Abstract
The efficacy of the Be82 gene product fused with glutathione S-transferase (GST/Be82) in an enzyme-linked immunosorbent assay (ELISA) for the diagnosis of Babesia equi infection was reported previously (H. Hirata et al., J. Clin. Microbiol. 40:1470-1474, 2002). However, the ELISA with the GST/Be82 antigen cross-reacted with Babesia caballi-infected horse sera, despite the high rate of detection of B. equi. These results suggested that GST/Be82 has an antigen in common with B. caballi or antigenicity similar to that of B. caballi. In the present study, we constructed a series of five clones with deletions in the Be82 gene product, each of which was fused with GST, and used them in ELISAs in order to overcome the cross-reactivity seen with B. caballi. One of the deletion clones, a clone with a deletion of the Be82 gene from positions 236 to 381 (Be82/236-381), specifically and sensitively detected B. equi-infected horse sera without cross-reactivity with B. caballi-infected horse sera. Assays with clones from which other gene products were deleted showed decreased sensitivities or remained nonspecific for the detection of B. equi-infected horse sera. These results suggest that the Be82/236-381 gene product is a novel antigen for the diagnosis of B. equi infection in horses.
Equine piroplasmosis is an economically important tick-borne protozoan disease of horses that has been reported worldwide. The disease is caused by blood parasites named Babesia equi and Babesia caballi (9). Babesia parasites destroy host erythrocytes and induce fever, anemia, and icterus in infected horses (6). These parasites are usually detectable in blood smears only during the acute stage of the infection. In contrast, horses that recover from disease continue to be parasite carriers, and these carriers as well as previously exposed animals should be identified serologically (3). Japan is considered free of equine piroplasmosis, but the number of imported horses has increased recently. Therefore, it is urgent that a highly specific and sensitive system for the diagnosis of equine piroplasmosis be developed to avoid the introduction of infected or carrier horses into Japan.
We recently reported on the efficacy of the Be82 gene product with glutathione S-transferase (GST/Be82) in an enzyme-linked immunosorbent assay (ELISA) for the diagnosis of B. equi infection (4). The ELISA with the Be82 gene antigen was shown to be highly specific only for B. equi antibodies when a cutoff value was set at a relatively high level (4). Although the number of serum samples tested was insufficient to evaluate whether the cross-reactivity of the GST/Be82 antigen with B. caballi-infected sera was authentic, there was a possibility that the B. equi Be82 protein might have an antigen in common with B. caballi or have antigenicity similar to that of B. caballi. Therefore, in this study we constructed a deletion series of the Be82 gene products and evaluated their sensitivities and specificities in ELISAs for the diagnosis of B. equi infection in horses.
MATERIALS AND METHODS
Construction of clones with Be82 deletions.
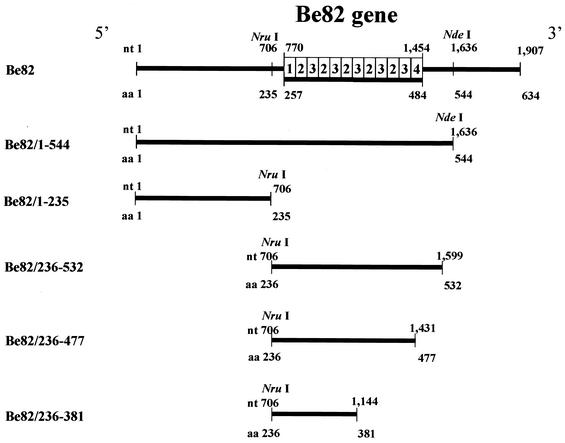
The Be82 gene was previously isolated and subcloned into the pGEX-4T-3 (Amersham Pharmacia Biotech, Little Chalfont, England) or pGEMEX-2 (Promega Corp., Madison, Wis.) vector (4). These plasmids were designated pGEX-4T-3/Be82 and pGEMEX-2/Be82, respectively. pGEX-4T-3/Be82 was digested with NruI and XhoI or NdeI and XhoI and then blunt ended with the Klenow fragment (Takara, Tokyo, Japan), followed by self-ligation with a ligation kit (Takara). The resultant plasmids were designated pGEX/Be82/1-235 and pGEX/Be82/1-544, respectively (Fig. 1). On the other hand, pGEMEX-2/Be82 was subjected to the generation of clones with deletions at the 3′ end by using a deletion kit (Takara), and three deletion clones were obtained. After digestion of each deletion clone with EcoRI and NotI, the deleted inserts were subcloned into the corresponding sites of pGEX-4T-3 in order to obtain pGEX/Be82/1-532, pGEX/Be82/1-477, and pGEX/Be82/1-381. The resultant inserts of these plasmids were checked by restriction enzyme analyses and sequenced with an ABI PRISM 3100 DNA sequencer (Perkin-Elmer, Foster, Calif.) with a big dye primer cycle sequencing ready reaction kit (Perkin-Elmer). Furthermore, plasmids pGEX/Be82/1-532, pGEX/Be82/1-477, and pGEX/Be82/1-381 were digested with BamHI and NruI, blunt ended with the Klenow fragment, and then self-ligated to obtain pGEX/Be82/236-532, pGEX/Be82/236-477, and pGEX/Be82/236-381, respectively (Fig. 1). Nucleic acid and protein homology searches were performed with the MacVector program (Oxford Molecular, Ltd., Oxford, United Kingdom) and the National Center for Biotechnology Information database.
