Abstract
The yeasts of the genus Candida are opportunistic pathogens associated with the rising incidence of life-threatening infections in immunocompromised individuals. Secretion of aspartic proteinases has been determined to be one of the virulence factors of the pathogenic Candida species. To analyze the extracellular proteolytic activities of a large number of Candida clinical isolates, we developed a screening system based on a solid medium containing hemoglobin as the sole nitrogen source. The cleavage of hemoglobin by the secreted proteinases results in formation of clearance zones. The visibility of such zones was enhanced by addition of an acid-base indicator. Using this system, we assessed 245 clinical isolates of Candida from patients in the hospital of the Faculty of Medicine, Palacky University, Olomouc, Czech Republic, for the presence of secreted aspartic proteases (Saps). We also used the test plates for rapid semiquantitative testing of Sap inhibitors. Most of the pepstatin analogs affected the formation of the zones of clearance as well as the growth of Candida albicans, C. tropicalis, and C. parapsilosis colonies. By contrast, the human immunodeficiency virus proteinase inhibitors saquinavir, ritonavir, nelfinavir, and indinavir had no effect on the Candida strains tested. These results are in agreement with the inhibition constants obtained for the individual inhibitors with purified Saps. Thus, the plates containing hemoglobin proved to be an appropriate tool for the rapid and reliable assessment of Sap production and inhibition.
The yeasts of the genus Candida are opportunistically invasive in individuals whose defense mechanisms are impaired. Pathogenic Candida species cause diseases ranging from superficial mycoses to disseminated and often fatal infections. The individuals at risk include intensive care and postsurgical patients, human immunodeficiency virus (HIV)-infected hosts, patients with hematological malignancies, elderly patients, and premature infants (4, 6, 18, 25, 28). Although Candida albicans is the most frequently isolated yeast associated with human infection, changing patterns of the Candida species detected among clinical isolates in the last decade are evident (5, 8, 24, 28). Therefore, rapid and reliable identification of Candida species producing certain virulence factors is important in routine clinical microbiology practice.
Virulence attributes of Candida species include adherence to host tissues, morphological changes, and secretion of hydrolases, e.g., phospholipases and proteinases (11). Secreted aspartic proteinases (Saps) of pathogenic Candida spp. have been studied extensively (13). C. albicans, C. tropicalis, C. dubliniensis, C. guilliermondii, and C. parapsilosis possess SAP gene families (7, 10, 13, 16, 30). The Saps of C. albicans, C. parapsilosis, C. tropicalis, and C. lusitaniae have been characterized (2, 9, 12, 14, 19, 22, 26), and their inhibitors have been tested as potential antimycotic drugs (for a review, see reference 27). Information on the extracellular proteolysis of other medically important Candida species (C. guilliermondii, C. stellatoidea, C. glabrata, C. krusei, C. kefyr) is limited (3, 8, 20, 21, 23).
In vitro, Sap production is elicited in culture medium containing exogenous protein (usually bovine serum albumin [BSA]) as a sole source of nitrogen. In addition to albumins, proteinase production can be stimulated by hemoglobin, keratin, collagen, or complex peptide mixtures, e.g., peptone and tryptone (15). Odds and Abbott (17) have developed a simple method for Sap detection that uses test plates containing minimal medium supplemented with BSA as a sole source of nitrogen. We have modified this method and tested a panel of clinical isolates of Candida for Sap production. Furthermore, we have examined the effects of proteinase inhibitors on Sap-producing strains of C. albicans, C. tropicalis, and C. parapsilosis.
MATERIALS AND METHODS
Yeast strains.
The study included 245 Candida strains from 207 patients collected from 1995 to 2001 (see Table 1). These isolates are deposited in the hospital of the Faculty of Medicine, Palacky University, Olomouc, Czech Republic. Multiple isolates from one patient were included only when the specimens were obtained from separate body sites. Identification of strains was performed by standard procedures including micromorphology on rice agar, biochemical assays with commercial kits (the Auxacolor system [Bio-Rad] and the ID 32C system [bioMérieux]), and fermentation of glucose, sucrose, lactose, maltose, and galactose.
TABLE 1.
