Abstract
Human granulocytic ehrlichiosis is an emerging infectious disease in the United States and Europe, and PCR methods have been shown to be effective for the diagnosis of acute infections. Numerous PCR assays and primer sets have been reported in the literature. The analytical sensitivities (limits of detection) of 13 published PCR primer sets were compared using DNA extracted from serial dilutions of Anaplasma phagocytophilum-infected HL-60 cells. The specificity of the assays that were able to detect ≤2.5 infected cells was tested by the use of template DNA extracted from Ehrlichia chaffeensis, Rickettsia rickettsii, and Bartonella henselae. The assays with the lowest limits of detection were shown to be a nested assay that amplifies the 16S rRNA gene (primer pairs ge3a-ge10 [primary] and ge9-ge3 [nested]; detects 0.25 infected cell), a direct assay that amplifies the major surface protein gene msp2 (primer pair msp2-3f-msp2-3r; detects 0.25 infected cell), and a direct assay that amplifies the 16S rRNA gene (primer pair ehr521-ehr790; detects 0.25 infected cell). The specificity and limit of detection of the MSP2 and 16S rRNA direct assays were further tested by use of A. phagocytophilum template DNA from both North America and Europe and from human, tick, white-footed mouse, equine, deer, bovine, and wood rat samples and of template DNA from closely related species (Anaplasma marginale, the white-tailed deer agent, and additional E. chaffeensis-positive samples). Three manufacturers' PCR kits were tested and showed distinct variations in the limit of detection, specificity, and nonspecific background amplification. The importance of these results for the molecular diagnosis of human granulocytic ehrlichiosis is discussed.
Human ehrlichiosis is an acute, febrile illness that is caused by at least three known agents in the United States. Ehrlichia chaffeensis infects mononuclear white blood cells, while Ehrlichia ewingii and Anaplasma phagocytophilum (previously Ehrlichia phagocytophila, the human granulocytic ehrlichiosis [HGE] agent) infect granulocytic-lineage leukocytes (2, 6-9, 11). HGE was first described in 1994, and the number of confirmed cases has increased to the point where it is now the predominant form of ehrlichiosis (3, 7, 20; J. McQuiston, personal communication) and second to Lyme disease as the most common tick-borne infection in the United States. Tetracyclines are efficacious for the treatment of all forms of ehrlichiosis; however, the nonspecific symptoms of infection can make diagnosis problematic. Serological methods are most commonly used for diagnosis, but these assays are often negative in the early stages of an acute infection, as antibody will typically be absent during the first week of infection (1). Diagnosis prior to seroconversion has utilized molecular methods that are based on PCR amplification of the DNA of the agent, and numerous PCR-based assays and primer sets have been reported (7, 13, 16, 19, 24-27, 29). The purpose of this study was to compare a selection of these assays in order to evaluate their specificities and limits of detection for A. phagocytophilum.
MATERIALS AND METHODS
Bacterial strains, propagation, and DNA extraction.
The Arkansas, St. Vincent, and Jax strains of E. chaffeensis were grown in DH-82 cells, and the USG3, WS27, and MRK strains of A. phagocytophilum were grown in HL-60 cells (2, 9, 22, 28). The Sheila Smith strain of Rickettsia rickettsii and the Houston-1 strain of Bartonella henselae were used for specificity testing. DNA was extracted from infected tissue culture cells, EDTA blood samples, and ticks by using the DNeasy (Qiagen, Valencia, Calif.) tissue kit following the manufacturer's protocol and eluted in 200 μl of sterile distilled water.
PCR amplification.
Tenfold serial dilutions of HL-60 cells infected with the USG3 strain of A. phagocytophilum were made in a background of uninfected human blood. DNA was extracted from each dilution, and 5 μl of the purified DNA was used as the template in each 50-μl PCR. All reactions also contained 200 μM concentrations of each deoxynucleoside triphosphate (dATP, dCTP, dGTP, and dTTP), 1.5 mM MgCl2, 2.5 U of Taq polymerase, and 0.5 μM concentrations of each primer. The initial limit of detection testing used reagents from the GeneAmp PCR kit with AmpliTaq DNA polymerase (PerkinElmer, Foster City, Calif.). Each reaction involved an initial 2-min denaturation at 95°C prior to thermal cycling and a 5-min extension at 72°C after cycling. Each cycle consisted of a 30-s denaturation at 94°C, a 30-s annealing, and a 1-min extension at 72°C. The number of cycles and the annealing temperatures were as listed in Table 1. Reactions were subsequently maintained at 4°C until analyzed by agarose gel electrophoresis.
TABLE 1.
