Abstract
Streptococcus dysgalactiae is classified by a combination of phenotypic and genotypic characteristics into Lancefield group C alpha-hemolytic Streptococcus dysgalactiae subsp. dysgalactiae and Lancefield group C, group G, and group L beta-hemolytic Streptococcus dysgalactiae subsp. equisimilis. In this study, we report the isolation of a catalase-negative, alpha-hemolytic, optochin- and bacitracin-resistant viridans group strain, which does not grow in 10 or 40% bile, on MacConkey agar or bile esculin agar, or in 6% NaCl, from the blood culture of a 73-year-old woman with pyomyositis and poststreptococcal reactive arthritis. Lancefield grouping revealed that the strain was a group G streptococcus. The Vitek system (GPI) showed that it was unidentified, and the API system (20 STREP) showed that it was 95.7% S. dysgalactiae subsp. dysgalactiae. 16S rRNA gene sequencing showed that it was a strain of S. dysgalactiae. Based on phylogenetic affiliation with 16S rRNA gene or GroEL amino acid (another bacterial gene, in addition to 16S rRNA gene, that is highly conserved) sequences, the strain is most closely related to Lancefield group C beta-hemolytic S. dysgalactiae subsp. equisimilis. PCR amplification and sequencing of the streptolysin S structural gene (sagA) and M protein gene (emm) hypervariable region showed the presence of these suspected primary virulence factors. Further studies would delineate whether the isolate is just a hemolysin-deficient variant of group G beta-hemolytic S. dysgalactiae subsp. equisimilis or a novel type of S. dysgalactiae. The present case showed that group G alpha-hemolytic S. dysgalactiae subsp. equisimilis can be associated with serious invasive infection and poststreptococcal sequelae.
Streptococcus dysgalactiae is classified by a combination of phenotypic and genotypic characteristics into four types: Lancefield group C alpha-hemolytic Streptococcus dysgalactiae subsp. dysgalactiae, Lancefield group C beta-hemolytic Streptococcus dysgalactiae subsp. equisimilis, Lancefield group G beta-hemolytic S. dysgalactiae subsp. equisimilis, and Lancefield group L beta-hemolytic S. dysgalactiae subsp. equisimilis (19). The natural reservoirs of group C alpha-hemolytic S. dysgalactiae subsp. dysgalactiae are animals such as cows and sheep. It mainly causes mastitis in cows and suppurative polyarthritis in lambs, but it only occasionally causes infections in humans (20). On the other hand, the natural reservoirs of S. dysgalactiae subsp. equisimilis are humans, and these streptococci are major causes of streptococcal infections in humans. In a recent study of 75 group G beta-hemolytic streptococcal strains isolated from the blood cultures of 66 patients, we discovered that all 75 strains were S. dysgalactiae subsp. equisimilis, with 16S rRNA gene sequencing as the “gold standard” (22).
Hemolysin production has always been described as associated with virulence, and it was shown that the beta-hemolytic phenotype of group G S. dysgalactiae subsp. equisimilis, produced by streptolysin S and encoded by a functional homologue of the 9-gene group A streptococcus sag operon, contributes to the pathogenesis of streptococcal necrotizing soft tissue infection (6). On the other hand, a hemolysin-deficient variant of group G S. dysgalactiae subsp. equisimilis has recently been recovered from the throat culture of a patient with pharyngitis, and it was suggested by the authors that these hemolysin-deficient variants may be overlooked as etiological agents of streptococcal infections (4). In this study, we report the isolation of a group G alpha-hemolytic streptococcus from the blood culture of a patient with pyomyositis and reactive arthritis. The strain, named HKU7, exhibited phenotypic characteristics that do not fit into patterns of any known species. 16S rRNA gene sequencing showed that there was more than 99% base identity between the 16S rRNA gene of HKU7 and those of other strains of S. dysgalactiae. Further phenotypic and genotypic characterization showed that this bacterium is a strain of Lancefield group G alpha-hemolytic S. dysgalactiae subsp. equisimilis.
CASE REPORT
A 73-year-old Chinese woman was admitted to the hospital in September 2000 because of bilateral hip, knee, and shoulder pain for 2 weeks. She had well-controlled diabetes mellitus, a history of carcinoma of the rectum, with abdominal perineal resection 5 years ago, and carcinoma of the cervix, with hysterectomy and radiotherapy 20 years ago. She was afebrile upon admission. An examination revealed left hip tenderness and limited range of movement in the bilateral shoulder and hip joints but no obvious swelling or effusion. Radiographs of the shoulders and hips showed only degenerative changes. The total white cell count was 11.9 × 109/liter, with a neutrophil count of 10.9 × 109/liter, a lymphocyte count of 0.7 × 109/liter, and a monocyte count of 0.3 × 109/liter. The hemoglobin level was 12.5 g/dl, the platelet count was 191 × 109/liter, the erythrocyte sedimentation rate was 81 mm/h, and the C-reactive protein level was 33.6 mg/dl. Liver and renal function tests were within normal limits, except for a low serum albumin level of 33 g/liter. She developed acute upper gastrointestinal bleeding 1 week after admission. Urgent upper endoscopy revealed multiple bleeding ulcers in the stomach and duodenum, which were related to the nonsteroidal anti-inflammatory drugs prescribed for her pain relief. The bleeding was settled, but she developed fever and septicemic shock 2 days later. Blood culture was performed, and treatment with empirical intravenous piperacillin-tazobactam was commenced. Tc-99m bone scintigraphy on the same day showed increase tracer uptake in the L1/L2, L5/S1, left sacroiliac, and left hip joints and in both elbow, knee, and ankle joints, which can be compatible with degeneration or arthritis.
