal input. Consequently, the number of resistant virions might have been relatively higher than that of the parental, drug-susceptible virus for a particular fraction. Figure 1 displays the results of the four experiments in which known NI-resistant mutants were mixed with associated parental wild-type viruses. Only the CL assay and FA-2 were studied, since our initial results showed that FA-1 was unlikely to be chosen for screening and resources were limited. Both assays showed marked shifts in inhibition when the susceptible parental virus was compared to 100% mutant virus (R292K, H274Y, and E119V) tested with oseltamivir carboxylate. Although initially recovered from a zanamivir-treated child, the influenza virus B R152K mutant demonstrated only about a 10-fold change with zanamivir in the CL assay (Fig. 1 and Table 4) but a greater difference with oseltamivir carboxylate in the CL assay (Table 4). Sigmoid curves were generated for all wild-type viruses by using both the CL assay and FA-2. Some resistant viruses (i.e., E119V and R152K) also produced sigmoid curves, whereas others (H274Y and R292K) did not do so within the concentration range of the NI tested. The CL assay was able to resolve each of the four different virus mixtures, while FA-2 could not do so when a lower mutant concentration was used in the E119V or R152K mixture.
FIG. 1.
Detection of NI-resistant influenza viruses in a mixture of wild-type and resistant viruses by the CL assay and FA-2. Wild-type and mutant viruses were mixed in the following ratios based on enzyme activity: 100:0, 80:20, 60:40, 40:60, 20:80, and 0:100. The results from one representative assay are displayed for each pair of viruses: R292K (A/H3N2), H274Y (A/H1N1), E119V (A/H3N2), R152K (B), and wild type (wt).
Effects of substrate concentrations.
Since substrate Km values vary by influenza virus NA type and subtype, as well as from isolate to isolate, the substrate concentration potentially has a direct effect on the sensitivity of the assay, as reflected by the observed IC50. We examined the effects of increasing the NA-STAR substrate concentration in assays with oseltamivir and zanamivir. Four mutant and wild-type isolate pairs were assayed a minimum of two times each at 100, 150, and 200 μM final NA-STAR concentrations. The concentration of substrate used when testing the wild-type influenza viruses had little effect on the mean IC50 values observed (data not shown). With oseltamivir, the IC50 values for the mutants increased approximately 1.5- to 3-fold when the concentration of substrate was increased from 100 to 200 μM. No important differences were observed with zanamivir across this concentration range (data not shown).
DISCUSSION
The purpose of these studies was to select an appropriate assay for high-throughput screening of clinical isolates of influenza virus in order to determine baseline susceptibility patterns and evaluate the possible emergence of NI-resistant isolates after the introduction of NI drugs into clinical practice. Since there are virus type, subtype, and drug-specific mutations, it is essential for the assay to be reproducible with N1, N2, and B NAs and the two drugs now in clinical use. We concluded that the assay that provided the most consistent results with both zanamivir and oseltamivir carboxylate and that was the easiest to use for testing large numbers of isolates was the CL assay. Consequently, it has been chosen for use by the NISN (21).
The CL assay demonstrated generally consistent IC50 values with low variability across repetitions for both NI drugs. A CV of greater than 100 is an arbitrary value used to denote instances where the SD is greater than the mean, a situation which may identify possible concerns regarding assay variability. With this criterion, only 2 of the 30 CVs across isolates, both for influenza virus A/H1N1 isolates tested with zanamivir, were greater than 100, a finding indicating that the inter- and intra-assay variabilities were low. Furthermore, all IC50 values fell within a narrow range, and the observed mean IC50 values were consistent among isolates of an influenza type or subtype. This background pattern of low variability should enhance the detection of possible resistant variants; the results obtained from testing of clinical isolates with defined resistance mutations confirmed this conclusion.
On average, higher IC50 values and ranges were obtained with FA-1 than with the CL assay. For almost every isolate, FA-1 yielded higher mean IC50 values than did the CL assay. This pattern was also reflected in greater variability and CVs for FA-1. When one considers the combined results with both drugs, fully one-half of the CVs were greater than 100 in FA-1, an observation which raises concerns about its variability. Consequently, the acceptability of this assay is dependent upon the user's awareness of the greater variability and of the necessity for incorporating appropriate control viruses with NAs having defined susceptibilities.