FIG. 1.
Construction of a series of clones with deletions in the Be82 gene. The five plasmids used for the E. coli expression system were constructed with the Deletion Kit (Takara). The resultant plasmids were labeled Be82/1-235, Be82/1-544, Be82/236-532, Be82/236-477, and Be82/236-381. In the schematic representation, nucleotide (nt) and amino acid (aa) positions are indicated by numbers based on the sequence. The column is a highly conserved region at the indicated positions of the Be82 gene, and the number in each box of the column is the group number.
Expression of Be82 genes with deletions in E. coli.
Plasmids pGEX/Be82, pGEX/Be82/1-235, pGEX/Be82/1-544, pGEX/Be82/236-532, pGEX/Be82/236-477, and pGEX/Be82/236-381 and control plasmid pGEX-4T-3 were used to transform competent strain Escherichia coli BL21 (Stratagene, La Jolla, Calif.) by standard techniques (8). After the transformation, these genes were expressed as glutathione S-transferase (GST) fusion proteins (GST/Be82, GST/Be82/1-544, GST/Be82/236-532, GST/Be82/236-477, GST/Be82/236-381, GST/Be82/1-235, and control GST, respectively). The purification of all proteins was performed as described previously (4, 10).
SDS-PAGE analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of the purified proteins was done as described previously (13).
ELISA.
Ninety-six-well microtitration plates (Nunc-Immuno Plate; Nunc, Roskilde, Denmark) were coated overnight at 4°C with 50 μl (0.5 ng/μl) of the purified series of proteins with GST/Be82 deletions or the GST protein as the control. ELISAs with these purified proteins were performed as described previously (4, 5, 12).
Data analysis.
For all tests an optical density at 415 nm (OD415) of 0.2 was defined as the cutoff value. OD415 readings above the cutoff value were considered positive. Sera from B. equi-infected horse with positive ELISA readings were considered true positives, and those with negative readings were considered false negatives, whereas sera from uninfected horses or B. caballi-infected horse with positive readings were considered false positives, and those with negative readings were considered true negatives. The following definitions were used to calculate the corresponding diagnostic parameters: sensitivity, tp × 100/(tp + fn); specificity, tn × 100/(tn + fp); and diagnostic efficiency, (tn + tp) × 100/(tp + fp + tn + fn), where tp is the number of samples with true-positive results, fn is the number of samples with false-negative results, fp is the number of samples with false-positive results, and tn is the number of samples with true-negative results (1, 2).
Serum samples.
Ten serum samples from uninfected horses, 13 serum samples from horses experimentally infected with B. equi, and 9 serum samples from horses experimentally infected with B. caballi were used for the ELISA. All experimental horse sera were collected from 30 days to 2 years after infection. The Equine Research Institute of the Japan Racing Association kindly donated all sera. They were kept at −80°C until use in the ELISA.
RESULTS
Construction and expression of a series of clones with deletions in the B. equi Be82 gene.
The Be82 gene was isolated from a B. equi cDNA library and identified as a truncated open reading frame of 1,907 bp comprising 634 amino acid residues (4). Four kinds of relatively conserved sequences were found to exist on the amino acid sequence of Be82, with tandemly repeated 19-residue periodicities occurring between amino acids 257 and 484 (Fig. 2). These four groups were arranged so that group 1 initiated the arrangement, groups 2 and 3 followed with five times the number of reduplications from amino acids 277 to 466, and group 4 ended the arrangement (Fig. 1). Five deletion clones were obtained by using a deletion kit. The resultant Be82 genes with deletions, Be82 with deletions from positions 1 to 544 (Be82/1-544), Be82/236-532, Be82/236-477, Be82/236-381, and Be82/1-235, consisted of 1,636, 893, 725, 438, and 706 bp, respectively (Fig. 1).