PCR assays tested including gene targets, primer designations, annealing and cycling conditions, limits of detection, and specificity results
| Target gene | Primer pair | Annealing temp (°C) | No. of cycles | Detection limita | Specificityb
|
Reference | ||
|---|---|---|---|---|---|---|---|---|
| Rr | Bh | Ec | ||||||
| 16S | ge9-ge10 | 55 | 40 | 2.5 | − | − | − | 7 |
| 16S | ec12-ec9 | 48, 52 | 3, 37 | 2.5 | − | − | − | 7 |
| Nested | ge9-ge10 | 55 | 30 | |||||
| 16S | ehr521-ehr747 | 60 | 50 | 0.25 | + | + | + | 24 |
| 16S | ehr521-ehr790 | 55 | 40 | 0.25 | − | − | + | 16 |
| 16S | per1-per2 | 45 | 40 | 2.5 | − | − | + | 13 |
| 16S | ger3-ger4 | 50 | 40 | UN | NT | NT | NT | 13 |
| 16S | ge3a-ge10 | 55 | 40 | 0.25 | − | − | − | 19 |
| Nested | ge9-ge2 | 55 | 30 | |||||
| groE | hs1-hs6 | 48, 52 | 3, 37 | 0.25 weak | − | − | + | 26 |
| Nested | hs43-hs45 | 55 | 30 | |||||
| ank | la6-la1 | 55 | 40 | 2.5 | − | − | − | 27 |
| msp2 | msp2-3f-msp2-3r | 55 | 40 | 0.25 | − | − | − | Massung, 1999c |
| 100 kDa | s7f-s7r | 55 | 40 | 25 weak | NT | NT | NT | 25 |
| 130 kDa | s22f-s22r | 55 | 40 | 2.5 weak | NT | NT | NT | 25 |
| Hsp-70 | b3f1-b3r | 55 | 40 | 25 | NT | NT | NT | 25 |
The estimated minimum number of A. phagocytophilum infected cells that could be detected per test sample. UN, could not be determined.
Bacteria used for specificity testing: Rr, R. rickettsii strain Sheila Smith; Bh, B. henselae strain Houston-1; Ec, E. chaffeensis strain Arkansas. NT, not tested.
R. F. Massung, presented at the EUWOG-ASR Joint Meeting, Marseille, France, 1999 (Program and Abstracts, p. 6), was subsequently published in reference 20.
Where indicated in the text, nested PCR was performed using 1 μl of the primary PCR product as template. Each nested amplification contained 200 μM concentrations of each deoxynucleoside triphosphate (dATP, dCTP, dGTP, and dTTP), 2.5 U of Taq polymerase, and 0.2 μM concentrations of each primer in a total reaction volume of 50 μl. Nested cycling conditions were as follows: an initial 2-min denaturation at 95°C, followed by 25 cycles with each cycle consisting of a 30-s denaturation at 94°C, a 30-s annealing at 55°C, and a 1-min extension at 72°C. These 25 cycles were followed by a 5-min extension at 72°C. Reactions were subsequently maintained at 4°C until analyzed by agarose gel electrophoresis.
All assays tested were screened for specificity with template DNA from R. rickettsii, B. henselae, and E. chaffeensis by using the GeneAmp PCR kit with AmpliTaq DNA polymerase (PerkinElmer). The assays that performed best in the initial limit of detection and specificity testing were subsequently tested with a battery of templates from closely related and more distantly related species. DNA was extracted from a second set of serial dilutions of A. phagocytophilum strain USG3 as described above. These extractions were used with the three optimal assays to compare the PCR amplification kits from three different manufacturers: the GeneAmp PCR kit with AmpliTaq DNA polymerase (PerkinElmer), Ready-To-Go PCR beads (Amersham Pharmacia Biotech, Piscataway, N.J.), and the Taq PCR Master Mix kit (Qiagen). All reactions were performed under the conditions described above and in Table 1 and stored at 4°C until analyzed by agarose gel electrophoresis.
Limit of detection quantification.
The limit of detection for each assay was evaluated with samples for which the number of infected cells per milliliter had previously been determined. DNA was extracted from 0.2 ml of each sample and eluted in 0.2 ml. Subsequently, 0.005 ml of the template DNA, or 1/40 of the total extracted DNA, was used per PCR. Therefore, in a sample with 50 infected cells/ml, DNA was extracted from 0.2 ml, or 10 infected cells (50 infected cells/ml × 0.2 ml = 10 infected cells). And the template for each PCR was 1/40 that of the total extracted DNA, or the equivalent of 0.25 infected cell (10 infected cells/40 PCRs = 0.25 infected cell/reaction).