In FA-2, three factors, day of assay, drug type, and virus subtype, influenced the results of repetitions. With zanamivir, the IC50 values for all isolates were closer in magnitude to those obtained with the CL assay than to those obtained with FA-1. FA-2 showed excellent reproducibility with oseltamivir carboxylate but significantly greater variability with zanamivir than did the CL assay, such that nearly two-thirds of the clinical isolates showed >10-fold differences between the minimum and maximum IC50 values (Table 2). In addition, there was variability across days for the testing of isolates with zanamivir, such that the second day of experimentation yielded higher values on average than did the first day (data not shown). For oseltamivir carboxylate, the IC50 values for influenza virus A/H1N1 and H3N2 isolates were consistent across repetitions, with low variability in a range similar to that for the CL assay. However, although variability was low for the influenza virus B isolates, the mean IC50 values were 15- to 20-fold higher than those observed with the CL assay and 2- to 4-fold higher than those observed with FA-1. The reasons for these differences are likely related to differences in the interactions of the substrates and oseltamivir around the active enzyme site of influenza virus B isolates (20). While the possible clinical significance of such differences are unclear, high baseline IC50 values in FA-2 could make the detection of a resistant influenza virus B variant difficult. These results suggest that further optimization and validation would need to be performed before this assay could be used widely for clinical isolates with zanamivir and for influenza virus B isolates with oseltamivir carboxylate.
It is important for any assay used to detect antiviral resistance to be able to consistently detect populations of isolates exhibiting relatively low levels of resistance (i.e., isolates with IC50 values near the cutoff) or the existence of a predominant resistant subpopulation. All three assays were able to detect clinical isolates with defined resistance mutations, as long as both drugs were included in the screen (Table 4). Our mixing experiments focused on NA activity rather than on the amount of NA, since variant NAs often show reduced enzyme activity or stability. Although a log transformation is commonly used for IC50 data, it was not used for the graphic presentations, as this method may mask important sources of variability. Attention must be paid to curve shape as well as to the derived IC50 value, especially with regard to complete inhibition (i.e., return to baseline) at the highest inhibitor concentration, so that mixed populations can be detected. For example, the inhibition curves resulting from mixtures involving the R292K and H274Y mutants did not return to zero (Fig. 1), whereas the inhibition curves resulting from mixtures involving the R152K and E119V mutants did (Fig. 1). It is noteworthy that the shape of the inhibition curves does not appear to be assay dependent but instead is dependent on the mutation present in the active enzyme site.
In these studies, we used low-passage clinical isolates to make their context as relevant as possible. Of note, NA enzyme activities in clinical specimens are generally inadequate for direct susceptibility testing, and isolates need to be expanded in cell cultures to provide enzyme activity levels within the quantification range for all three assays that we tested. One limitation of this approach is that viruses with mutant NAs may have compromised replication in cell cultures and thus may be more difficult to detect after passage. However, although recognized variants with NA mutations show reduced enzyme activity or stability (8, 9, 12, 17, 18), most grow as well as their drug-susceptible parent in MDCK cells, making detection in a mixed population even more challenging.
Each of the three assays accurately measured IC50 values within virus type or subtype and drug type. All could be acceptably performed within reasonably well-designed influenza virus reference or research laboratories (Table 1). A wide selection of instrumentation is available for CL and fluorometric assays at greatly different costs. Because the CL substrate is a flash emitter with a half-life of 5 min, optimal instrumentation for the CL assay includes a Luminometer with injectors and fast read times (i.e., 1 min/96-well assay plate); otherwise, signal intensity will be significantly decreased. Instrumentation may be a limitation to the use of the CL assay due to its expense, although instruments less expensive than the Luminometer used in this study are available. Such instrumentation, however, is not an absolute requirement, as improved technical competence can overcome some time constraints.
The primary expenses of the assays are incurred in labor and the cost of substrates, MUNANA for FA-1 and FA-2 and NA-STAR for the CL assay. The reagents for FA-2 are more expensive than are those for FA-1 due to the increased final substrate concentration used in the former assay, but this substrate cost is offset by the elimination of the viral titration step. FA-1 and the CL assay are approximately equal in cost on a per-sample basis. MUNANA is widely available through a number of suppliers worldwide, whereas NA-STAR is supplied at present by a single commercial source (Applied Biosystems/Tropix, Foster City, Calif.). Ease of assay completion is essentially equivalent for all of the assays, each requiring a similar level of technical competence. Including the inoculum titration and incubation times, FA-1 requires a total performance time of 2 h 30 min, and the CL assay requires 1 h. FA-2 requires a total time of 2 h 45 min.