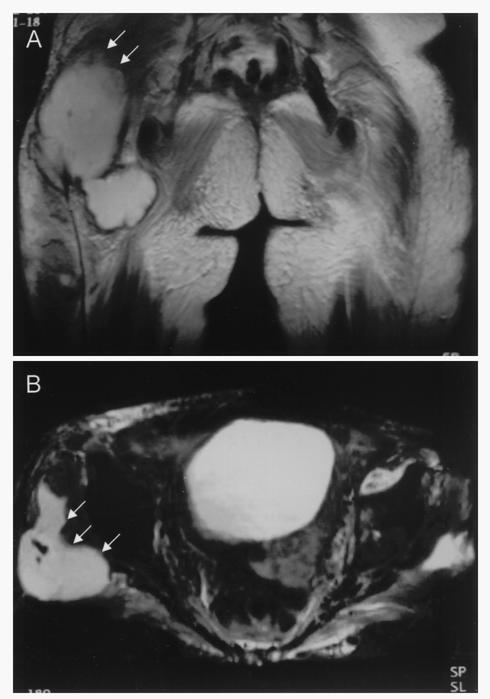
On day 1 postincubation, the aerobic blood culture bottle turned positive for a gram-positive coccus (HKU7). Her fever and sepsis initially responded to piperacillin-tazobactam. Two days later, she developed progressive shortness of breath. A chest radiograph showed the new onset of a globular heart and bilateral pleural effusion. A transthoracic echocardiogram confirmed pericardial effusion with a tamponade effect. Urgent pericardiocentesis was performed, and 50 ml of blood-stained pericardial fluid was aspirated. The fluid was sent for bacterial culture and cytology, but the results were negative. The pleural and pericardial effusions resolved, but the joint pain persisted. Despite maintenance oral antibiotics, she developed a new swelling over the posterior aspect of her right thigh. There was also acute synovitis and arthritis of both shoulders and knees with prominent effusions. The total white cell count was 10.1 × 109/liter, the erythrocyte sedimentation rate was 107 mm/h, and the C-reactive protein level was 9.6 mg/dl. The levels of C3 and C4 in serum were within normal limits. The rheumatoid factor was negative, and the anti-nuclear antibody titer was 1/80 with a speckled pattern. Aspiration of the right thigh mass yielded 2 ml of thick pus. A Gram smear of the pus showed numerous white blood cells, but no bacteria, fungi, or mycobacteria were recovered. All Gram smears and cultures from left knee and left shoulder aspirates were also negative. Owing to the progressive right thigh swelling, a magnetic resonance imaging scan was performed and showed a large abscess of 15 by 7 by 6 cm, involving the muscles and subcutaneous tissue of the right buttock and thigh (Fig. 1). The abscess was drained, and intravenous azithromycin and cefazolin were administered. Although the arthritis and abscess of the right thigh gradually subsided, small subcutaneous abscesses subsequently appeared over her right buttock and left thigh. Drainage was performed, but microbiological studies of the drained pus from all abscesses were negative. The final diagnosis was streptococcal bacteremia, pyomyositis, and reactive arthritis. The patient responded to oral salazopyrin and hydroxychloroquine and remained asymptomatic, with no recurrence of abscess or arthritis, up to the time of writing, 1 year from discharge.
FIG. 1.
Coronal (A) and axial (B) T1- and T2-weighted magnetic resonance imaging of the pelvis showing a multiloculated abscess (arrows) in the subcutaneous area of the right buttock.
MATERIALS AND METHODS
Patient and microbiological methods.
All clinical data were collected prospectively as described in a previous publication (14). The BACTEC 9240 blood culture system (Becton Dickinson, Sparks, Md.) was used. The bacterium was identified by standard conventional biochemical methods (15). Lancefield serogrouping was performed by using Streptex (Murex Biotech Ltd., Dartford, United Kingdom) according to the manufacturer's instructions. All tests were performed in triplicate with freshly prepared media on separate occasions. In addition, the Vitek system (GPI) (bioMerieux Vitek, Hazelwood, Mo.) and the API system (20 STREP) (bioMerieux Vitek) were used for the identification of the bacterial isolate in this study. Antimicrobial susceptibility was tested by E-test for penicillin and by the Kirby Bauer disk diffusion method for the other antibiotics, and results were interpreted according to the NCCLS criteria for alpha-hemolytic streptococci.