RESULTS
The cycling conditions used and the results obtained from the testing of various reported PCR assays and primer sets are shown in Table 1. These assays were all performed by using the PerkinElmer GeneAmp PCR reagent kit and core reagents with AmpliTaq DNA polymerase. The limits of detection of the assays ranged from 0.25 to 25 A. phagocytophilum strain USG3-infected HL-60 cells, although the limit of detection of one assay could not be determined because of a high background (16S assay with primer pair ger3-ger4). Four of the assays (16S assay with primer pair ehr521-ehr747, 16S assay with primer pair ehr521-ehr790, 16S-nested assay with primer pairs ge3a-ge10 and ge9-ge2, and msp2 assay with primer pair msp2-3f-msp2-3r) showed the lowest limit of detection, 0.25 infected cell. The specificity of the nine assays that were able to detect ≤2.5 infected cells was tested with R. rickettsii, B. henselae, and E. chaffeensis (strain Arkansas) genomic DNAs as PCR templates. Four of the assays amplified E. chaffeensis DNA in addition to the targeted A. phagocytophilum DNA, and one of the assays (16S with primer set ehr521-ehr747) amplified DNA from E. chaffeensis, R. rickettsii, and B. henselae. The results of the limit of detection testing for the ehr521-ehr790 and msp2 assays are shown in Fig. 1; both assays had detection limits equivalent to those for the 16S-rRNA-nested assay using ge3a-ge10 for the primary amplification and ge9-ge2 for the nested reaction (19) (Table 1) and had a low background.
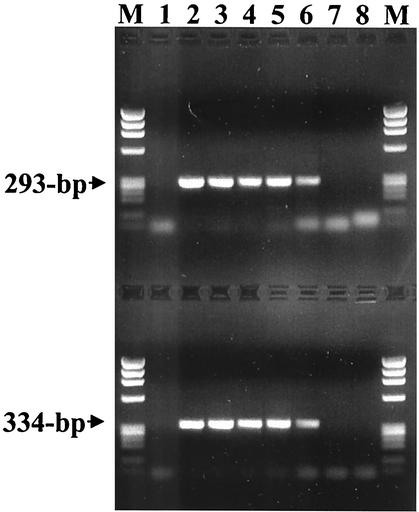
FIG. 1.
Limit of detection of PCR assays for A. phagocytophilum strain USG3 DNA with primer pairs ehr521-ehr790 (upper lanes) and msp2-3f-msp2-3r (lower lanes). Lanes 1 show amplification products from the DNA extracted from uninfected human blood as negative controls. Lanes 2 through 8 represent amplifications from a 10-fold dilution series ranging from 5 × 105 infected cells/ml to 0.5 infected cell/ml. Phage ϕX174 DNA digested with HaeIII was run in the lanes labeled M. The sizes of the amplified products are indicated to the left.
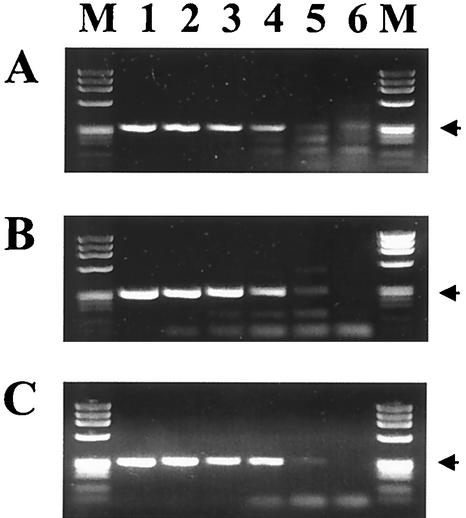
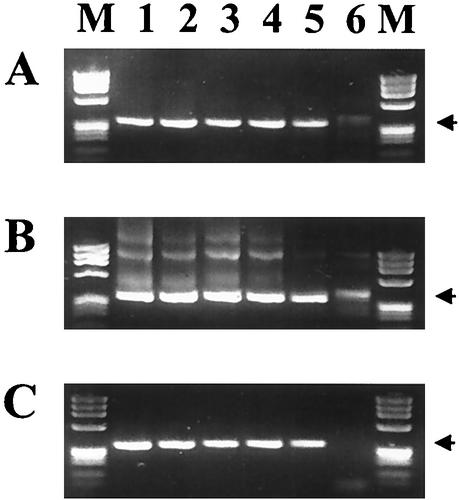
The two nonnested assays that detected 0.25 A. phagocytophilum-infected cell and did not amplify the DNA of either R. rickettsii or B. henselae, the ehr521-ehr790 and msp2 assays, were further evaluated with a series of clinical and nonclinical samples that had previously been confirmed as positive or negative using the ge3a-ge10 and ge9-ge2 16S-nested assay (Fig. 2). Both assays detected each of the confirmed positives (lanes 1 through 6), although the lane 4 samples gave only a weak positive with the erh521-ehr790 assay. All other samples, which included HGE-negative samples (lanes 7 through 11) and E. chaffeensis-positive samples (lanes 12 through 15), were negative, although some background banding was seen in lane 13 with one E. chaffeensis sample on the ehr521-ehr790 assay only. Another set of DNA samples extracted from the blood of various animals, including equine, bovine, deer, white-tailed mice (Peromyscus leucopus), and dusky-footed wood rat (Neotoma fuscipes); Ixodes scapularis ticks; and tissue-culture-grown E. chaffeensis, B. henselae, R. rickettsii, Anaplasma marginale, and A. phagocytophilum, was also tested with each assay (Fig. 3). Each of these DNA samples was previously shown to be positive with the 16S-nested ge3a-ge10 and ge9-ge2 assay except for the culture-grown E. chaffeensis, B. henselae, and R. rickettsii (lanes 9 through 11). Both assays amplified products from an Ehrlichia equi (now A. phagocytophilum)-infected horse (lane 1), an E. phagocytophila (now A. phagocytophilum)-infected cow from Sweden (lane 2), a white-footed mice from Connecticut (lanes 5 and 6), a dusky-footed wood rat from California (lane 7), an I. scapularis tick from Connecticut (lane 8), and the A. phagocytophilum-positive control sample (lane 13) (10). The ehr521-ehr790 assay also detected the white-tailed deer agent from deer blood (lanes 3 and 4), A. marginale (lane 12), and was weakly positive for E. chaffeensis (lane 9), samples of which were not detected with the msp2 assay (8). There were no samples that tested positive by the msp2 assay that were not also detected by the erh521-ehr790 assay, although one of the I. scapularis samples from Connecticut (lane 5) produced significantly more product with the msp2 assay.
