Abstract
Fifty-eight imipenem-nonsusceptible (MIC ≥ 8 μg/ml) Pseudomonas aeruginosa strains isolated during May 2001 in 15 Greek hospitals were studied. Thirty-six isolates derived from nine hospitals carried VIM-type metallo-β-lactamase genes, as found by PCR. In 34 isolates, blaVIM was associated with class 1 integrons of various sizes. DNA sequencing indicated the presence of blaVIM-2 gene cassettes in a variety of integron structures. Random amplified polymorphic DNA typing suggested diversity of the blaVIM-positive strains. Synergy between 2-mercaptoacetic acid and imipenem indicated carbapenemase activity in 26 blaVIM-positive strains.
Carbapenems exhibit potent antipseudomonal activity. Intensive use of these antibiotics, however, has facilitated the emergence of resistance in Pseudomonas aeruginosa. The mechanisms include decreased outer membrane permeability, overproduction of AmpC, up-regulation of multidrug efflux pumps (7), and production of carbapenem-hydrolyzing metallo-β-lactamases (MBLs) that belong to two types, IMP and VIM (8). The spread of MBL-producing P. aeruginosa strains has been reported mostly in the Far East (6, 17) and the Mediterranean region (3, 5, 12-14, 18).
In 2000 an outbreak of VIM-2-producing P. aeruginosa was described in a hospital in Thessaloniki, Greece (10, 18). Also, data from the National Surveillance System for Antimicrobial Resistance (WHONET-Greece) indicated an average frequency of imipenem-nonsusceptible (IPM-NS) (MIC ≥ 8 μg/ml, or ≤15-mm zone diameter in the disk diffusion test) P. aeruginosa isolates of 12% for the year 2000 (www.mednet.gr/whonet). This study was undertaken to assess the contribution of MBLs in this resistance. For this purpose, 18 hospital laboratories participating in WHONET-Greece were asked to contribute all IPM-NS P. aeruginosa clinical strains isolated during May 2001.
MICs of ceftazidime (CAZ), aztreonam (ATM), piperacillin-tazobactam (PTZ), IPM, and meropenem (MER) were determined by the Etest method (AB Biodisk, Solna, Sweden). Susceptibility to other antibiotics was assessed by disk diffusion (11). P. aeruginosa ATCC 27853 was used as a control.
VIM-type genes were detected by PCR using the primers VIM-F (5′-AGTGGTGAGTATCCGACAG-3′) and VIM-R (5′-ATGAAAGTGCGTGGAGAC-3′) (10). Detection of blaIMP by PCR was performed as described previously (16). Class 1 integrons were detected by PCR using the 5′CS and 3′CS oligonucleotides (15). Association of integrons with MBL genes was confirmed by PCR using combinations of bla- and integron-specific primers. Partial nucleotide sequences of selected PCR products were determined with an ABI Prism 377 DNA sequencer (Perkin Elmer, Applied Biosystems Division, Foster City, Calif.) using 5′CS, 3′CS and blaVIM-specific primers.
A synergy test using disks of CAZ (30 μg) and IPM (10 μg) combined with in-house-prepared disks containing mercaptoacetic acid (MAA) (3 μl per disk) was employed to detect strains producing MBLs as described previously (1).
Molecular typing was carried out by random amplified polymorphic DNA (RAPD) fingerprinting (9). Genomic DNA was extracted as described previously (4) and amplified by PCR using the oligonucleotide primer 208 (5′-AGCGGGCCAA-3′). Amplification products were separated in 1.5% agarose. RAPD patterns were compared as suggested by Campbell et al. (2).
Frequencies of IPM-NS P. aeruginosa isolates in the participating hospitals during 2001 ranged from 7 to 53%. Fifty-eight IPM-NS P. aeruginosa strains isolated in May 2001 were obtained from 15 hospitals. Three hospitals reported that no IPM-NS isolates were recovered during the study period (Table 1).