Bacterial DNA extraction, PCR amplification, and 16S rRNA gene sequencing.
Bacterial DNA extraction, PCR amplification, and DNA sequencing of the 16S rRNA genes were performed according to methods described in previous publications (3, 10-12, 21-35). LPW200 (5′-GAGTTGCGAACGGGTGAG-3′) and LPW205 (5′-CTTGTTACGACTTCACCC-3′) (Gibco BRL, Rockville, Md.) were used as the PCR primers, and LPW200, LPW205, LPW99 (5′-TTATTGGGCGTAAAGCGA-3′), and LPW273 (5′-TTGCGGGACTTAACCCAAC-3′) were used as the sequencing primers. The sequences of the PCR products were compared with known 16S rRNA gene sequences in the GenBank database by multiple-sequence alignment with the CLUSTAL W program (17).
Streptolysin S structural gene (sagA) sequencing.
PCR amplification and DNA sequencing of the sagA gene of HKU7 were performed according to the method described in a previous publication (6), with LPW614 (5′-ATKARAAAGAAAGGGTTTACAT-3′) and LPW615 (5′-CATATAGTAATTAGCAGGTAC-3′) as the PCR and sequencing primers.
M protein gene (emm) hypervariable region sequencing.
PCR amplification and DNA sequencing of the emm gene hypervariable region of HKU7 were performed according to the method described in a previous publication (16), with LPW616 (5′-ATAAGGAGCATAAAAATGGCT-3′) and LPW617 (5′-AGCTTAGTTTTCTTCTTTGCG-3′) as the PCR and sequencing primers.
Cloning and sequencing of groEL genes.
Cloning and sequencing of the groEL genes of HKU7, Lancefield group C alpha-hemolytic S. dysgalactiae subsp. dysgalactiae (ATCC 43078), Lancefield group C beta-hemolytic S. dysgalactiae subsp. equisimilis (ATCC 35666), Lancefield group G beta-hemolytic S. dysgalactiae subsp. equisimilis (ATCC 12394), and Lancefield group L beta-hemolytic S. dysgalactiae subsp. equisimilis (CIP55-123) were performed according to the following protocol. The bacterial DNA extracts were amplified with 0.5 μM primers (LPW368 [5′-ATTTTCAKCAGATGCNMG-3′] and LPW369 [5′-ACDACDGCTTCKGTDGTYA-3′]) (Gibco BRL). The PCR mixture (50 μl) contained bacterial DNA, PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 2 mM MgCl2, and 0.01% gelatin), 200 μM concentrations of each deoxynucleoside triphosphate, and 1.0 U of Taq polymerase (Boehringer Mannheim, Mannheim, Germany). The mixtures were amplified in 40 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 2 min, and a final extension at 72°C for 10 min in an automated thermal cycler (Perkin-Elmer Cetus, Gouda, The Netherlands). The PCR products were gel purified and sequenced as described previously (35), with LPW368, LPW369, LPW385 (5′-ATGGTCCACAGACAATGAA-3′), LPW513 (5′-ATACTGTTGCAGTTGTGG-3′), and LPW514 (5′-GCTCTTGAGCTTGATGGC-3′) as the sequencing primers.
Phylogenetic characterization.
The phylogenetic relationships between strain HKU7 and the other Streptococcus species and subspecies were determined by using PileUp and the neighbor-joining method with GrowTree (Genetics Computer Group, Inc.). A total of 1,373 nucleotide positions of the 16S rRNA genes were included in the analysis.
Nucleotide sequence accession number.
The 16S rRNA and groEL gene sequences of HKU7, Lancefield group C alpha-hemolytic S. dysgalactiae subsp. dysgalactiae (ATCC 43078), Lancefield group C beta-hemolytic S. dysgalactiae subsp. equisimilis (ATCC 35666), Lancefield group G beta-hemolytic S. dysgalactiae subsp. equisimilis (ATCC 12394), and Lancefield group L beta-hemolytic S. dysgalactiae subsp. equisimilis (CIP55-123) have been logged within the GenBank sequence database under accession no. AF433167, AY121359, AY121360, AY121361, AY121362, AY121367, AY121363, AY121364, AY121365, and AY121366, respectively.
RESULTS
Phenotypic characteristics.
Strain HKU7 is a gram-positive, non-spore-forming coccus arranged in chains. It grows on sheep blood agar as alpha-hemolytic, gray colonies of 0.5 to 1 mm in diameter after 24 h of incubation at 37°C in ambient air. No enhancement of growth is observed in 5% CO2. It also grows on chocolate agar in a microaerophilic or anaerobic environment, but not in 10 or 40% bile, on MacConkey agar or bile esculin agar, or in 6% NaCl. It was catalase negative, alpha-hemolytic, and optochin resistant. Lancefield grouping of the strain showed that it belonged to Lancefield group G. It is resistant to optochin and bacitracin and is nonmotile at both 25 and 37°C. The biochemical profile of strain HKU7 is shown in Table 1. The Vitek system (GPI) showed that it was unidentified, and the API system (20 STREP) showed that it was 95.7% identical to S. dysgalactiae subsp. dysgalactiae and 4.1% identical to S. dysgalactiae subsp. equisimilis. It is sensitive to penicillin (MIC = 0.012 μg/ml), cefepime, clindamycin, erythromycin, tetracycline, and vancomycin.