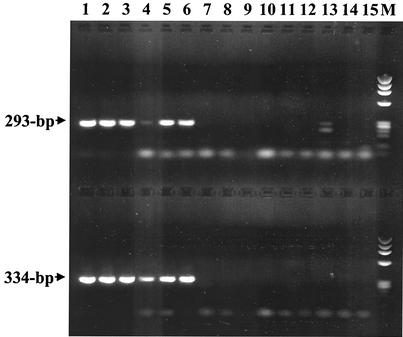
FIG. 2.
PCR assay testing on DNA extracted from human EDTA blood samples with primer pairs ehr521-ehr790 (upper lanes) and msp2-3f-msp2-3r (lower lanes). Lanes 1 through 6 show amplification products from samples previously confirmed as A. phagocytophilum positive by 16S-rRNA-nested PCR. Lanes 7 through 11 are amplification products from samples previously shown to be negative for A. phagocytophilum and E. chaffeensis. Lanes 12 through 15 show amplification products from samples previously confirmed as E. chaffeensis positive. Phage ϕX174 DNA digested with HaeIII was run in the lane labeled M. The sizes of the amplified products are indicated to the left.
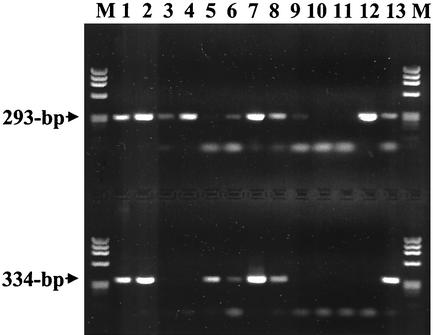
FIG. 3.
PCR assay testing on DNA extracted from veterinary, tick, and cultured bacterial samples with primer pairs ehr521-ehr790 (upper lanes) and msp2-3f-msp2-3r (lower lanes). Samples represented are E. equi (now A. phagocytophilum) from an experimentally infected horse (lane 1), E. phagocytophila (now A. phagocytophilum) from a Swedish cow (lane 2), white-tailed deer agent from white-tailed deer (lanes 3 and 4), A. phagocytophilum-positive I. scapularis ticks from Connecticut (lanes 5 through 7), E. equi (now A. phagocytophilum) from a wood rat from California (lane 8), E. chaffeensis Arkansas strain (lane 9), B. henselae Houston-1 strain (lane 10), R. rickettsii Sheila Smith strain (lane 11), A. marginale (lane 12), and A. phagocytophilum USG3 strain (lane 13). Phage ϕX174 DNA digested with HaeIII was run in the lanes labeled M. The sizes of the amplified products are indicated to the left.
A second dilution series was used for additional testing of the three assays that were able to detect 0.25 infected cell (Table 1): the 16S-rRNA-nested assay using ge3a-ge10 and ge9-ge2, the 16S rRNA assay using ehr521-ehr790, and the msp2 assay using msp2-3f-msp2-3r. Three commercially available PCR amplification kits were used for this comparison, although each of the kits used AmpliTaq DNA polymerase and the same magnesium concentration (1.5 mM). The nested-16S assay showed little difference when tested with the three kits (Fig. 4), and each clearly detected 0.25 infected cell. The 16S assay using ehr521-ehr790 produced faint products from 0.25 infected cell with each kit (Fig. 5). However, spurious background bands were significantly higher with both the PerkinElmer (panel A) and Amersham (panel B) kits. The PerkinElmer kit showed a faint product, which could be misinterpreted and result in a false positive, in the lane (Fig. 5A, lane 6) where the control template was DNA extracted from uninfected human blood. The msp2 assay, shown in Fig. 6, strongly detected 0.25 infected cell with each kit, but both the PerkinElmer (panel A) and Amersham (panel B) kits amplified bands from the uninfected human control (lane 6) that migrated close to the correct product size and could again be interpreted as false positives. However, the msp2 assay with the Qiagen kit (Fig. 6C) produced a strong positive band for the 0.25-infected-cell sample and had a minimal background in both the positive (lanes 1 through 5) and negative (lane 6) samples. Overall, the Qiagen kit performed best for all assays (Fig. 4 through 6).