Growth and extracellular proteolytic activities of clinical yeast isolates determined on hemoglobin-containing medium
| Species | No. of strains tested | No. of strains with:
|
|
|---|---|---|---|
| Growth | Extracellular proteolysisa | ||
| C. albicans | 59 | 59 | 59 |
| C. parapsilosis | 74 | 74 | 74 |
| C. tropicalis | 48 | 48 | 48 |
| C. lusitaniae | 7 | 0 | 0 |
| C. krusei | 39 | 39 | 0 |
| C. glabrata | 9 | 0 | 0 |
| C. gullermondii | 9 | 0 | 0 |
Nineteen strains of three species were found to be highly proteolytic: C. albicans (n = 14), C. parapsilosis (n = 3), and C. tropicalis (n = 2).
The following strains were used for the inhibition study: C. albicans CA 345/IDE96, isolated from blood; C. parapsilosis CP 386/IDE98, isolated from an ear; and C. tropicalis CT30/HK, isolated from blood. Yeast isolates were stored in 20% (wt/vol) BBL Skim Milk Powder (Becton Dickinson) at −70°C until they were tested. Then, the strains were streaked onto Sabouraud dextrose agar and replated onto the hemoglobin test plates after approximately 24 h of incubation at 30°C.
Media.
Test plates for the assessment of Sap production contained agar (Himedia Ltd.), yeast carbon base (YCB; Difco), bovine hemoglobin (Sigma), and bromphenol blue (Lachema). The plates were prepared as follows: the agar and YCB were suspended in water, so that the final concentrations were 4.5 and 1.2%, respectively. After sterilization, the suspension was cooled to 55°C and maintained at that temperature with a water bath. Dissolved hemoglobin was filter sterilized and added to the agar-YCB up to a final concentration of 0.08%. Then, bromphenol blue was added to a concentration of 0.02 ppm in the medium (the stock solution consisted of 1.6 g of bromphenol blue/100 ml of 50% ethanol). The suspension was mixed, and its pH was adjusted to 4 or 4.5. A steam-sterilizable pH electrode was used for the pH measurements. Then, the suspension was immediately poured into the plates. Each 90-mm test plate contained approximately 17 ml of the suspension.
Assessment of extracellular proteolytic activity.
For observation of culture growth and extracellular proteolytic activity, one colony of each strain was streaked onto the test plates. The plates were incubated at 30°C. The streaks were checked for growth, zones of clarification, and macromorphological features at 3, 5, and 7 days. The isolates that did not form visible zones of clarification over the 7 days were recorded as nonproteolytic. The isolates were considered highly proteolytic if the zones of clarification surrounding the streaks were 3 mm wide after 3 days of incubation.
Testing of Sap inhibitors.
The inhibition studies were performed as follows: 20 μl of a suspension of the individual cultures (optical density at 550 nm, ∼0.25) was plated onto the hemoglobin plates. Two sterile filter paper disks (diameter, 5 mm) were laid on each plate. The inhibitors were dissolved in dimethyl sulfoxide (DMSO), and 5 μl of the stock solution (5 mg/ml) or further diluted solutions was pipetted onto the disks. The concentrations of inhibitors on the disks, determined by amino acid analysis, ranged usually from 10 μM to 1.2 mM. When necessary, the concentration range was adapted. The plates were cultivated at 30°C for 3 days. Then, the diameter of the zone without clearance was measured. To compare the efficiencies of the inhibitors in the environment of Candida cultures, we have introduced the value ICP50, which is the concentration of an inhibitor which reduces the area of hydrolysis one-half after 3 days of cultivation at 30°C and which was calculated by the following equation: ICP50 = {[(Dmax × [I])]/d} − [I], where d is the diameter of nonhydrolyzed zone, [I] is the concentration of an inhibitor, and Dmax is the maximal theoretical diameter of the nonhydrolyzed area. All inhibition tests were performed in triplicate, and then the average values and the deviations were calculated by using Excel 2000 software.
Inhibitors.
All pepstatin analogs were synthesized by an established solution method (1). Crude inhibitors were purified by preparative reversed-phase high-pressure liquid chromatography (RP-HPLC); and their homogeneities were confirmed by isocratic RP-HPLC, amino acid analysis, and high-resolution fast-atom-bombardment mass spectrometry.