TABLE 1.
Biochemical profile of strain HKU7 by conventional biochemical tests and Vitek GPI and API 20 STREP systems
| Biochemical reactions or enzyme | Result with testa
|
||
|---|---|---|---|
| Conventional | Vitek GPIb | API 20 STREPc | |
| Catalase | − | − | |
| Resistance to bacitracin | + | + | |
| Resistance to optochin | + | + | |
| Growth in 6% NaCl | − | − | |
| Growth in 10% bile | − | ||
| Growth in 40% bile | − | − | |
| Esculin hydrolysis | − | − | − |
| Hippurate hydrolysis | − | − | |
| Arginine hydrolysis | + | + | + |
| Lysine decarboxylase | − | ||
| Ornithine decarboxylase | − | ||
| Urease | − | − | |
| Voges-Proskauer test | − | − | |
| Tetrazolium reduction | + | ||
| Resistance to novobiocin | + | + | |
| Resistance to polymyxin B | + | ||
| Utilization of: | |||
| Hemicellulase | + | ||
| Dextrose | + | ||
| Lactose | + | + | + |
| Mannitol | − | − | − |
| Raffinose | − | − | − |
| Salicin | + | + | |
| Sorbitol | − | − | − |
| Sucrose | + | + | |
| Trehalose | + | + | + |
| Arabinose | − | − | − |
| Pyruvate | − | ||
| Pullulan | + | ||
| Inulin | − | − | − |
| Melibiose | − | − | |
| Melezitose | − | ||
| Cellobiose | + | + | |
| Ribose | + | + | + |
| Xylose | − | − | |
| d-Glucose | + | ||
| d-Mannose | + | ||
| Maltose | + | ||
| Starch | + | ||
| Glycogen | − | ||
| Pyrrolidonylarylamidase | − | ||
| α-Galactosidase | − | ||
| β-Glucuronidase | + | ||
| β-Galactosidase | − | ||
| Leucine arylamidase | + | ||
| Nitrate reduction | − | ||
| Alkaline phosphatase | + | ||
−, negative; +, positive.
The Vitek GPI System did not identify the strain.
The API 20 STREP system identified the strain as 95.7% S. dysgalactiae subsp. dysgalactiae and 4.1% S. dysgalactiae subsp. equisimilis.
Molecular characterization by 16S rRNA gene and groEL gene sequencing and phylogenetic characterization.
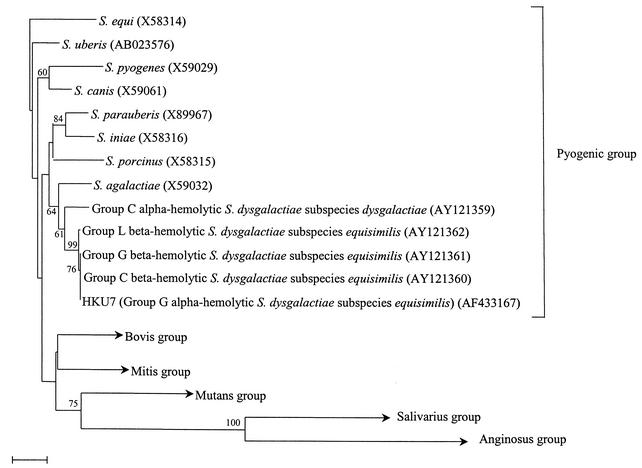
PCR of the 16S rRNA gene of strain HKU7 showed a band at about 1,470 bp. There was no difference between the 16S rRNA gene sequence of strain HKU7 and that of Lancefield group C beta-hemolytic S. dysgalactiae subsp. equisimilis (GenBank accession number AY121360) or Lancefield group G beta-hemolytic S. dysgalactiae subsp. equisimilis (GenBank accession number AY121361), there were 3 base differences between the 16S rRNA gene sequence of strain HKU7 and that of Lancefield group L beta-hemolytic S. dysgalactiae subsp. equisimilis (GenBank accession number AY121362), and there were 14 base differences between the 16S rRNA gene sequence of strain HKU7 and that of Lancefield group C alpha-hemolytic S. dysgalactiae subsp. dysgalactiae (GenBank accession number AY121359), indicating that the isolate was a strain of S. dysgalactiae. As for the GroEL amino acid sequences, there was no difference between the GroEL amino acid sequence of strain HKU7 and that of Lancefield group C beta-hemolytic S. dysgalactiae subsp. equisimilis (GenBank accession number AY121360) but there was 1 amino acid difference between the GroEL amino acid sequence of strain HKU7 and that of Lancefield group G beta-hemolytic S. dysgalactiae subsp. equisimilis (GenBank accession number AY121361), Lancefield group L beta-hemolytic S. dysgalactiae subsp. equisimilis (GenBank accession number AY121362), and Lancefield group C alpha-hemolytic S. dysgalactiae subsp. dysgalactiae (GenBank accession number AY121359). Based on phylogenetic affiliation with the 16S rRNA gene (Fig. 2) and GroEL amino acid sequences, HKU7 is most closely related to Lancefield group C beta-hemolytic S. dysgalactiae subsp. equisimilis.