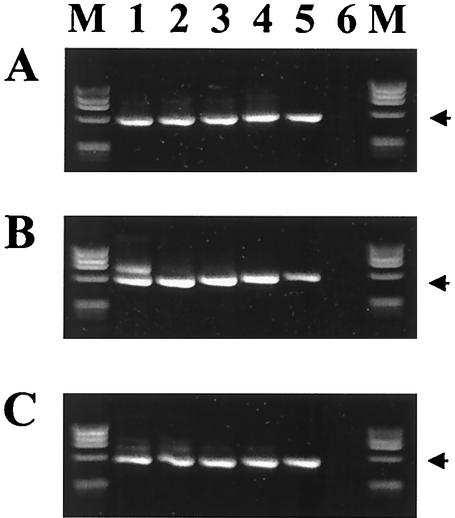
FIG. 4.
Comparison of the 16S-rRNA-nested-PCR assay with primer pairs ge3a-ge10 and ge9-ge2 with commercially available kits from three manufacturers: (A) PerkinElmer GeneAmp PCR reagent kit with AmpliTaq DNA polymerase; (B) Amersham Pharmacia Ready-To-Go PCR beads; (C) Qiagen Taq PCR Master Mix kit. Lanes 1 through 5 represent amplifications from a 10-fold dilution series ranging from 5 × 105 A. phagocytophilum-infected cells/ml to 50 infected cells/ml, and lane 6 contains DNA from uninfected human blood as a negative control. Phage ϕX174 DNA digested with HaeIII was run in the lanes labeled M. The arrows indicate the position of the 546-bp amplified products.
FIG. 5.
Comparison of the 16S rRNA assay with primer pair ehr521-ehr790 by using commercially available kits from three manufacturers: (A) PerkinElmer GeneAmp PCR reagent kit with AmpliTaq DNA polymerase; (B) Amersham Pharmacia Ready-To-Go PCR beads; (C) Qiagen Taq PCR Master Mix kit. Lanes are as described in the legend to Fig. 4. The arrows indicate the position of the 293-bp amplified products.
FIG. 6.
Comparison of the msp2 gene PCR assay using commercially available kits from three manufacturers: (A) PerkinElmer GeneAmp PCR reagent kit with AmpliTaq DNA polymerase; (B) Amersham Pharmacia Ready-To-Go PCR beads; (C) Qiagen Taq PCR Master Mix kit. Lanes are as described in the legend to Fig. 4. The arrows indicate the position of the 334-bp amplified products.
DISCUSSION
PCR-based molecular assays are powerful tools for the diagnosis of infectious diseases and for the detection of viral and bacterial agents in both clinical and environmental samples. These assays were instrumental in the identification of E. chaffeensis and A. phagocytophilum as the etiologic agents of human monocytic ehrlichiosis (HME) and HGE, respectively (2, 7, 12). Since the identification of the HGE agent by Chen et al. (7), numerous PCR amplifications assays have been described for the detection of the agent. In this report, we have attempted to systematically evaluate and compare a number of these assays, along with several commercially available PCR kits, to serve as a guide for laboratories considering establishing molecular testing for the HGE agent.