The following compounds were prepared: Boc-Val-Val-Pst-Ala-Pst-OH, Boc-Val-Val-Pst-Ala-Pst-OMe Boc-Val-Val-Pst-Ala-Pst-NH2, Boc-Val-Val-Pst-Ala-(3R,4R)Pst-OMe, Boc-Val-Val-(3R,4R)Pst-Ala-Pst-OMe, and Boc-Val-Val-Pst-Ala-OMe. Pepstatin A (i.e., isovaleryl-Val-Val-Sta-Ala-Sta-OH) was purchased from Amersham Life Sciences.
RESULTS
Development and optimization of medium.
The test medium used by Odds and Abbott (17) contains BSA as the only source of nitrogen. The cleavage of BSA by Saps results in zones of clearance. When hemoglobin was used instead of BSA, proteolytic Candida strains formed yellowish zones surrounding colonies or streaks. The zones were easy to read and compare.
In liquid protein-supplemented cultures of C. albicans, the characteristic drop in pH associated with Sap production has been observed by others (29). To examine possible pH changes during Candida culture growth on hemoglobin plates, we added bromphenol blue to the medium. Bromphenol blue did not affect the growth of Candida because all strains tested grew similarly on the plates with and without the indicator. The plates containing the indicator were deep blue, but light yellow zones surrounding the streaks of proteolytic Candida strains (n = 181) appeared. Thus, the addition of bromphenol blue further enhanced the visibility of the proteolytic zones. No color change was caused by nonproteolytic strains (n = 64). A C. albicans mutant strain lacking the SAP1, SAP2, and SAP3 genes displayed limited growth and did not form any proteolytic zones.
Screening of Candida isolates for Sap production.
We analyzed 245 clinical isolates of Candida species (Table 1). We examined the growth of these isolates and their ability to produce extracellular proteinase on the hemoglobin-supplemented medium. We focused on the species that are known to produce extracellular aspartic proteinases, i.e., C. albicans, C. parapsilosis, and C. tropicalis (11). As C. lusitaniae was found to produce proteinase in liquid protein-supplemented media (12, 19), we included also seven strains of this species. Isolates of C. krusei, C. glabrata, and C. guilliermondii were included, as they are allegedly nonproteolytic (21).
Nonproteolytic strains.
The isolates of C. guilliermondii (n = 9), C. glabrata (n = 9), and C. lusitaniae (n = 7) grew poorly on the hemoglobin test plates and produced a very low extracellular proteolytic activity that was detectable only after prolonged cultivation (i.e., longer than 7 days).
C. krusei isolates (n = 39) were able to grow on the hemoglobin-supplemented medium. However, the proteolytic zones surrounding the C. krusei streaks were not visible within the 7 days of cultivation. We hypothesized that C. krusei can grow on the nitrogen-limited medium due to an ability to utilize the nitrogen pool from previous passages. To test this, we prepared plates containing only agar-YCB, agar-YCB-bromphenol blue, or agar-YCB-BSA (pH 4). The growth of the C. krusei strains on these media was similar to that on the hemoglobin test plates. C. krusei grew even when it was replated from Sabouraud dextrose agar to a plate with only agar at pH 4. The growth ceased only after the third passage on agar.
C. lusitaniae secreted proteinase in a liquid medium containing BSA as a sole source of nitrogen. Thus, the lack of proteolysis on the hemoglobin test plates is puzzling. In several cases, C. lusitaniae formed filaments of a slightly different shape than those formed by C. krusei. As opposed to C. krusei, however, replating of C. lusitaniae onto agar-YCB or only agar resulted in a complete loss of growth. The C. lusitaniae isolates also grew poorly on the hemoglobin plates without bromphenol blue, as well as on the plates containing BSA instead of hemoglobin. Hence, growth of C. lusitaniae on either solid or liquid medium may affect the production of Sap.
All 48 strains of C. tropicalis analyzed in this study were found to possess extracellular proteolytic activity. However, five isolates produced zones of clearance 3 mm in diameter only at day 7 of incubation. These isolates were obtained from sputum (n = 3), stool (n = 1), and a central venous catheter (n = 1).
Highly proteolytic strains.