In conclusion, all three assays are suitable for quantifying the NA activity of influenza virus isolates and to detect resistant variants. FA-1 was more variable in these experiments but may be a reasonable choice for use given the acceptance of greater variability. Further optimization and validation of FA-2 are recommended prior to its use for testing of isolates with zanamivir and possibly influenza virus B isolates with oseltamivir carboxylate. The CL assay provided the most consistent results with the overall lowest variability. Based on these data, ease of automation, shorter assay time, and greater sensitivity, the NISN has selected the CL assay for future evaluation of the susceptibility of influenza virus isolates circulating globally.
Acknowledgments
J. L. McKimm-Breschkin, M. Zambon, and F. G. Hayden are members of the Neuraminidase Inhibitor Susceptibility Network. Other members include M. Aymard, Université Claude Bernard, Lyon, France; A. Hampson, WHO Collaborating Centre for Reference and Research on Influenza, Melbourne, Victoria, Australia; A. Hay, WHO Collaborating Center for Reference and Research on Influenza, National Institute for Medical Research, Mill Hill, London, United Kingdom; A. Klimov, WHO Collaborating Center for Surveillance, Epidemiology and Control of Influenza, Influenza Branch, Centers for Disease Control and Prevention, Atlanta, Ga.; A. Monto, University of Michigan School of Public Health, Ann Arbor, Mich.; M. Tashiro, WHO Collaborating Center for Reference and Research on Influenza, National Institute of Infectious Diseases, Tokyo, Japan; and R. Webster, St. Jude Children's Research Hospital, Memphis, Tenn.
REFERENCES
- 1.Babu, Y. S., P. Chand, S. Bantia, P. L. Kotian, A. Dehghani, Y. El-Kattan, T. Lin, T. L. Hutchinson, A. J. Elliott, C. D. Parker, S. L. Ananth, L. L. Horn, G. W. Laver, and J. A. Montgomery. 2000. BCX-1812 (RWJ-270201): discovery of a novel, highly potent, orally active, and selective influenza neuraminidase inhibitor through structure-based drug design. J. Med. Chem. 43:3482-3486. [DOI] [PubMed]
- 2.Barnett, J. M., A. Cadman, D. Gor, M. Dempsey, M. Walters, A. Candlin, M. Tisdale, P. J. Morley, I. J. Owens, R. J. Fenton, A. P. Lewis, E. C. J. Claas, G. F. Rimmelzwaan, R. De Groot, and A. D. M. E. Osterhaus. 2000. Zanamivir susceptibility monitoring and characterization of influenza virus clinical isolates obtained during phase II clinical efficacy studies. Antimicrob. Agents Chemother. 44:78-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burmeister, W. P., and P. M. Colman. 1992. The 2.2 A resolution crystal structure of influenza B neuraminidase and its complex with sialic acid. EMBO J. 11:49-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buxton, R. C., B. Edwards, R. R. Juo, J. C. Voyta, M. Tisdale, and R. C. Bethell. 2000. Development of a sensitive chemiluminescent neuraminidase assay for the determination of influenza virus susceptibility to zanamivir. Anal. Biochem. 280:291-300. [DOI] [PubMed] [Google Scholar]
- 5.Gubareva, L. V., M. N. Matrosovich, M. K. Brenner, R. C. Bethell, and R. G. Webster. 1998. Evidence for zanamivir resistance in an immunocompromised child infected with influenza B virus. J. Infect. Dis. 178:1257-1262. [DOI] [PubMed] [Google Scholar]
- 6.Gubareva, L. V., L. Kaiser, and F. G. Hayden. 2000. Influenza virus neuraminidase inhibitors. Lancet 355:827-835. [DOI] [PubMed] [Google Scholar]
- 7.Guvareva, L. V., L. Kaiser, M. N. Matrosovich, Y. Soo-Hoo, and F. G. Hayden. 2001. Selection of influenza virus mutants in experimentally infected volunteers treated with oseltamivir. J. Infect. Dis. 183:523-531. [DOI] [PubMed] [Google Scholar]