FIG. 2.
Phylogenetic tree showing the relationships of Lancefield group G alpha-hemolytic S. dysgalactiae subsp. equisimilis to other types of S. dysgalactiae and other streptococci. The tree was inferred from 16S rRNA data by the neighbor-joining method, and bootstrap values were calculated from 1,000 trees. The scale bar indicates the estimated number of substitutions per 100 bases with the Jukes-Cantor correction. Names and accession numbers (in parentheses) are given as cited in the GenBank database.
Streptolysin S structural gene (sagA) sequencing.
PCR of the sagA gene of HKU7 showed a band at about 500 bp. There was no difference between the predicted amino acid sequence of the product of the sagA gene of strain HKU7 and that of S. dysgalactiae subsp. equisimilis (GenBank accession number AY033399) (6).
M protein gene (emm) hypervariable region sequencing.
PCR of the emm gene hypervariable region of HKU7 showed a band at about 1,600 bp. There was no difference between the nucleotide sequence of the emm gene hypervariable region of HKU7 and that of a novel emm gene described recently (GenBank accession number AF485842).
DISCUSSION
In 1936, S. dysgalactiae subsp. equisimilis was first proposed for the description of a group of Lancefield group C beta-hemolytic streptococci isolated from the pharynx, nose, skin, and vagina of humans (5). Subsequently, S. dysgalactiae subsp. dysgalactiae, which was shown to be similar to S. dysgalactiae subsp. equisimilis except for the absence of beta-hemolysis, was recovered from cattle (2). In the next 40 years, numerous strains of a combination of Lancefield serogroups C, G, and L and alpha-hemolysis, beta-hemolysis, and no hemolysis were recovered from humans and animals. Vandamme et al. (in 1996) and Vieira et al. (in 1998) employed a combination of 16S rRNA gene sequencing, DNA-DNA hybridization, and multilocus enzyme electrophoresis to elucidate the relationships of these streptococci (18, 19). This tremendously improves our understanding of the epidemiology of this phenotypically heterogeneous group of streptococci. Using 16S rRNA gene sequence data, members of this heterogeneous group of streptococci were all classified under the species S. dysgalactiae, within the pyogenic group of the genus Streptococcus (9). With the help of additional information derived from DNA-DNA hybridization and multilocus enzyme electrophoresis, S. dysgalactiae was further classified into 2 subspecies and 4 types: Lancefield group C alpha-hemolytic S. dysgalactiae subsp. dysgalactiae, Lancefield group C beta-hemolytic S. dysgalactiae subsp. equisimilis, Lancefield group G beta-hemolytic S. dysgalactiae subsp. equisimilis, and Lancefield group L beta-hemolytic S. dysgalactiae subsp. equisimilis (19).
We report the isolation of HKU7, a Lancefield group G alpha-hemolytic S. dysgalactiae isolate, from the blood culture of a Chinese patient with pyomyositis and reactive arthritis, although the bacterium was not recovered from the abscess, probably due to the administration of antibiotics 48 h before the operative collection of the abscess pus. Further characterization showed that the suspected primary virulence factors (sagA and emm genes) were also present. Similar to other patients with group G beta-hemolytic streptococcal bacteremia (22), our patient also had major underlying diseases, including diabetes mellitus, carcinoma of the rectum, and carcinoma of the cervix. Our patient developed a nonmigratory sterile arthritis and synovitis shortly after the bacteremia, which responded poorly to nonsteroidal anti-inflammatory agents but well to more-potent antirheumatic agents, salazopyrine and hydroxychloroquine. This is compatible with the well-recognized syndrome of poststreptococcal reactive arthritis that typically follows Lancefield group A streptococcal throat infections (1, 7, 13). Although a similar syndrome has also been reported after throat infections with Lancefield group C and group G streptococci, all of the culture-proven cases were due to beta-hemolytic streptococci (8). The present report represents the first case of poststreptococcal reactive arthritis that follows an alpha-hemolytic streptococcal infection. While hemolysins of beta-hemolytic streptococci were shown to contribute to their virulence, the overwhelming clinical manifestations of the present case suggest that alpha-hemolytic streptococci are able to cause similar clinical syndromes.