Extracellular proteolysis of the majority of C. albicans, C. parapsilosis, and C. tropicalis isolates was evaluated as average, as they formed zones of clarification smaller than 3 mm in diameter during the 5 days of incubation. Among the 245 Candida isolates tested, 19 were found to be highly proteolytic. Fourteen of them were C. albicans strains; and these were isolated from the nasopharynx (n = 7), vagina (n = 2), sputum (n = 2), skin (n = 1), oral cavity (n = 1), and urine (n = 1). Six of the nasopharyngeal isolates were obtained from infants with very low birth weights. One highly proteolytic strain was isolated from the nasopharynx of an HIV-positive adult female. The highly proteolytic vaginal isolates were obtained from women diagnosed with acute vaginitis. The sputum isolates were obtained from elderly patients suffering from different diseases. Altogether, about one-third of the highly proteolytic isolates comprised C. albicans isolates from the nasopharynges of premature infants.
Three isolates of C. parapsilosis and two isolates of C. tropicalis were found to be highly proteolytic. The sites of collection of these isolates differed, as did the ages and diagnoses for the patients from whom they were recovered.
pH requirements.
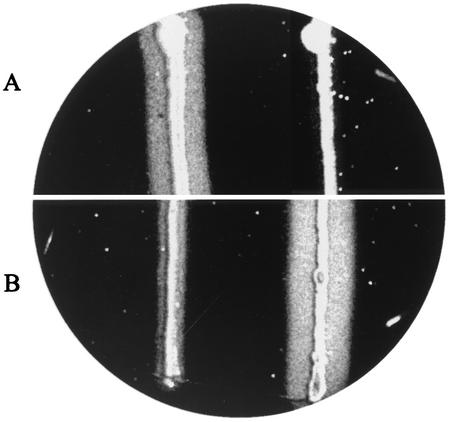
In the assays of Odds and Abbott (17), the pH of the medium was 3.5. Using three strains of each of the Candida species, we found that the optimum pH for substrate hydrolysis and culture growth ranges from 4 to 4.5 on the hemoglobin test plates. Limited culture growth and no hemoglobin hydrolysis were observed at pHs lower than 3.5 or higher than 5 for all of the strains tested. All 245 strains were cultivated on media adjusted to pH 4 and 4.5. Isolates of C. parapsilosis were able to grow readily at both pH 4 and 4.5, but the latter was preferred (Fig. 1A). Most of the C. albicans strains grew better and displayed higher levels of extracellular proteolysis at pH 4 (Fig. 1B). C. tropicalis isolates grew and produced Saps at both pH 4 and pH 4.5 without substantial differences.
FIG. 1.
Assessment of extracellular proteolytic activity (corresponding to the zone of clearance) of C. parapsilosis (left) and C. albicans (right) growing for 5 days at 30°C on solid YCB supplemented with 0.08% hemoglobin and bromphenol blue at 0.02 ppm and pH 4.5 (A) and pH 4.0 (B).
The pH measurements obtained with the electrode for semisolid materials confirmed that the Sap-producing strains acidify their environment. The pHs of the yellowish proteolytic zones surrounding the yeast culture ranged from 3.5 to 3.8. Thus, the pHs in the proximity of C. parapsilosis streaks dropped by 0.7 to 1 units.
Testing of Sap inhibitors.
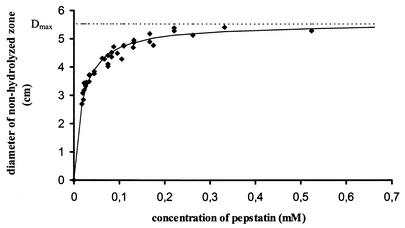
We have selected two series of compounds, one comprising pepstatin A and its analogs (Table 2) and the second consisting of four HIV proteinase inhibitors used in clinical practice as anti-AIDS drugs (indinavir, nelfinavir, ritonavir, and saquinavir). Both inhibitor series were tested with C. albicans, C. parapsilosis, and C. tropicalis strains that displayed average levels of extracellular proteolysis. The inhibitors applied on filter paper disks diffused into the medium and blocked the hydrolysis of hemoglobin and subsequent culture growth. Figure 2 shows that a plot of the diameter of the inhibition zone versus the inhibitor concentration provides a sufficiently accurate curve for semiquantitative evaluation of the inhibitor efficiency.
TABLE 2.
