Abstract
Direct heteroduplex analysis and a universal heteroduplex generator assay were performed to detect rifampin resistance rapidly in Turkish Mycobacterium tuberculosis isolates. Cross-resistance to rifapentine, rifabutin, and rifalazil was investigated. A relationship between specific mutations and resistance patterns, which can guide the choice of an appropriate therapeutic regimen for tuberculosis patients, was identified.
Rifampin is a key component of short-course multidrug antituberculosis therapy. It binds to the β subunit of the DNA-dependent RNA polymerase encoded by the gene rpoB and therefore inhibits transcription (7, 8). Ninety-six percent of rifampin-resistant isolates have missense mutations, deletions, or insertions in the 81-bp rifampin resistance-determining region of the rpoB gene coding for amino acids 507 through 533. Codons 531, 526, and 516 are reported as the most frequent mutation sites, with codon 531 mutations being the most common (7).
The emergence of rifampin-resistant strains has led to the use of structural analogs of rifampin. Rifapentine, rifabutin, and rifalazil are new rifamycin derivatives and are tested in rifampin-resistant isolates. Recently, several studies showed a correlation between specific mutations in rpoB and the level of resistance to rifamycin derivatives (6, 11, 14).
In this study we amplified the resistance-determining region of the rpoB gene by PCR and performed direct heteroduplex analysis (1, 15) and a universal heteroduplex generator (UHG) assay (13) for rapid detection of rifampin resistance in clinical isolates of Mycobacterium tuberculosis. We performed DNA sequencing for certain isolates and determined the rifamycin cross-resistance in rifampin-resistant isolates and showed the correlation between antimycobacterial activities of rifamycins and mutations in the rpoB gene.
Ninety-seven rifampin-resistant and 21 rifampin-susceptible isolates of M. tuberculosis were included in this study. All isolates were isolated from different patients in the microbiology laboratory of Atatürk Chest Diseases and Chest Surgery Center, Ankara, Turkey. The rifampin-susceptible strain ATCC 25177 H37Ra was used as a negative control.
Susceptibility testing was performed by the proportion method (2, 3, 4) with Middlebrook 7H10 agar. Each drug (rifampin, rifapentine, rifabutin, and rifalazil) was added at a concentration of 1 μg/ml. Isolates with growth on drug-containing media that was >1% greater than growth on control media were considered resistant.
DNA extracts from clinical isolates and from the rifampin-susceptible control strain H37Ra of M. tuberculosis, already grown on Löwenstein-Jensen media, were prepared by the boiling method as described previously (5).
A 305-bp region of the rpoB gene covering the 81-bp rifampin resistance-determining region was amplified by using TbRif-1 and TbRif-2 primers, and DNA duplexes were obtained as previously described by Willams et al. (9). They were loaded on a 20-cm-long 1× mutation detection enhancement (FMC Bioproducts) gel containing 15% urea. The electrophoresis was run at 300 V for 24 h with 1× Tris-boric acid-EDTA buffer.
The UHG was a gift from Diana Williams, Hansen's Disease Research Laboratory, Louisiana State University. The heteroduplex generator is a synthetic (Genelab) PCR-amplified (double-stranded) 181-bp DNA fragment. It mimics the genomic DNA and covers the 81-bp rifampin resistance-determining region. It has four 3-bp deletions and three 2-bp substitutions (10). We performed the UHG assay by using primers rpo105 and rpo273 and the protocol described by Williams et al. (10, 12).
We performed DNA sequencing for 14 strains showing different drug susceptibility and heteroduplex patterns. The PCR products were separated from unincorporated nucleic acids and primers with PCR Preps DNA purification system (Promega) according to the manufacturer's instructions. DNA-sequencing reactions were performed with a DNA sequencing kit (Silver Sequence DNA sequencing system; Promega). Primers TbRif-1 and TbRif-2 and a new primer, TbRif-0 (5′ AAC CGA CGA CAT CGA CCA CT 3′), designed to show frequent mutation sites more clearly, were used for PCR and DNA-sequencing reactions.
Fifty-one of 97 (52.6%) rifampin-resistant isolates were resistant to all four rifamycin derivatives tested. Thirty-five (36%) isolates were resistant to rifampin and rifapentine, and nine (9.3%) isolates were resistant to rifampin only (Table 1). All rifampin-susceptible strains were susceptible to other rifamycin derivatives.
TABLE 1.