FIG. 2.
Deduced amino acid sequence of the Be82 coding region from residues 276 to 485. This region was classified as an amino acid sequence with four groups of 19 amino acid residues. Highly conserved residues are underlined.
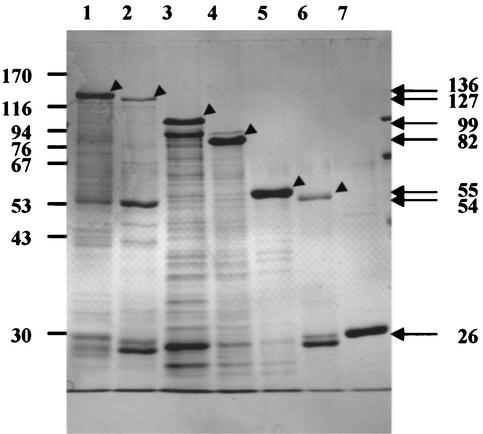
Six kinds of gene products, GST/Be82, GST/Be82/1-544, GST/Be82/236-532, GST/Be82/236-477, GST/Be82/236-381, and GST/Be82/1-235, were purified and observed by SDS-PAGE analysis to have molecular sizes of 136, 127, 99, 82, 55, and 54 kDa, respectively (Fig. 3). All of these gene products contained a GST tag with a molecular size of 26 kDa. The entire GST/Be82 gene product was larger than the estimated size, as determined by computer-aided analysis as described previously (4). The molecular sizes of the gene products from the GST/Be82/1-544, GST/Be82/236-532, GST/Be82/236-477, and GST/Be82/236-381 deletion clones calculated by SDS-PAGE analyses were still larger than those calculated by computer-aided analysis. The discrepancies in molecular sizes decreased when the amino acid of Be82 was deleted from the C terminus. On the other hand, the molecular size of the GST/Be82/1-235 gene product was the same as that estimated by computer-aided analysis.
FIG. 3.
Expression of Be82 gene product, the five Be82 clones with deletions, and the pGEX-4T-3 gene product in E. coli and purification of these recombinant proteins with GST-Sepharose 4B beads. Results are shown for the GST/Be82 (lane 1), GST/Be82/1-544 (lane 2), GST/Be82/236-532 (lane 3), GST/Be82/236-477 (lane 4), GST/Be82/236-381 (lane 5), GST/Be82/1-235 (lane 6), and GST (lane 7) proteins stained with Coomassie blue. The molecular sizes of the markers (in kilodaltons) are shown on the left.
Evaluation of ELISAs with a deletion series of the Be82 gene products.
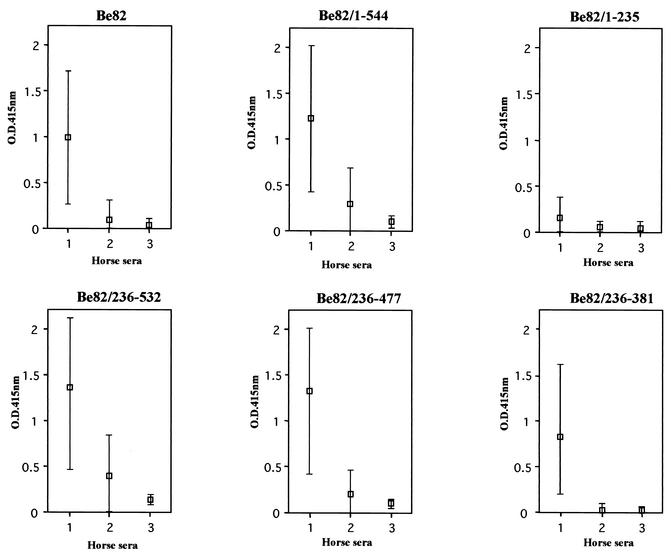
In order to improve the specificity of the GST/Be82 protein, all of the deletion antigens constructed were subjected to ELISA, and their sensitivities and specificities were evaluated. The reactivities of the GST/Be82, GST/Be82/1-544, GST/Be82/236-532, GST/Be82/236-477, GST/Be82/236-381, and GST/Be82/1-235 antigens with each of the serum samples in the ELISAs are shown in Fig. 4 and Table 1. The sensitivity of the ELISA with the GST/Be82/1-235 antigen was decreased in comparison to that of the ELISA with the GST/Be82 antigen, although its specificity was high. The sensitivities of the ELISAs with the GST/Be82/1-544 and GST/Be82/236-531 antigens were increased for all horse sera compared with that of the ELISA with the GST/Be82 antigen; however, the specificities of the ELISAs with those antigens were decreased. The ELISA with the GST/Be82/236-477 antigen had a higher sensitivity and a relatively higher specificity in comparison to those of the ELISA with the GST/Be82 antigen. On the other hand, among the ELISAs with the series of six GST/Be82 antigens, the ELISA with the GST/Be82/236-381 antigen showed the highest sensitivity and specificity. These results indicate that the ELISA with the GST/Be82/236-381 antigen provided a highly specific and sensitive system for the diagnosis of B. equi infection.