Activities of pepstatin A and its analogs against isolates of C. albicans, C. tropicalis, and C. parapsilosis determined on hemoglobin-containing medium
| Compound no. and inhibitora |
C. albicans
|
C. tropicalis
|
C. parapsilosis
|
|||
|---|---|---|---|---|---|---|
| ICP50 (μM) | Ki (nM) | ICP50 (μM) | Ki (nM) | ICP50 (μM) | Ki (nM) | |
| 1. Iva-Val-Val-Sta-Ala-Sta-OH (pepstatin A) | 7 ± 3 | 1.4 | 31 ± 10 | 5.1 | 19 ± 5 | 0.3 |
| 2. Boc-Val-Val-Pst-Ala-Pst-OH | 25 ± 10 | 2.9 | 40 ± 15 | 1.8 | 37 ± 15 | 6.7 |
| 3. Boc-Val-Val-Pst-Ala-Pst-OMe | 18 ± 7 | 3.9 | 45 ± 19 | 0.5 | 25 ± 7 | 0.3 |
| 4. Boc-Val-Val-Pst-Ala-Pst-NH2 | 34 ± 12 | 1.1 | 55 ± 26 | 1 | 69 ± 16 | 0.1 |
| 5. Boc-Val-Val-Pst-Ala-(3R,4R)Pst-OMe | 6 ± 0.9 | 1.3 | 24 ± 8 | 0.25 | 16 ± 2 | 0.4 |
| 6. Boc-Val-Val-(3R,4R)Pst-Ala-Pst-OMe | NI | 610 | NI | 256 | NI | 174 |
| 7. Boc-Val-Val-Pst-Ala-OMe | NI | 83 | NI | 479 | NI | 14.6 |
The calculated ICP50 are compared to the Ki constants obtained with purified Saps (19). NI, no inhibition observed; Iva, isovaleryl; Sta, statine; Pst, phenylstatine, (3S,4S)-4-amino-3-hydroxy-5-phenylpentanoic acid; Boc, tert-butoxycarbonyl; OMe, methoxycarbonyl.
FIG. 2.
Plot of the inhibition zone diameter versus the inhibitor (pepstatin A) concentration. A total of 20 μl of the C. parapsilosis suspensions (optical density at 550 nm, ∼0.25) was spread onto hemoglobin-supplemented agar medium (pH 4.5), filter paper disks containing 0.01 to 0.7 mM pepstatin A were applied to each plate, and the plates were incubated at 30°C. The diameter of nonhydrolyzed area was measured after 48 h of incubation.
The diameter of the inhibition zone is affected by several factors such as the pH of the medium, the temperature, and the cultivation time. Therefore, the pH was adjusted to the optimum value for the growth of individual Candida species on the hemoglobin-supplemented medium (i.e., pH 4 for C. albicans and C. tropicalis and pH 4.5 for C. parapsilosis). Incubation periods longer than 3 days resulted in depletion of the inhibitor applied to the test plate and, thus, in a decreased accuracy of reading of the zone diameters. The growth of the colonies in the zones of inhibition was not blocked completely. However, the colonies affected by the presence of inhibitor were much smaller and less frequent than those growing on a control plate without inhibitor. Moreover, potent inhibitors prevented the drop of the pH in the environment of the Candida colonies.
To confirm that the diffusion of inhibitors from the filter paper disks proceeds quantitatively, the disks with inhibitors were placed on test plates containing no yeast culture. After 3 days, the disks were removed, hydrolyzed, and subjected to amino acid analysis. The analysis confirmed that no detectable amount of inhibitors remained on the filter paper disks.
DMSO does not affect growth and extracellular proteolysis of C. albicans, C. parapsilosis, and C. tropicalis strains. This was proved in a separate experiment, in which only DMSO was pipetted onto a filter paper disk placed on a test plate with Candida cultures.
We used the inhibitors tested here in our previous study (19) to map the substrate specificities of purified secreted proteinases of C. albicans, C. parapsilosis, C. tropicalis, and C. lusitaniae. Inhibitors with nanomolar and subnanomolar Ki values (Table 2, compounds 1 to 5) efficiently inhibited the proteolysis and growth of all Candida species tested on hemoglobin-supplemented medium. Of all the compounds tested here, inhibitor 5, which contained a Pst residue in the 3R,4R configuration, was the most effective one for all three Candida species. This is in a good agreement with the results obtained with purified proteases. The less effective inhibitors (inhibitors 6 and 7) had Ki values ranging between 83 and 640 nM and did not inhibit the cleavage of hemoglobin in the agar. Interestingly, the compounds tested here were more efficient against C. albicans than against the two other Candida species. The average error for the ICP50s obtained from three independent experiments was ±33%.