Lancefield grouping of viridans group streptococci may uncover important streptococcal isolates. In clinical microbiology laboratories, for alpha-hemolytic streptococci, Lancefield grouping is only performed on Streptococcus bovis (Lancefield group D), Streptococcus suis (Lancefield group R which may cross-react with Lancefield group D), members of the Streptococcus milleri group (Lancefield group A, C, F, or G), and the enterococci (Lancefield group D). Lancefield grouping is not performed for other viridans group streptococci. This would miss not only the isolate described in the present study but also some other important streptococcal isolates, such as the hemolysin-deficient group A, C, and G streptococci (4). A study of Lancefield grouping of clinical isolates of alpha-hemolytic streptococci would reveal the usefulness of routine Lancefield grouping of the viridans group streptococci.
Further studies could be performed to delineate whether HKU7 is just a hemolysin-deficient variant of group G beta-hemolytic S. dysgalactiae subsp. equisimilis or a novel type of S. dysgalactiae. For the hemolysin-deficient group G S. dysgalactiae subsp. equisimilis described recently, the strain was identified both phenotypically by the API system (20 STREP) and genotypically by 16S rRNA gene sequencing as S. dysgalactiae subsp. equisimilis. Genotypically, the 16S rRNA gene of HKU7 exhibited more than 99% nucleotide identity with the 16S rRNA gene of all previously described bacterial strains of S. dysgalactiae. Based on both 16S rRNA gene sequence data and GroEL amino acid sequence data, the type most closely related to HKU7 is Lancefield group C beta-hemolytic S. dysgalactiae subsp. equisimilis, and it is clustered with the other types of S. dysgalactiae subsp. equisimilis. On the other hand, besides hemolysis on sheep blood agar and Lancefield serogrouping, HKU7 exhibited additional phenotypic characteristics that are different from the other types of S. dysgalactiae subsp. equisimilis and S. dysgalactiae subsp. dysgalactiae (Table 2). Lancefield group L beta-hemolytic S. dysgalactiae subsp. equisimilis, but not the other types, hydrolyzes hippurate and produces acid from glycogen. Lancefield group C alpha-hemolytic S. dysgalactiae subsp. dysgalactiae, but not the other types, produces acid from sorbitol. Lancefield groups C, G, and L beta-hemolytic S. dysgalactiae subsp. equisimilis, but not HKU7 or Lancefield group C alpha-hemolytic S. dysgalactiae subsp. dysgalactiae, produce β-d-galactosidase. HKU7 and Lancefield groups C and G beta-hemolytic S. dysgalactiae subsp. equisimilis, but not Lancefield group C alpha-hemolytic S. dysgalactiae subsp. dysgalactiae or Lancefield group L beta-hemolytic S. dysgalactiae subsp. equisimilis, are resistant to bacitracin. In fact, HKU7 was identified as S. dysgalactiae subsp. dysgalactiae by the API 20 STREP kit, but none of the 66 Lancefield group G beta-hemolytic S. dysgalactiae subsp. equisimilis strains that were reported previously was identified as S. dysgalactiae subsp. dysgalactiae by the API 20 STREP kit. Although whether HKU7 represents a novel type of S. dysgalactiae remains to be determined, the present case showed that group G alpha-hemolytic S. dysgalactiae subsp. equisimilis can be associated with serious invasive infection and poststreptococcal sequelae.
TABLE 2.
Comparison of characteristics of HKU7, Lancefield group C alpha-hemolytic S. dysgalactiae subsp. dysgalactiae, Lancefield group C beta-hemolytic S. dysgalactiae subsp. equisimilis, Lancefield group G beta-hemolytic S. dysgalactiae subsp. equisimilis, and Lancefield group L beta-hemolytic S. dysgalactiae subsp. equisimilis
| Characteristic | Resultsa for:
|
||||
|---|---|---|---|---|---|
| HKU7 | Lancefield group C alpha-hemolytic S. dysgalactiae subsp. dysgalactiae | Lancefield group C beta-hemolytic S. dysgalactiae subsp. equisimilis | Lancefield group G beta-hemolytic S. dysgalactiae subsp. equisimilis | Lancefield group L beta-hemolytic S. dysgalactiae subsp. equisimilis | |
| Hemolysis type | Alpha | Alpha | Beta | Beta | Beta |
| Lancefield group | G | C | C | G | Lb |
| Hippurate hydrolysis | − | − | − | − | + |
| Acid from glycogen | − | − | − | − | + |
| Acid from sorbitol | − | + | − | − | − |
| β-d-Galactosidase | − | − | + | + | + |
| Bacitracin resistance | + | − | + | + | − |
−, negative; +, positive.
Cross-reacts with Lancefield group A antiserum.
Acknowledgments
This work is partly supported by the University Development Fund, University Research Grant Council, and the Committee for Research and Conference Grants, The University of Hong Kong.
REFERENCES
- 1.Ayoub, E. M., and H. A. Majeed. 2000. Postreptococcal reactive arthritis. Curr. Opin. Rheumatol. 12:306-310. [DOI] [PubMed] [Google Scholar]
- 2.Breed, R. S., E. G. D. Murray, and A. P. Hitchens. 1948. Manual of determinative bacteriology, 6th ed., p. 318-319. Williams and Wilkins, Baltimore, Md.