The results of testing these various assays were surprising in several regards. For example, three of the single-step amplification assays (16S with primer pair ehr521-ehr790, 16S with primer pair ehr521-ehr747, and msp2 with primer pair msp2-3f-msp2-3r) showed a limit of detection comparable to that in the 16S-nested assay (primer pairs ge3a-ge10 and ge9-ge2) that was previously shown to detect <2 copies of genomic DNA (19). One likely reason for the excellent analytical sensitivity of the msp2 assay is that the primers for the assay were designed to amplify each of the multiple copies on the msp2 gene that are present in the A. phagocytophilum genome (14, 21, 30). The msp2 assay was also the most species-specific assay tested, as it did not amplify products from samples containing the DNA of the white-tailed deer agent A. marginale or from any sample other than the ones previously confirmed as A. phagocytophilum positive (10). One of these three assays, the 16S assay with primer pair ehr521-ehr747, also amplified the 16S rRNA gene of E. chaffeensis, R. rickettsii, and B. henselae. While this assay can and has been used for analyzing sterile clinical samples, any laboratory using this assay must be aware of the potential of this assay to detect the non-HGE agents tested in this report, and the assay will likely amplify many other bacteria that we have not tested. Certainly, the ehr521-ehr747 assay should not be used for testing samples such as I. scapularis ticks, the vector of the HGE agent in the northeastern and upper Midwestern United States, because the assay may detect bacterial endosymbionts within the tick that are closely related to Rickettsia species (23) or other bacterial environmental contaminants. Neither should this assay be used for screening P. leucopus or other rodent samples because it has been well documented that numerous Bartonella species are found in rodents (4, 17, 18). Two of the assays tested also amplified the E. chaffeensis 16S rRNA gene but not the 16S rRNA gene of R. rickettsii or B. henselae: the 16S assay using ehr521-ehr790 and the groE-nested assay. Similar to the 16S-rRNA-nested assay with ge3a-ge10 and ge9-ge2, the erh521-ehr790 assay will also amplify DNA from the white-tailed deer agent and can thereby be used to screen for this agent (10). The amplification of E. chaffeensis DNA by the groE assay was expected because the assay was not designed as a specific assay to be used for diagnostics but rather as a general assay that could be used for screening various samples for most ehrlichial species (26). Several of the primer sets tested (assays 100 kDa, 130 kDa, Hsp-70) were not designed for use in a diagnostic assay and were not optimized for analytical sensitivity or specificity (25). So it is not surprising that each showed a relatively poor limit of detection. If these assays had been optimized, they may have performed significantly better.
The 16S assay using primer pair ehr521-ehr790 performed quite well in our tests, although it detected E. chaffeensis DNA. While A. phagocytophilum (HGE agent) and E. chaffeensis (HME agent) are transmitted by different ticks and the areas of endemicity for each disease in the United States have little overlap (20), the lack of specificity of the erh521-ehr790 assay may be problematic, particularly for reference laboratories that do not have access to patient information, and could result in HME cases being misdiagnosed as HGE. The incorrect detection and diagnosis of HME would be problematic for epidemiologic studies and potentially dangerous if used for patient management. Doxycycline has been recommended as the drug of choice for treating HGE and HME in both adults and children, so even a misdiagnosis in a diagnostic setting should lead to the appropriate treatment of most patients (8, 11). However, in patients where tetracyclines are contraindicated (pregnant women and young children), the misdiagnosis of an HME infection as HGE could lead to the patient being improperly treated with rifamycins or quinolones, drugs for which in vitro testing has shown that A. phagocytophilum is susceptible but E. chaffeensis is resistant (5, 15).
The two assays that provided the highest analytical sensitivity and specificity were the 16S-nested assay (ge3a-ge10 and ge9-ge2) and the msp2 assay. Both assays detected the equivalent of 0.25 infected cell, and neither detected E. chaffeensis, R. rickettsii, or B. henselae. However, dramatic differences were noted between these two assays when they were evaluated using three different commercially available PCR kits. Whereas the results of the 16S-nested assay were not affected by the PCR kit that was used, the msp2 assay and every other assay showed variable limits of detection and/or specificity (nonspecific banding patterns) based on which manufacturer's kit was used. For example, of the kits tested in this report, the msp2 assay should be used with only the Qiagen kit because both of the other kits resulted in banding patterns that could be interpreted as false positives. Relative to the other kits tested, the PerkinElmer kit provided fairly good specificity but decreased analytical sensitivity, the Amersham kit showed high analytical sensitivity but poor specificity, and the Qiagen kit provided high analytical sensitivity and specificity.
There are a large number of factors involved in optimizing a PCR assay that has a low limit of detection and is also specific and reproducible. Some of these variables include the PCR kit and the enzyme used; Mg2+, template, and primer concentrations; optimization factors specific to the assay, such as the annealing temperature and number of cycles; the method used for extraction; and the thermal cycler used for amplification. While we attempted to control for many of these factors, the large number of potential variables makes it difficult to predict the performance of a given assay in an outside laboratory or whether significant improvement in the performance of single assays could be achieved by optimizing assay conditions. Each laboratory should determine the efficacy of any PCR assay within their local environment. This includes providing an appropriate workspace free of contaminants and properly trained personnel. The assay should be well controlled with positive and negative controls used for both DNA extraction and PCR amplification. Reproducibility and proficiency should be confirmed by the use of replicates. DNA sequencing should be used to authenticate that the expected product has been amplified. These general guidelines, coupled with the use of appropriate assays, should permit most laboratories to perform quality PCR assays for the molecular detection of A. phagocytophilum and the diagnosis of HGE infections.