Inhibitors of the HIV type 1 proteinase used in clinical practice are weak inhibitors of Saps, probably because of insufficient chain length (19). Indinavir, nelfinavir, ritonavir, and saquinavir did not exert any effect on the cultures of C. albicans, C. parapsilosis, or C. tropicalis growing on the hemoglobin test plates.
DISCUSSION
The increased frequency of fungal infections in immunocompromised hosts has resulted in the need to develop new antifungal compounds and diagnostic methods. The aim of this work was to develop a simple screening method and to use it in a prototype study of virulence factor production and inhibition. Since secretion of aspartic proteinases is one of the important determinants associated with the pathogenicity of Candida species, we focused on testing the extracellular proteolytic activities of clinical Candida isolates. We modified solid medium for semiquantitative testing of Sap activity. The advantage of the present method is the increased reproducibility of reading of the proteolytic zones compared to the reproducibilities of tests with BSA or gelatin. The critical factor is the pH, because relatively small pH shifts can cause changes in the growth and extracellular proteolysis of the strains tested. The presence of an acid-base indicator in the medium facilitates the detection of pH changes both in the medium and in the microenvironment of the yeast culture.
We introduced this novel screening method into the routine operation of a clinical microbiological laboratory of a hospital in Olomouc, Czech Republic, and analyzed 245 Candida isolates from 207 patients. Most of the C. albicans, C. parapsilosis, and C. tropicalis isolates grew and readily displayed proteolysis in our test system, as expected. With few exceptions, a low level of Sap activity was detected. Experiments are under way in our laboratory to elucidate how the regulation of extracellular proteolysis is changed in these strains and if there is any compensating mechanism for the decreased Sap activity.
Isolates of C. glabrata, C. guilliermondii, C. krusei, and C. lusitaniae were classified as nonproteolytic. Interestingly, C. guilliermondii is known to possess homologues of C. albicans SAP genes (16), and C. lusitaniae was found to produce Sap in a liquid protein-supplemented medium (12, 19). It appears that agar supplemented with YCB and hemoglobin at pH 4 to 4.5 is not a suitable environment for Sap induction in these Candida species. C. krusei isolates were categorized as nonproteolytic. In comparison with other nonproteolytic species, however, C. krusei displayed a greater ability to survive on the nitrogen-limited medium.
There was no significant relationship among the diagnoses for patients harboring Candida strains with a certain level of extracellular proteolysis. However, highly proteolytic C. albicans strains were preferentially isolated from the nasopharynges of low-birth-weight infants. One of the highly proteolytic C. albicans strains was isolated from an HIV-positive patient. Three other HIV-positive hosts harbored C. albicans as well, but the strains isolated displayed average levels of extracellular proteolysis.
The hemoglobin test plates also provided a tool for semiquantitative assessment of the efficiencies of the inhibitors in the presence of growing yeast cells. The accuracy of this method depends on many aspects including the moisture content and the thickness of the plates, the diffusion rates of individual compounds, etc. Although we made efforts to control these problems, it is not surprising that the deviations calculated for the ICP50s often exceeded 40%. Despite this, the compounds that proved to be good inhibitors of purified Saps also displayed good inhibitory activities in the test plate system. The weak inhibitors of purified Saps were also evaluated as inefficient by the present method. The ICP50s are not precisely proportional to the Ki values obtained with purified enzymes. Therefore, one can assume that the inhibitors may also interact with other molecules secreted by the Candida spp. Moreover, the presence of inhibitors that resulted in the lack of Sap activity may cause changes in the expression of other genes, which in turn may trigger a response of the Candida colonies to the peptidomimetic structures. The most obvious effect is that there is no change in the pH of the environment of the colonies and the sizes of the colonies are reduced in the presence of potent inhibitors. It is evident that the data obtained with the hemoglobin test plates and by the introduction of ICP50 constants are in agreement with the results of kinetic characterization of the inhibitors with purified Saps (19).
The system presented here can be used to detect one of the virulence factors of Candida species and to screen potential antimycotics targeted against Saps.