FIG. 4.
ELISA results showing the reactivities with equine sera of the Be82 gene product and the gene products of the series of clones with Be82 gene deletions. Minimum and maximum values (bars) and median values (□) from the ELISA with the Be82 gene product and the gene products of the series of clones with Be82 gene deletions are shown. The bars represent the OD415 of B. equi-infected horse sera (column 1), B. caballi-infected horse sera (column 2), and uninfected sera (column 3).
TABLE 1.
Diagnostic performance of the deletion series of Be82 products
| ELISA antigena | Sensitivity (%) | Specificity (%) | Diagnostic efficiency (%) |
|---|---|---|---|
| Be82 | 92 | 79 | 84 |
| Be82/1-544 | 100 | 53 | 72 |
| Be82/1-235 | 31 | 100 | 72 |
| Be82/236-532 | 100 | 37 | 63 |
| Be82/236-477 | 100 | 84 | 91 |
| Be82/236-381 | 100 | 100 | 100 |
The concentrations of ELISA antigens were adjusted to 0.5 ng/μl, respectively.
DISCUSSION
We previously isolated a Be82 cDNA clone of 1,953 bp by immunoscreening B. equi-infected horse serum (4). The Be82 gene has a 1,907-bp truncated open reading frame lacking the 5′-terminal sequence. Furthermore, an ELISA with a GST/Be82 antigen proved to be highly sensitive for B. equi antibodies, but it suggested that the Be82 gene shares a region that causes a cross-reaction with B. caballi-infected horse sera (4). Therefore, it was necessary to determine the region in the GST/Be82 antigen specific for B. equi to avoid nonspecific reactions as much as possible. In the present study, we constructed a series of five clones with deletions in the Be82 gene product, each of which was fused with GST, and used it in ELISAs in order to overcome the cross-reaction against B. caballi. We developed a novel ELISA with the GST/Be82/236-381 antigen, and it proved to have a higher specificity and a higher sensitivity for B. equi antibodies than those of the ELISA with the complete GST/Be82 antigen.
The molecular size of GST/Be82 was found to be about 40 kDa larger than the theoretically calculated size (4). These discrepancies in molecular size were gradually reduced when the C terminus of the amino acid sequence of Be82 was deleted. Furthermore, the discrepancies in the molecular sizes of the GST/Be82/1-235 protein disappeared. These results indicated that the discrepancies in molecular sizes were caused by the presence of the highly conserved and charged region between amino acids 257 and 484 (Fig. 1). Similar findings have been reported for the pf322 gene of Plasmodium falciparum and the 200-kDa protein of Babesia bigemina (7, 11).
We observed that purified fractions of all recombinant antigens had contaminants (Fig. 3). In the ELISAs, however, none of the antigens had nonspecific reactivity with normal horse sera. Furthermore, among the three antigens GST/Be82/1-544, GST/Be82/1-235, and GST/Be82/236-532, GST/Be82/1-544 and GST/Be82/236-532 showed cross-reactivities with B. caballi-infected horse sera similar to that of the Be82 antigen and GST/Be82/1-235 showed the lowest level of specific reactivity to all three groups of horse sera evaluated in the present study. These results suggest that the cross-reactivity of the ELISA with B. caballi-infected horse sera is mainly caused by the region from amino acids 236 to 532 of Be82. This possibility was further confirmed by the fact that GST/Be82/236-381 showed the highest level of specific reactivity to B. equi-infected horse sera in the ELISAs with GST/Be82/236-477 and GST/Be82/236-381, which were made by deletion of amino acids from the C terminus of GST/Be82/236-532. Therefore, we concluded that the region between amino acids 236 and 381 has a highly immunodominant region with antigenicity for B. equi and that another region between amino acids 382 and 532 causes a cross-reaction with B. caballi-infected horse sera. Further analysis such as whether the conserved region between amino acids 382 and 532 of Be82 may be highly charged and cause changes in the three-dimensional structure will be required to determine the reason for the cross-reactivity of this region.