Acknowledgments
We thank Anneli Bjöersdorff, William Nicholson, Russell Regnery, Victoria Solberg, Jacqueline Dawson, Kirby Stafford, and James Childs for providing the samples used in this study. We are grateful to the Biotechnology Core Facility of the National Center for Infectious Diseases for the synthesis of oligonucleotides. We thank Cheryl Murphy for providing several of the A. phagocytophilum gene sequences prior to publication and Gregory Dasch for reviewing the manuscript and contributing numerous useful suggestions.
REFERENCES
- 1.Aguero-Rosenfeld, M. E., F. Kalantarpour, M. Baluch, H. W. Horowitz, D. F. McKenna, J. T. Raffalli, T.-Z. Hsieh, J. Wu, J. S. Dumler, and G. P. Wormser. 2000. Serology of culture-confirmed cases of human granulocytic ehrlichiosis. J. Clin. Microbiol. 38:635-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, B. E., J. E. Dawson, D. C. Jones, and K. H. Wilson. 1991. Ehrlichia chaffeensis, a new species associated with human ehrlichiosis. J. Clin. Microbiol. 29:2838-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakken, J. S., J. S. Dumler, S.-M. Chen, M. R. Eckman, L. L. Van Etta, and D. H. Walker. 1994. Human granulocytic ehrlichiosis in the upper Midwest United States. A new species emerging? JAMA 272:212-218. [PubMed] [Google Scholar]
- 4.Birtles, R. J., T. G. Harrison, and D. H. Molyneux. 1994. Grahamella in small woodland mammals in the U.K.: isolation, prevalence and host specificity. Ann. Trop. Med. Parasitol. 88:317-327. [DOI] [PubMed] [Google Scholar]
- 5.Brouqui, P., and D. Raoult. 1992. In vitro susceptibility of the newly recognized agent of ehrlichiosis in humans, Ehrlichia chaffeensis. Antimicrob. Agents Chemother. 36:2799-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buller, R. S., M. Arens, S. P. Hmiel, C. D. Paddock, J. W. Sumner, Y. Rikihisa, A. Unver, M. Gaudreault-Keener, F. A. Manian, A. M. Liddell, N. Schmulewitz, and G. A. Storch. 1999. Ehrlichia ewingii, a newly recognized agent of human ehrlichiosis. N. Engl. J. Med. 341:148-155. [DOI] [PubMed] [Google Scholar]
- 7.Chen, S.-M., J. S. Dumler, J. S. Bakken, and D. H. Walker. 1994. Identification of a granulocytotropic Ehrlichia species as the etiologic agent of human disease. J. Clin. Microbiol. 32:589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Childs, J. E., and C. D. Paddock. 2002. Ehrlichia species (the ehrlichioses), p. 913-916. In S. S. Long, L. K. Pickering, and C. G. Prober (ed.), Principles and practice of pediatric infectious diseases, 2nd ed. Churchill Livingstone, Philadelphia, Pa.
- 9.Dawson, J. E., B. E. Anderson, D. B. Fishbein, J. L. Sanchez, C. S. Goldsmith, K. L. Wilson, and C. W. Duntley. 1991. Isolation and characterization of an Ehrlichia sp. from a patient diagnosed with human ehrlichiosis. J. Clin. Microbiol. 29:2741-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawson, J. E., C. K. Warner, V. Baker, S. A. Ewing, D. E. Stallknecht, W. R. Davidson, A. A. Kocan, J. M. Lockhart, and J. G. Olson. 1996. Ehrlichia-like 16S rDNA sequence from white-tailed deer (Odocoileus virginianus). J. Parasitol. 82:52-58. [PubMed] [Google Scholar]
- 11.Dumler, J. S., and J. S. Bakken. 1995. Ehrlichial diseases of humans: emerging tick-borne infections. Clin. Infect. Dis. 20:1102-1110. [DOI] [PubMed] [Google Scholar]
- 12.Dumler, J. S., A. F. Barbet, C. P. J. Bekker, G. A. Dasch, G. H. Palmer, S. C. Ray, Y. Rikihisa, and F. R. Rurangirwa. 2001. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ′HGE agent' as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. E vol. Microbiol. 51:2145-2165. [DOI] [PubMed] [Google Scholar]
- 13.Goodman, J. L., C. Nelson, B. Vitale, J. E. Madigan, J. S. Dumler, T. J. Kurtti, and U. G. Munderloh. 1996. Direct cultivation of the causative agent of human granulocytic ehrlichiosis. N. Engl. J. Med. 334:209-215. [DOI] [PubMed] [Google Scholar]
- 14.Ijdo, J. W., W. Sun, Y. Zhang, L. A. Magnarelli, and E. Fikrig. 1998. Cloning of the gene encoding the 44-kilodalton antigen of the agent of human granulocytic ehrlichiosis and characterization of the humoral response. Infect. Immun. 66:3264-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein, M. B., C. M. Nelson, and J. L. Goodman. 1997. Antibiotic susceptibility of the newly cultivated agent of human granulocytic ehrlichiosis: promising activity of quinolones and rifamycins. Antimicrob. Agents Chemother. 41:76-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolbert, C. 1996. Detection of the agent of human granulocytic ehrlichiosis by PCR, p.106-111. In D. Persing (ed.), PCR protocols for emerging infectious diseases, ASM Press, Washington, D.C.