TABLE 1.
Isolation frequencies of IPM-NS P. aeruginosa strains in 18 hospitals during 2001
| Hospital (locationa) | No. of isolates in:
|
|||
|---|---|---|---|---|
| January-December 2001
|
May 2001
|
|||
| Total | IPM-NS (%) | Studied | blaVIM positivec | |
| A (Patra, SW) | NDb | ND | 10 | 7 |
| B (Ioannina, NW) | 317 | 114 (36) | 10 | 10 |
| C (Athens) | ND | ND | 5 | 5 |
| D (Alexandroupolis, NE) | 65 | 25 (38) | 1 | 0 |
| E (Thessaloniki) | 411 | 219 (53) | 5 | 5 |
| F (Athens) | 389 | 143 (37) | 2 | 1 |
| G (Athens) | 165 | 45 (27) | 5 | 0 |
| H (Heraklion, S) | ND | ND | 3 | 3 |
| I (Thessaloniki) | 43 | 11 (26) | 2 | 2 |
| J (Athens) | 403 | 76 (19) | 2 | 0 |
| K (Athens) | 222 | 20 (9) | 2 | 0 |
| L (Athens) | 129 | 33 (26) | 5 | 0 |
| M (Volos, E) | 88 | 16 (18) | 2 | 2 |
| N (Xanthi, N) | 92 | 12 (13) | 2 | 0 |
| O (Athens) | 180 | 55 (31) | 3 | 1 |
| P (Thessaloniki) | 85 | 9 (11) | 0 | |
| Q (Thessaloniki) | 29 | 2 (7) | 0 | |
| R (Athens) | 38 | 7 (18) | 0 | |
SW, southwest; NW, northwest; NE, northeast; S, south; E, east; N, north.
ND, not determined.
As determined by blaVIM-specific PCR assays.
Etest confirmed the reported status of susceptibility to IPM (all MICs were ≥8 μg/ml). In 53 isolates, levels of resistance to IPM and to MER were similar (differences of ≤2 doubling dilutions). For four isolates, MICs of IPM were significantly higher than those of MER; one isolate was more resistant to MER than to IPM. Frequencies of resistance to CAZ and ATM were 71 and 64%, respectively. Only nine (16%) and three (5%) isolates were susceptible to PTZ and ticarcillin-clavulanate, respectively. Extensive cross-resistance to non-β-lactam drugs was also observed. Fifty-one (88%) isolates were resistant to ciprofloxacin, and 53 (91%) were resistant to at least one aminoglycoside.
Of the 58 IPM-NS isolates, 36 (62%), derived from nine hospitals, were blaVIM positive, producing an amplicon of the expected size (260 bp). blaIMP-positive isolates were not detected. The presence of class 1 integrons was confirmed in 34 of the blaVIM-positive isolates. The sizes of the regions encompassed by the 5′ and 3′ conserved sequences ranged from 1.3 to 3.0 kb (Table 2). For 22 isolates, a single product of 1.5 kb was observed. For seven isolates, the sizes of the products were 1.8 to 2.0 kb. For three isolates, a product of approximately 3.0 kb was observed. Two amplicons, 1.0 and 1.3 kb, were found in each of the remaining two isolates. By combining the primer 5′CS with VIM-R and 3′CS with VIM-F, colinearity of blaVIM genes with class 1 integrons was indicated in all 34 integron-positive isolates. The sizes of the blaVIM-carrying integrons are in Table 2.
TABLE 2.