- 3.Cheuk, W., P. C. Y. Woo, K. Y. Yuen, P. H. Yu, and J. K. C. Chan. 2001. Intestinal inflammatory pseudotumor with regional lymph node involvement: identification of a new bacterium as the etiologic agent. J. Pathol. 192:289-292. [DOI] [PubMed] [Google Scholar]
- 4.Dierksen, K. P., and J. R. Tagg. 2000. Haemolysin-deficient variants of Streptococcus pyogenes and S. dysgalactiae subsp. equisimilis may be overlooked as aetiological agents of pharyngitis. J. Med. Microbiol. 49:811-816. [DOI] [PubMed] [Google Scholar]
- 5.Frost, W. D., and M. A. Engelbrecht. 1940. The streptococci. Willdorf Book Co., Madison, Wis.
- 6.Humar, D., V. Datta, D. J. Bast, B. Beall, J. C. De Azavedo, and V. Nizet. 2002. Streptolysin S and necrotising infections produced by group G streptococcus. Lancet 359:124-129. [DOI] [PubMed] [Google Scholar]
- 7.Iglesias-Gamarra, A., E. A. Mendez, M. L. Cuellar, J. H. Ponce de Leon, C. Jimenez, C. Canas, J. Restrepo, M. Pena, R. Valle, and L. R. Espinoza. 2001. Poststreptococcal reactive arthritis in adults: long-term follow-up. Am. J. Med. Sci. 321:173-177. [DOI] [PubMed] [Google Scholar]
- 8.Jansen, T. L., M. Janssen, and A. J. de Jong. 1998. Reactive arthritis associated with group C and group G beta-hemolytic streptococci. J. Rheumatol. 25:1126-1130. [PubMed] [Google Scholar]
- 9.Kawamura, Y., X. G. Hou, F. Sultana, H. Miura, and T. Ezaki. 1995. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int. J. Syst. Bacteriol. 45:406-408. [DOI] [PubMed] [Google Scholar]
- 10.Lau, S. K. P., P. C. Y. Woo, B. Y. L. Chan, A. M. Y. Fung, T. L. Que, and K. Y. Yuen. 2002. Haemophilus segnis polymicrobial and monomicrobial bacteremia identified by 16S ribosomal RNA gene sequencing. J. Med. Microbiol. 51:635-640. [DOI] [PubMed] [Google Scholar]
- 11.Lau, S. K. P., P. C. Y. Woo, G. K. S. Woo, and K. Y. Yuen. 2002. Catheter-related Microbacterium bacteremia identified by 16S ribosomal RNA gene sequencing. J. Clin. Microbiol. 40:2681-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau, S. K. P., P. C. Y. Woo, J. L. L. Teng, K. W. Leung, and K. Y. Yuen. 2002. Identification by 16S ribosomal RNA gene sequencing of Arcobacter butzleri bacteraemia in a patient with acute gangrenous appendicitis. Mol. Pathol. 55:182-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, E. K. 2000. Rheumatic disorders associated with streptococcal infections. Baillieres Best Pract. Res. Clin. Rheumatol. 14:559-578. [DOI] [PubMed] [Google Scholar]
- 14.Luk, W. K., S. S. Wong, K. Y. Yuen, P. L. Ho, P. C. Y. Woo, R. A. Lee, and P. Y. Chau. 1998. Inpatient emergencies encountered by an infectious disease consultative service. Clin. Infect. Dis. 26:695-701. [DOI] [PubMed] [Google Scholar]
- 15.Murray, P. R., E. J. Baro, M. A. Pfaller, F. C. Tenover, and. R. H. Yolken (ed.). 1999. Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 16.Podbielski, A., B. Melzer, and R. Lutticken. 1991. Application of the polymerase chain reaction to study the M protein(-like) gene family in beta-hemolytic streptococci. Med. Microbiol. Immunol. 180:213-227. [DOI] [PubMed] [Google Scholar]
- 17.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vandamme, P., B. Pot, E. Falsen, K. Kersters, and L. A. Devriese. 1996. Taxonomic study of lancefield streptococcal groups C, G, and L (Streptococcus dysgalactiae) and proposal of S. dysgalactiae subsp. equisimilis subsp. nov. Int. J. Syst. Bacteriol. 46:774-781. [DOI] [PubMed] [Google Scholar]
- 19.Vieira, V. V., L. M. Teixeira, V. Zahner, H. Momen, R. R. Facklam, A. G. Steigerwalt, D. J. Brenner, and A. C. D. Castro. 1998. Genetic relationships among the different phenotypes of Streptococcus dysgalactiae strains. Int. J. Syst. Bacteriol. 48:1231-1243. [DOI] [PubMed] [Google Scholar]
- 20.Wilson, C. D., G. F. H. Salt, F. A. Skinner, and L. B. Quesnel. (ed.). 1978. Streptococci, p. 143-156. Academic Press, New York, N.Y.