In conclusion, we have provided convincing data demonstrating the specificity of the GST/Be82/236-381 protein in the detection of B. equi infection. This highly specific recombinant protein shows promise for use in the diagnosis of B. equi infection by ELISA.
Acknowledgments
We thank T. Kanemaru of the Equine Research Institute of the Japan Racing Association for providing horse sera.
This study was supported by a Grant-in-Aid for Scientific Research (A) from the Japan Society for the Promotion of Science and by a grant of Gakujutsu-Frontier Cooperative Research in Rakuno-Gakuen University.
REFERENCES
- 1.Barbieri, M., V. Fernandez, G. Gonzalez, V. Martinez Luaces, and A. Nieto. 1998. Diagnostic evaluation of a synthetic peptide derived from a novel antigen B subunit as related to other available peptides and native antigens used for serology of cystic hydatidosis. Parasite Immunol. 20:51-61. [DOI] [PubMed] [Google Scholar]
- 2.Gonzalez-Sapienza, G., C. Lorenzo, and A. Nieto. 2000. Improved immunodiagnosis of cystic hydatid disease by using a synthetic peptide with higher diagnostic value than that of its parent protein, Echinococcus granulosus antigen B. J. Clin. Microbiol. 38:3979-3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halbrook, A. A. 1969. Biology of equine piroplasmosis. Am. J. Vet. Med. Assoc. 155:453-454. [PubMed] [Google Scholar]
- 4.Hirata, H., H. Ikadai, N. Yokoyama, X. Xuan, K. Fujisaki, N. Suzuki, and I. Igarashi. 2002. Cloning of a truncated Babesia equi gene encoding an 82-kilodalton protein and its potential use in an enzyme-linked immunosorbent assay. J. Clin. Microbiol. 40:1470-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ikadai, H., X. Xuan, I. Igarashi, T. Kanemaru, H. Nagasawa, K. Fujisaki, N. Suzuki, and T. Mikami. 1999. Cloning and expression of a 48-kilodalton Babesia caballi merozoite rhoptry protein and potential use of the recombinant antigen in an enzyme-linked immunosorbent assay. J. Clin. Microbiol. 37:3475-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knowles, D. P. 1996. Control of Babesia equi parasitemia. Parasitol. Today 12:195-198. [DOI] [PubMed] [Google Scholar]
- 7.Mattei, D., and A. Scherf. 1992. The Pf 332 gene of Plasmodium falciparum codes for a giant protein that is translocated from the parasite to the membrane of infected erythrocytes. Gene 110:71-79. [DOI] [PubMed] [Google Scholar]
- 8.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 9.Schein, E. 1988. Equine babesiosis, p. 197-208. In M. Ristic (ed.), Babesiosis of domestic animals and man. CRC Press, Inc., Boca Raton, Fla.
- 10.Smith, D. B., and K. S. Johonson. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67:31-40. [DOI] [PubMed] [Google Scholar]
- 11.Tebele, N., A. R. Skilton, J. Katende, W. C. Wells, V. Nene, T. Mcelwain, P. S. Morzaria, and J. A. Musoke. 2000. Cloning, characterization, and expression of a 200-kilodalton diagnostic antigen of Babesia bigemina. J. Clin. Microbiol. 38:2240-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xuan, X., A. Larsen, H. Ikadai, T. Tanaka, I. Igarashi, H. Nagasawa, K. Fujisaki, Y. Toyoda, N. Suzuki, and T. Mikami. 2001. Expression of Babesia equi merozoite antigen 1 in insect cells by recombinant baculovirus and evaluation of its diagnostic potential in an enzyme-linked immunosorbent assay. J. Clin. Microbiol. 39:705-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xuan, X., K. Maeda, T. Mikami, and H. Otsuka. 1996. Characterization of canine herpesvirus glycoprotein C expressed in insect cells. Virus Res. 46:57-64. [DOI] [PubMed] [Google Scholar]