- 17.Kosoy, M. Y., R. L. Regnery, T. Tzianabos, E. L. Marston, D. C. Jones, D. Green, G. O. Maupin, J. G. Olson, and J. Childs. 1997. Distribution, diversity, and host specificity of Bartonella in rodents from the southeastern United States. Am. J. Trop. Med. Hyg. 57:578-588. [DOI] [PubMed] [Google Scholar]
- 18.Kosoy, M. Y., E. K. Saito, D. Green, E. L. Marston, D. C. Jones, and J. E. Childs. 2000. Experimental evidence of host specificity of Bartonella infection in rodents. Comp. Immunol. Microbiol. Infect. Dis. 23:221-238. [DOI] [PubMed] [Google Scholar]
- 19.Massung, R. F., K. Slater, J. H. Owens, W. L. Nicholson, T. N. Mather, V. B. Solberg, and J. G. Olson. 1998. Nested PCR assay for detection of granulocytic ehrlichiae. J. Clin. Microbiol. 36:1090-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McQuiston, J. H., C. D. Paddock, R. C. Holman, and J. E. Childs. 1999. The human ehrlichioses in the United States. Emerg. Infect. Dis. 5:635-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy, C. I., J. R. Storey, J. Recchia, L. A. Doros-Richert, C. Gingrich-Baker, K. Munroe, J. S. Bakken, R. T. Coughlin, and G. A. Beltz. 1998. Major antigenic proteins of the agent of human granulocytic ehrlichiosis are encoded by members of a multigene family. Infect. Immun. 66:3711-3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicholson, W. L., J. A. Comer, J. W. Sumner, C. Gingrich-Baker, R. T. Coughlin, L. A. Magnarelli, J. G. Olson, and J. E. Childs. 1997. An indirect immunofluorescence assay using a cell culture-derived antigen for detection of antibodies to the agent of human granulocytic ehrlichiosis. J. Clin. Microbiol. 35:1510-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noda, H., U. K. Munderloh, and T. J. Kurtti. 1997. Endosymbionts of ticks and their relationship to Wolbachia spp. and tick-borne pathogens of humans and animals. Appl. Environ. Microbiol. 63:3926-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pancholi, P., C. P. Kolbert, P. D. Mitchell, K. D. Reed, J. S. Dumler, J. S. Bakken, S. R. Telford, and D. H. Persing. 1995. Ixodes dammini as a potential vector of human granulocytic ehrlichiosis. J. Infect. Dis. 172:1007-1012. [DOI] [PubMed] [Google Scholar]
- 25.Storey, J. R., L. A. Doros-Richert, C. Gingrich-Baker, K. Munroe, T. N. Mather, R. T. Coughlin, G. A. Beltz, and C. I. Murphy. 1998. Molecular cloning and sequencing of three granulocytic Ehrlichia genes encoding high-molecular-weight immunoreactive proteins. Infect. Immun. 66:1356-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sumner, J. W., W. L. Nicholson, and R. F. Massung. 1997. PCR amplification and comparison of nucleotide sequences from the groESL heat shock operon of Ehrlichia species. J. Clin. Microbiol. 35:2087-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walls, J. J., P. Caturegli, J. S. Bakken, K. M. Asanovich, and J. S. Dumler. 2000. Improved sensitivity of PCR for diagnosis of human granulocytic ehrlichiosis using epank1 genes of Ehrlichia phagocytophila-group ehrlichiae. J. Clin. Microbiol. 38:354-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeh, M.-T., T. N. Mather, R. T. Coughlin, C. Gingrich-Baker, J. W. Sumner, and R. F. Massung. 1997. Serologic and molecular detection of granulocytic ehrlichiosis in Rhode Island. J. Clin. Microbiol. 35:944-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeidner, N. S., T. R. Burkot, R. Massung, W. L. Nicholson, M. C. Dolan, J. S. Rutherford, B. J. Biggerstaff, and G. O. Maupin. 2000. Transmission of the agent of human granulocytic ehrlichiosis by Ixodes spinipalpis ticks: evidence of an enzootic cycle of dual infection with Borrelia burgdorferi in Northern Colorado. J. Infect. Dis. 182:616-619. [DOI] [PubMed] [Google Scholar]
- 30.Zhi, N., N. Ohashi, Y. Rikihisa, H. W. Horowitz, G. P. Wormser, and K. Hechemy. 1998. Cloning and expression of the 44-kilodalton major outer membrane protein gene of the human granulocytic ehrlichiosis agent and application of the recombinant protein to serodiagnosis. J. Clin. Microbiol. 36:1666-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]