Comparison of three methods for determining the drug resistance of M. tuberculosis isolates
| Drug(s) | No. of rifampin-resistant isolates found to be resistant to indicated drug(s) by:
|
||
|---|---|---|---|
| Proportion method | Direct heteroduplex analysis | UHG analysis | |
| Rifampin + rifapentine + rifabutin + rifalazil | 51 | 50 | 48 |
| Rifampin + rifapentine + rifabutin | 1 | 1 | 1 |
| Rifampin + rifapentine + rifalazil | 1 | 0 | 1 |
| Rifampin + rifapentine | 35 | 30 | 31 |
| Rifampin | 9 | 9 | 7 |
| Total | 97 | 90 | 88 |
Among 97 rifampin-resistant isolates, 90 were classified as resistant to rifampin by direct heteroduplex analysis. All 21 rifampin-susceptible isolates were classified as susceptible to rifampin by direct heteroduplex analysis. The sensitivity and specificity of direct heteroduplex analysis, compared to the sensitivity and specificity of conventional drug susceptibility testing, were 92.7 and 100%, respectively. The proportion of agreement was 94%.
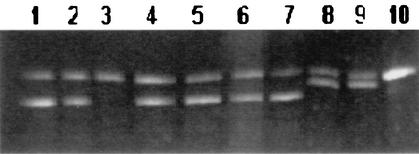
M. tuberculosis H37Ra and rifampin-susceptible isolates were expected to form a single band belonging to homoduplex DNA, and resistant isolates were expected to form extra bands due to heteroduplex DNA with mismatches moving at different speeds than the homoduplex DNA in electrophoresis (Fig. 1).
FIG. 1.
Electrophoretic patterns obtained by direct heteroduplex analysis of rpoB from M. tuberculosis isolates. Lanes 1, 2, and 4 to 9, rifampin-resistant isolates; lane 3, rifampin-susceptible isolate; lane 10, rifampin-susceptible control strain H37Ra.
The UHG assay detected 88 of 97 rifampin-resistant strains. All 21 rifampin-susceptible isolates produced the electrophoretic patterns expected from susceptible strains with the UHG assay. The sensitivity and specificity of this method, compared to those of the proportion plate method, were 90.7 and 100%, respectively. The proportion of agreement was 92.4%.
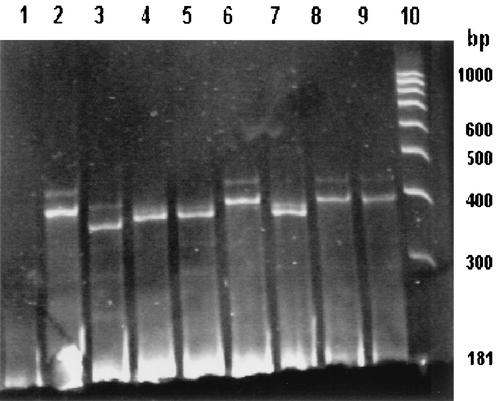
Figure 2 shows rifampin-resistant and -susceptible isolates. M. tuberculosis H37Ra forms a four-band pattern containing homoduplexes at 181 and 193 bp and heteroduplexes running in approximately the range of electrophoresis corresponding to homoduplex double-stranded DNA with sizes of 400 and 500 bp. The isolates showing the same band pattern as that of M. tuberculosis H37Ra were considered susceptible, and isolates showing a band pattern different from that of M. tuberculosis H37Ra were considered rifampin resistant. A single band at 181 bp indicates that there is no M. tuberculosis in the sample.
FIG. 2.
Electrophoretic patterns obtained by UHG assay from rpoB genes. Lane 1, UHG; lane 2, M. tuberculosis H37Ra; lanes 3 to 5 and 7 to 9, rifampin-resistant isolates; lane 6, rifampin-susceptible isolate; lane 10, molecular weight standard (100-bp ladder).
A comparison of the two heteroduplex analysis methods is shown in Table 2. The difference in detection of rifampin-resistant isolates by the two methods wasn't statistically significant (P = 0.791).
TABLE 2.