Acknowledgments
We thank Elena Dolejší and Jitka Cankařová for excellent technical assistance. We are grateful to Bernhard Hube for the SAP1-, SAP2-, and SAP3-negative C. albicans strain.
This work was supported by the Ministry of Health of the Czech Republic (grants NI/6485-3 and NI/6190-3) and the Grant Agency of the Czech Republic (grant 303/01/0831). This work was part of research project Z4 055 905.
REFERENCES
- 1.Bodanszky, M., and A. Bodanszky. 1984. The practice of peptide synthesis. Springer-Verlag, Berlin, Germany.
- 2.Brožková, K., I. Křížová, L. Pavlíčková, M. Hradilek, M. Fusek, T. Ruml, M. Souček, and I. Pichová. 1999. Peptidomimetic inhibitors of extracellular aspartic proteinases of Candida albicans and Candida tropicalis. Collect. Czech. Chem. Commun. 64:130-137. [Google Scholar]
- 3.Capobianco, J. O., C. G. Lerner, and R. C. Goldman. 1992. Application of a fluorogenic substrate in the assay of proteolytic activity and in the discovery of a potent inhibitor of Candida albicans aspartic proteinase. Anal. Biochem. 204:96-102. [DOI] [PubMed] [Google Scholar]
- 4.Caugant, D. A., and P. Sandven. 1993. Epidemiological analysis of Candida albicans strains by multilocus enzyme electrophoresis. J. Clin. Microbiol. 31:215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dan, M., F. Poch, and D. Levin. 2002. High rate of vaginal infection caused by non-C. albicans Candida species among asymptomatic women. Med. Mycol. 40:383-386. [DOI] [PubMed] [Google Scholar]
- 6.Dean, D. A., and K. W. Burchard. 1996. Fungal infection in surgical patients. Am. J. Surg. 171:374-382. [DOI] [PubMed] [Google Scholar]
- 7.De Viragh, P. A., D. Sanglard, G. Togni, R. Falchetto, and M. Monod. 1993. Cloning and sequencing of two Candida parapsilosis genes encoding acid proteases. J. Gen. Microbiol. 139:335-342. [DOI] [PubMed] [Google Scholar]
- 8.Fidel, P. L., Jr., J. A. Vazquez, and J. D. Sobel. 1999. Candida glabrata: review of epidemiology, pathogenesis and clinical disease with comparison to C. albicans. Clin. Microbiol. Rev. 12:80-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fusek, M., E. A. Smith, M. Monod, B. Dunn, and S. I. Foundling. 1994. Extracellular aspartic proteinases from Candida albicans, Candida tropicalis and Candida parapsilosis yeast differ substantially in their specificities. Biochemistry 33:9791-9799. [DOI] [PubMed] [Google Scholar]
- 10.Gilfillan, G. D., D. J. Sullivan, K. Haynes, T. Parkinson, D. C. Coleman, and N. A. R. Gow. 1998. Candida dubliniensis: phylogeny and putative virulence factors. Microbiology 144:829-838. [DOI] [PubMed] [Google Scholar]
- 11.Haynes, K. 2001. Virulence in Candida species. Trends Microbiol. 9:591-596. [DOI] [PubMed] [Google Scholar]
- 12.Hrušková-Heidingsfeldová, O., J. Dostál, P. Hamal, J. Pazlarová, T. Ruml, and I. Pichová. 2001. Enzymological characterization of secreted proteinases from Candida parapsilosis and Candida lusitaniae. Collect. Czech. Chem. Commun. 66:1707-1719. [Google Scholar]
- 13.Hube, B., and J. Naglik. 2001. Candida albicans proteinases: resolving the mystery of a gene family. Microbiology 147:1997-2005. [DOI] [PubMed] [Google Scholar]
- 14.Koelsch, G., J. Tang, J. A. Loy, M. Monod, K. Jackson, S. I. Foundling, and X. Lin. 2000. Enzymic characteristic of secreted aspartic proteases of Candida albicans. Biochim. Biophys. Acta 1480:117-131. [DOI] [PubMed] [Google Scholar]
- 15.Lerner, C. G., and R. C. Goldman. 1993. Stimuli that induce production of Candida albicans extracellular aspartyl proteinase. J. Gen. Microbiol. 139:1643-1651. [DOI] [PubMed] [Google Scholar]