Characteristics of 36 blaVIM-containing IMP-NS P. aeruginosa isolates
| Hospital | Isolate | Result of MAA test | VIM integron size (kb) | RAPD type | β-Lactam resistance phenotypea
|
||||
|---|---|---|---|---|---|---|---|---|---|
| IPM | MER | CAZ | ATM | PTZb | |||||
| A | A1 | + | 1.5 | 1 | R | R | R | R | R |
| A2 | + | 1.5 | 1 | R | R | R | R | R | |
| A3 | + | 1.5 | 1 | R | R | R | R | R | |
| A4 | − | 1.5 | 1 | R | R | R | R | R | |
| A5 | + | 1.5 | 2 | R | R | R | I | R | |
| A6 | + | 1.5 | 3 | R | R | I | S | R | |
| A7 | + | 1.5 | 3 | R | R | S | S | S | |
| B | B1 | + | 1.8 | 3 | R | R | I | S | R |
| B2 | + | 1.8 | 3 | R | R | S | S | R | |
| B3 | + | 1.8 | 3 | I | R | R | I | R | |
| B4 | + | 1.8 | 3 | R | R | S | S | R | |
| B5 | + | 1.8 | 3 | R | R | S | S | R | |
| B6 | − | 1.8 | 3 | R | R | S | S | R | |
| B7 | + | 1.5 | 4 | R | R | I | S | R | |
| B8 | + | 1.5 | 4 | R | R | I | S | S | |
| B9 | + | 1.5 | 4 | R | R | S | I | S | |
| B10 | + | 1.5 | 5 | R | R | S | S | R | |
| C | C1 | + | 1.5 | 6 | R | R | I | S | R |
| C2 | + | 1.5 | 6 | R | R | I | S | S | |
| C3 | + | NDc | 3 | R | R | I | I | S | |
| C4 | − | 1.5 | 7 | R | R | R | I | R | |
| C5 | − | 1.5 | 8 | R | R | R | I | R | |
| E | E1 | − | 1.5 | 9 | R | I | S | I | R |
| E2 | − | 1.5 | 10 | I | I | I | I | R | |
| E3 | − | ND | 11 | R | S | I | S | S | |
| E4 | + | 2.0 | 12 | R | R | I | S | R | |
| E5 | − | 1.5 | 13 | R | I | R | I | R | |
| F | F1 | − | 3.0 | ND | I | R | S | I | R |
| H | H1 | + | 1.5 | 10 | R | R | R | I | R |
| H2 | + | 1.5 | 10 | R | R | R | R | R | |
| H3 | + | 1.5 | 10 | R | R | R | R | R | |
| I | I1 | + | 3.0 | 14 | R | S | I | S | R |
| I2 | + | 3.0 | 14 | R | R | I | S | R | |
| M | M1 | + | 1.3 | 15 | R | R | R | I | R |
| M2 | + | 1.3 | 15 | R | R | R | I | R | |
| O | O1 | − | 1.5 | 16 | R | R | S | S | R |
R, resistant; I, intermediate; S, susceptible.
The inhibitor was fixed at 4 μg/ml.
ND, not determined.
Nucleotide sequencing of the 1.3-kb product from isolate M1 showed that the 5′CS-3′CS region included a single gene cassette identical to blaVIM-2 of integron In56 (14) (GenBank accession no. AF191564). Partial sequencing of the 1.5-kb amplicons derived from isolates A4 and C5 indicated that both also contained blaVIM-2 preceded by an aacA29 gene cassette. This structure resembles part of the VIM-2-encoding integron In59 (GenBank accession no. AF263519), found recently in a P. aeruginosa clinical strain in France (13).
Results of the synergy test using CAZ and 2-MAA were equivocal and not reproducible for most IPM-NS isolates, including the blaVIM-negative ones. The combination of IPM with 2-MAA performed better, giving clear synergy images for 26 of the 36 blaVIM-carrying isolates (sensitivity, 72%) (Table 2); all 22 isolates that did not contain blaVIM were negative in this test (specificity, 100%).