- 21.Woo, P. C. Y., A. M. Y. Fung, S. K. P. Lau, and K. Y. Yuen. 2002. Identification by 16S ribosomal RNA gene sequencing of Lactobacillus salivarius bacteremic cholecystitis. J. Clin. Microbiol. 40:265-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woo, P. C. Y., A. M. Y. Fung, S. K. P. Lau, S. S. Y. Wong, and K. Y. Yuen. 2001. Group G beta-hemolytic streptococcal bacteremia characterized by 16S ribosomal RNA gene sequencing. J. Clin. Microbiol. 39:3147-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woo, P. C. Y., A. M. Y. Fung, S. S. Y. Wong, H. W. Tsoi, and K. Y. Yuen. 2001. Isolation and characterization of a Salmonella enterica serotype Typhi variant and its clinical and public health implications. J. Clin. Microbiol. 39:1190-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woo, P. C. Y., A. M. Y. Fung, S. K. P. Lau, E. Hon, and K. Y. Yuen. 2002. Diagnosis of pelvic actinomycosis by 16S ribosomal RNA gene sequencing and its clinical significance. Diagn. Microbiol. Infect. Dis. 43:113-118. [DOI] [PubMed] [Google Scholar]
- 25.Woo, P. C. Y., A. S. P. Leung, K. W. Leung, and K. Y. Yuen. 2001. Identification of slide-coagulase positive, tube-coagulase negative Staphylococcus aureus by 16S ribosomal RNA gene sequencing. Mol. Pathol. 54:244-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woo, P. C. Y., C. Y. Lo, S. K. Lo, H. Siau, J. S. M. Peiris, S. S. Y. Wong, W. K. Luk, T. M. Chan, W. W. Lim, and K. Y. Yuen. 1997. Distinct genotypic distributions of cytomegalovirus (CMV) envelope glycoprotein in bone marrow and renal transplant recipients with CMV disease. Clin. Diagn. Lab. Immunol. 4:515-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woo, P. C. Y., D. M. W. Tam, K. W. Leung, S. K. P. Lau, J. L. L. Teng, M. K. M. Wong, and K. Y. Yuen. 2002. Streptococcus sinensis sp. nov., a novel Streptococcus species isolated from a patient with infective endocarditis. J. Clin. Microbiol. 40:805-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woo, P. C. Y., E. Y. L. Cheung, K. W. Leung, and K. Y. Yuen. 2001. Identification by 16S ribosomal RNA gene sequencing of an Enterobacteriaceae species with ambiguous biochemical profile from a renal transplant recipient. Diagn. Microbiol. Infect. Dis. 39:85-93. [DOI] [PubMed] [Google Scholar]
- 29.Woo, P. C. Y., H. W. Tsoi, K. W. Leung, P. N. L. Lum, A. S. P. Leung, C. H. Ma, K. M. Kam, and K. Y. Yuen. 2000. Identification of Mycobacterium neoaurum isolated from a neutropenic patient with catheter-related bacteremia by 16S rRNA sequencing. J. Clin. Microbiol. 38:3515-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woo, P. C. Y., J. H. C. Li, W. M. Tang, and K. Y. Yuen. 2001. Acupuncture mycobacteriosis. N. Engl. J. Med. 345:842-843. [DOI] [PubMed] [Google Scholar]
- 31.Woo, P. C. Y., K. T. K. Chong, K. W. Leung, T. L. Que, and K. Y. Yuen. 2001. Identification of Arcobacter cryaerophilus isolated from a traffic accident victim with bacteraemia by 16S ribosomal RNA gene sequencing. Diagn. Microbiol. Infect. Dis. 40:125-127. [DOI] [PubMed] [Google Scholar]
- 32.Woo, P. C. Y., K. W. Leung, H. W. Tsoi, S. S. Y. Wong, J. L. L. Teng, and K. Y. Yuen. 2002. Thermo-tolerant Campylobacter fetus bacteraemia identified by 16S ribosomal RNA gene sequencing: an emerging pathogen in immunocompromised patients. J. Med. Microbiol. 51:740-746. [DOI] [PubMed] [Google Scholar]
- 33.Woo, P. C. Y., K. W. Leung, S. S. Y. Wong, K. T. K. Chong, E. Y. L. Cheung, and K. Y. Yuen. 2002. Relative alcohol-resistant mycobacteria are emerging pathogens in patients receiving acupuncture treatment. J. Clin. Microbiol. 40:1219-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woo, P. C. Y., P. K. L. Leung, K. W. Leung, and K. Y. Yuen. 2000. Identification by 16S ribosomal RNA gene sequencing of an Enterobacteriaceae species from a bone marrow transplant recipient. Mol. Pathol. 53:211-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuen, K. Y., P. C. Y. Woo, J. L. L. Teng, K. W. Leung, M. K. M. Wong, and S. K. P. Lau. 2001. Laribacter hongkongensis gen. nov., sp. nov., a novel gram-negative bacterium isolated from a cirrhotic patient with bacteremia and empyema. J. Clin. Microbiol. 39:4227-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]