- 16.Monod, M., G. Togni, B. Hube, and D. Sanglard. 1994. Multiplicity of genes encoding secreted aspartic proteinases in Candida species. Mol. Microbiol. 13:357-368. [DOI] [PubMed] [Google Scholar]
- 17.Odds, F. C., and A. B. Abbott. 1980. A simple system for the presumptive identification of Candida albicans and differentiation of strains within the species. Sabouraudia 18:301-317. [PubMed] [Google Scholar]
- 18.Pfaller, M., R. N. Jones, G. V. Doern, H. S. Sader, R. J. Hollis, and S. A. Messer for the SENTRY Participant Group. 1998. International surveillance of bloodstream infections due to Candida species: frequency of occurrence and antifungal susceptibilities of isolates collected in 1997 in the United States, Canada, and South America for the SENTRY Program. J. Clin. Microbiol. 36:1886-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pichová, I., L. Pavlíčková, J. Dostál, E. Dolejší, O. Hrušková-Heidingsfeldová, J. Weber, T. Ruml, and M. Souček. 2001. Secreted aspartic proteinases of Candida albicans, Candida tropicalis, Candida parapsilosis and Candida lusitaniae: inhibition with peptidomimetic inhibitors. Eur. J. Biochem. 268:2669-2677. [DOI] [PubMed] [Google Scholar]
- 20.Ray, T. L., and C. D. Payne. 1990. Comparative production and rapid purification of Candida acid proteinase from protein-supplemented cultures. Infect. Immun. 58:508-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruchel, R., F. De Bernardis, T. L. Ray, P. A. Sullivan, and T. G. Cole. 1992. Candida acid proteinases. J. Med. Vet. Mycol. 30:123-132. [PubMed] [Google Scholar]
- 22.Ruchel, R. 1981. Properties of a purified proteinase from the yeast Candida albicans. Biochim. Biophys. Acta 659:99-113. [DOI] [PubMed] [Google Scholar]
- 23.Ruchel, R., K. Uhlemann, and B. Boning. 1983. Secretion of aspartic proteinases by different species of the genus Candida. Zentbl. Bacteriol. Microbiol. Hyg. 255:537-548. [PubMed] [Google Scholar]
- 24.Safdar, A., V. Chaturvedi, E. W. Cross, S. Park, E. M. Bernard, D. Armstrong, and D. S. Perlin. 2001. Prospective study of Candida species in patients at a comprehensive cancer center. Antimicrob. Agents Chemother. 45:2129-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saiman, L., E. Ludington, M. Pfaller, S. Rangel-Frausto, R. T. Wiblin, J. Dawson, H. M. Blumberg, J. E. Patterson, M. Rinaldi, J. E. Edwards, R. P. Wenzel, and W. Jarvis. 2000. Risk factors for candidemia in neonatal intensive care unit patients. Pediatr. Infect. Dis. J. 19:319-324. [DOI] [PubMed] [Google Scholar]
- 26.Smolenski, G., P. A. Sullivan, S. M. Cutfield, and J. F. Cutfield. 1997. Analysis of secreted aspartic proteinases from Candida albicans: purification and characterization of individual Sap1, Sap2 and Sap3 isoenzymes. Microbiology 143:349-356. [DOI] [PubMed] [Google Scholar]
- 27.Stewart, K., and C. Abad-Zapatero. 2001. Candida proteases and their inhibition: prospects for antifungal therapy. Curr. Med. Chem. 8:941-948. [DOI] [PubMed] [Google Scholar]
- 28.Verduyn Lunel, F. M., J. F. Meis, and A. Voss. 1999. Nosocomial fungal infections: candidemia. Diagn. Microbiol. Infect. Dis. 34:213-220. [DOI] [PubMed] [Google Scholar]
- 29.Wu, T., L. P. Samaranayake, W. K. Leung, and P. A. Sullivan. 1999. Inhibition of growth and secreted aspartyl proteinase production in Candida albicans by lysozyme. J. Med. Microbiol. 48:721-730. [DOI] [PubMed] [Google Scholar]
- 30.Zaugg, C., M. Borg-von Zepelin, U. Reichard, D. Sanglard, and M. Monod. 2001. Secreted aspartic proteinase family of Candida tropicalis. Infect. Immun. 69:405-412. [DOI] [PMC free article] [PubMed] [Google Scholar]