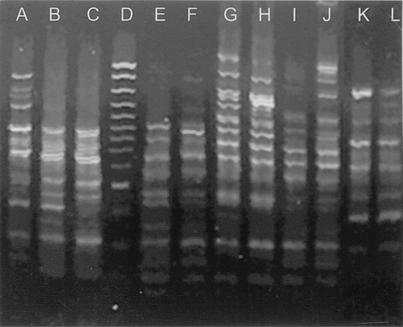
blaVIM-positive isolates were typed by RAPD fingerprinting. The discriminatory power and reproducibility of the method were satisfactory. Sixteen distinct RAPD patterns were observed; six of them are presented in Fig. 1. RAPD typing did not indicate any significant spread of epidemic clones, though strains exhibiting similar patterns (RAPD types 3 and 10) were observed in more than one hospital (Table 2). However, patients' records did not provide indications for interhospital spread. Notably, in four hospitals (A, B, C, and E) there were more than two blaVIM-carrying strains exhibiting distinct RAPD patterns (Table 2).
FIG. 1.
RAPD patterns of P. aeruginosa isolates carrying blaVIM. Lane A, RAPD type 6 (Table 2); lanes B and C, RAPD type 4; lanes E and F, RAPD type 10; lanes G and H, RAPD type 14; lanes I and J, RAPD type 1; lanes K and L, RAPD type 3. Molecular weight markers (100-bp DNA Ladder Plus; MBI Fermentas) are in lane D. The RAPD pattern similarity of the isolates in lanes I and J was confirmed in repeated experiments.
The diversity of RAPD types found here suggests the spread of blaVIM genes among genetically distinct P. aeruginosa strains. This spread is likely facilitated by the carriage of the blaVIM genes by integrons, which, though not mobile themselves, are frequently parts of transposons and/or transferable plasmids. DNA sequencing indicated that the predominant MBL gene type is blaVIM-2. It is not known if the observed differences in the size and structure of the integrons reflect evolution of an index integron or acquisition of blaVIM cassettes by different integrons. Studies on the structure of these elements are under way.
There were no phenotypic characteristics suggestive of blaVIM carriage. The majority of blaVIM-positive and blaVIM-negative isolates were resistant to all tested antibiotics. Additionally, no consistent quantitative differences in the levels of resistance to β-lactams, including carbapenems, were observed between the two groups. The MAA synergy test exhibited low sensitivity. Its specificity, however, appeared to be adequate. This test may be a useful adjunct to trace blaVIM-containing P. aeruginosa in this setting.
The relatively small number of isolates examined did not allow a reliable estimation of the prevalence of the blaVIM-containing P. aeruginosa. Also, there were sampling differences between hospitals. Finally, the possibility that blaVIM genes may be present among IPM-susceptible strains cannot be excluded. These resistance determinants are carried by integrons, and thus, their expression and subsequently their levels of resistance to carbapenems may vary significantly. Nevertheless, this preliminary study shows that blaVIM genes have spread not only in the large tertiary-care hospitals of Athens and Thessaloniki but also in district hospitals throughout the country.
Acknowledgments
The National Surveillance System for Antimicrobial Resistance is sponsored by the Hellenic Center for Infectious Disease Control (KEEL), Ministry of Health.
We thank Argiro Meni for excellent technical assistance. We also thank CANA SA Pharmaceutical Laboratories for providing Etest strips.