- 8.Ives, J. A. L., J. A. Carr, D. B. Mendel, C. Y. Tai, R. Lambkin, L. Kelly, J. S. Oxford, F. G. Hayden, and N. A. Roberts. 2002. The H274Y mutation in the influenza A/H1N1 neuraminidase active site following oseltamivir phosphate treatment leave virus severely compromised both in vitro and in vivo. Antivir. Res. 55:307-317. [DOI] [PubMed] [Google Scholar]
- 9.Jackson, H. C., N. Roberts, M. Z. Wang, and R. Belshe. 2000. Management of influenza; use of new antivirals and resistance perspective. Clin. Drug Investig. 20:447-454. [Google Scholar]
- 10.Kati, W. M., D. Montgomery, R. Carrick, L. Gubareva, C. Maring, K. McDaniel, K. Steffy, A. Molla, F. Hayden, D. Kempf, and W. Kohlbrenner. 2002. In vitro characterization of A-315675, a highly potent inhibitor of A and B strain influenza virus neuraminidases and influenza virus replication. Antimicrob. Agents Chemother. 46:1014-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKimm-Breschkin, J. L., T. J. Blick, A. Sahasrabudhe, T. Tiong, D. Marshall, G. J. Hart, R. C. Bethell, and C. R. Penn. 1996. Generation and characterization of variants of NWS/G70C influenza virus after in vitro passage in 4-amino-Neu5Ac2en and 4-guanidino-Neu5Ac2en. Antimicrob. Agents Chemother. 40:40-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKimm-Breschkin, J. L. 2000. Resistance of influenza viruses to neuraminidase inhibitors—a review. Antivir. Res. 47:1-17. [DOI] [PubMed] [Google Scholar]
- 13.Mendel, D. B., C. Y. Tai, P. A. Escarpe, W. Li, R. W. Sidwell, J. H. Huffman, C. Sweet, K. J. Jakeman, J. Merson, S. A. Lacy, W. Lew, M. A. Williams, L. Zhang, M. S. Chen, N. Bischofberger, and C. U. Kim. 1998. Oral administration of a prodrug of the influenza virus inhibitor GS 4071 protects mice and ferrets against influenza infection. Antimicrob. Agents Chemother. 42:640-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Potier, M., L. Mameli, M. Bélisle, L. Dallaire, and S. B. Melançon. 1979. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-α-d-N-acetylneuraminate) substrate. Anal. Biochem. 94:287-296. [DOI] [PubMed] [Google Scholar]
- 15.Reyes, M., N. S. Subedar, J. M. Graber, R. Nisenbaum, N. T. Wetherall, K. Fukuda, W. C. Reeves, et al. 2003. Acyclovir-resistant genital herpes among persons attending STD and HIV clinics. Arch. Int. Med. 163:76-80. [DOI] [PubMed]
- 16.Roberts, N. 2001. Treatment of influenza with neuraminidase inhibitors: virological implications. Philos. Trans. R. Soc. Lond. B 356:1895-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tai, C. Y., P. A. Escarpe, R. W. Sidwell, M. A. Williams, W. Lew, H. Wu, C. U. Kim, and D. B. Mendel. 1998. Characterization of human influenza virus variants selected in vitro in the presence of the neuraminidase inhibitor GS 4071. Antimicrob. Agents Chemother. 42:3234-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tisdale, M. 2000. Monitoring of viral susceptibility: new challenges with the development of influenza NA inhibitors. Rev. Med. Virol. 10:45-55. [DOI] [PubMed] [Google Scholar]
- 19.Varghese, J. N., and P. M. Colman. 1991. Three-dimensional structure of the neuraminidase of influenza virus A/Tokyo/3/67 at 2.2 A resolution. J. Mol. Biol. 221:473-486. [DOI] [PubMed] [Google Scholar]
- 20.von Itzstein, M., W. Y. Wu, G. B. Kok, M. S. Pegg, J. C. Dyason, B. Jin, T. Van Phan, M. L. Smythe, H. F. White, S. W. Oliver, P. M. Colman, J. N. Varghese, D. M. Ryan, J. M. Woods, R. C. Bethell, V. J. Hotham, J. M. Cameron, and C. R. Penn. 1993. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature 363:401-402. [DOI] [PubMed] [Google Scholar]
- 21.Zambon, M., and F. G. Hayden. 2001. Position statement: global neuraminidase inhibitor susceptibility network. Antivir. Res. 49:147-156. [DOI] [PubMed] [Google Scholar]