REFERENCES
- 1.Arakawa, Y., N. Shibata, K. Shibayama, H. Kurokawa, T. Yagi, H. Fujiwara, and M. Goto. 2000. Convenient test for screening metallo-β-lactamase-producing gram-negative bacteria by using thiol compounds. J. Clin. Microbiol. 38:40-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campbell, M., E. Mahenthiralingam, and D. P. Speert. 2000. Evaluation of random amplified polymorphic DNA typing of Pseudomonas aeruginosa. J. Clin. Microbiol. 38:4614-4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cornaglia, G., A. Mazzariol, L. Lauretti, G. M. Rossolini, and R. Fontana. 2000. Hospital outbreak of carbapenem-resistant Pseudomonas aeruginosa producing VIM-1, a novel transferable metallo-β-lactamase. Clin. Infect. Dis. 31:1119-1125. [DOI] [PubMed] [Google Scholar]
- 4.Johnson, J. L. 1994. Similarity analysis of DNAs, p. 655-682. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 5.Lauretti, L., M. L. Riccio, A. Mazzariol, G. Cornaglia, G. Aminosante, R. Fontana, and G. M. Rossolini. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43:1584-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee, K., J. B. Lim, J. H. Yum, D. Yong, Y., Chong, J. M. Kim, and D. M. Livermore. 2002. blaVIM-2 cassette-containing novel integrons in metallo-β-lactamase-producing Pseudomonas aeruginosa and Pseudomonas putida isolates disseminated in a Korean hospital. Antimicrob. Agents Chemother. 46:1053-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Livermore, D. M. 2001. Of Pseudomonas, porins, pumps and carbapenems. J. Antimicrob. Chemother. 47:247-250. [DOI] [PubMed] [Google Scholar]
- 8.Livermore, D. M., and N. Woodford. 2000. Carbapenemases: a problem in waiting? Curr. Opin. Microbiol. 3:489-495. [DOI] [PubMed] [Google Scholar]
- 9.Mahenthiralingam, E., M. E. Campbell, J. Foster, J. S. Lam, and D. P. Speert. 1996. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J. Clin. Microbiol. 34:1129-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mavroidi, A., A. Tsakris, E. Tzelepi, S. Pournaras, V. Loukova, and L. S. Tzouvelekis. 2000. Carbapenem-hydrolysing VIM-2 metallo-β-lactamase in Pseudomonas aeruginosa from Greece. J. Antimicrob. Chemother. 46:1041-1042. [DOI] [PubMed] [Google Scholar]
- 11.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial disk susceptibility tests, 6th ed. Approved standard M2-A6 (M100-S7). National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 12.Pallecchi, L., M. L. Riccio, J.-D. Docquier, R. Fontana, and G. M. Rossolini. 2001. Molecular heterogeneity of blaVIM-2-containing integrons from Pseudomonas aeruginosa plasmids encoding the VIM-2 metallo-β-lactamase. FEMS Microbiol. Lett. 195:145-150. [DOI] [PubMed] [Google Scholar]
- 13.Poirel, L., T. Lambert, S. Turkoglu, E. Ronco, J.-L. Gaillard, and P. Nordmann. 2001. Characterization of class 1 integrons from Pseudomonas aeruginosa that contain the blaVIM-2 carbapenem-hydrolyzing β-lactamase gene and of two novel aminoglycoside resistance gene cassettes. Antimicrob. Agents Chemother. 45:546-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poirel, L., T. Naas, D. Nicolas, L. Collet, S. Bellais, J. D. Cavallo, and P. Nordmann. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 45:891-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandvang, D., F. M. Aarestrup, and L. B. Jensen. 1998. Characterisation of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol. Lett. 160:37-41. [DOI] [PubMed] [Google Scholar]
- 16.Senda, K., Y. Arakawa, S. Ichiyama, K. Nakashima, H. Ito, S. Ohsuka, K. Shimokata, N. Kato, and M. Ohta. 1996. PCR detection of metallo-β-lactamase gene (blaIMP) in gram-negative rods resistant to broad-spectrum β-lactams. J. Clin. Microbiol. 34:2909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Senda, K., Y. Arakawa, K. Nakashima, H. Ito, S. Ichiyama, K. Shimokata, N. Kato, and M. Ohta. 1996. Multifocal outbreaks of metallo-β-lactamase-producing Pseudomonas aeruginosa resistant to broad-spectrum β-lactams, including carbapenems. Antimicrob. Agents Chemother. 40:349-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsakris, A., S. Pournaras, N. Woodford, M.-F. I. Palepou, G. S. Babini, J. Douboyas, and D. M. Livermore. 2000. Outbreak of infections caused by Pseudomonas aeruginosa producing VIM-1 carbapenemase in Greece. J. Clin. Microbiol. 38:1290-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]