Comparison of two methods of heteroduplex analysis
| Direct heteroduplex analysis | No. of isolates with UHG analysis result that was:
|
||
|---|---|---|---|
| Resistant | Susceptible | Total | |
| Resistant | 82 | 8 | 90 |
| Susceptible | 6 | 22 | 28 |
| Total | 88 | 30 | 118 |
Among 14 resistant isolates, seven different mutations were identified (Table 3). All were single nucleotide mutations involving codons 531, 526, 516, and 513, which constitute the sites where mutations are most frequently encountered. While the isolates with mutations in codons 531 and 513 were resistant to all four rifamycin derivatives; the isolates with mutations in codon 516 were resistant to rifampin and rifapentine but susceptible to rifabutin and rifalazil. The isolates with mutations in codon 526 were resistant to either two or four rifamycin derivatives depending on the amino acid change. The isolates with glutamic acid and leucine substitutions were resistant to rifampin and rifapentine but susceptible to rifabutin and rifalazil. Tyrosine substitutions created resistance to all four rifamycin derivatives.
TABLE 3.
Relationship between mutations in rpoB and resistance to rifamycins
| Mutation | Rifamycin derivatives to which resistance is gained |
|---|---|
| 513 Glycine→leucine | Rifampin plus rifapentine plus rifabutin plus rifalazil |
| 516 Aspartic acid→valine | Rifampin plus rifapentine |
| 516 Aspartic acid→tyrosine | Rifampin plus rifapentine |
| 526 Histidine→glutamic acid | Rifampin plus rifapentine |
| 526 Histidine→leucine | Rifampin plus rifapentine |
| 526 Histidine→tyrosine | Rifampin plus rifapentine plus rifabutin plus rifalazil |
| 531 Serine→leucine | Rifampin plus rifapentine plus rifabutin plus rifalazil |
In this study, 46% of rifampin-resistant isolates were susceptible to rifabutin, which is available on the market and may be an important alternative to rifampin.
In conclusion, both heteroduplex methods can be used for rapid diagnosis of rifampin resistance, because their sensitivity and specificity are high. Detection of specific mutations in the rpoB gene by DNA sequencing may be very useful for deter-mining the rifamycin resistance phenotypes, offering patients infected with rifampin-resistant isolates the option to be treated with another rifamycin derivative.
Acknowledgments
We are grateful to Diana Williams, Molecular Biology Research Department, Laboratory Research Branch, National Hansen's Disease Programs at the School of Veterinary Medicine, Louisiana State University, Baton Rouge, for providing the UHG.
We thank Atatürk Chest Diseases and Chest Surgery Center, Ankara, Turkey, for providing clinical isolates of M. tuberculosis.
REFERENCES
- 1.Cotton, R. G. H. 1993. Current methods of mutation detection. Mutat. Res. 285:125-144. [DOI] [PubMed] [Google Scholar]
- 2.Heifets, L., T. Sanchez, J. Vanderkolk, and V. Pham. 1999. Development of rifapentine susceptibility tests for Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 43:25-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heifets, L. B. 1991. Drug susceptibility tests in the management of chemotherapy of tuberculosis, p. 89-120. In L. B. Heifets (ed.), Drug susceptibility in the chemotherapy of mycobacterial infections. CRC Press, Boca Raton, Fla.
- 4.Inderlied, C. B. 1991. Antimycobacterial agents: In vitro susceptibility testing, spectrums of activity, mechanisms of action and resistance, and assays for activity in biological fluids, p. 134-197. In V. Lorian (ed.), Antibiotics in laboratory medicine, 3rd ed. Williams & Wilkins, Baltimore, Md.
- 5.Kocagöz, T., E. Yılmaz, Ş. Özkara, S. Kocagöz, M. Hayran, M. Sachedeva, and H. F. Chambers. 1993. Detection of Mycobacterium tuberculosis in sputum samples by polymerase chain reaction using a simplified procedure. J. Clin. Microbiol. 31:1435-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moghazeh, S. L., X. Pan, T. Arain, C. D. Stover, J. M. Musser, and B. N. Kreiswirth. 1996. Comparative antimycobacterial activities of rifampin, rifapentine, and KRM-1648 against a collection of rifampin-resistant Mycobacterium tuberculosis isolates with known rpoB mutations. Antimicrob. Agents Chemother. 40:2655-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramaswamy, S., and J. M. Musser. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 79:3-29. [DOI] [PubMed] [Google Scholar]
- 8.Telenti, A., P. Imboden, F. Marchesi, D. Lowrie, S. Cole, M. J. Colston, L. Matter, K. Schopfer, and T. Bodmer. 1993. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet 341:647-650. [DOI] [PubMed] [Google Scholar]
- 9.Williams, D. L., C. Waguespack, K. Eisenach, J. T. Crawford, F. Portaels, M. Salfinger, C. M. Nolan, C. Abe, V. Sticht-Groh, and T. P. Gillis. 1994. Characterization of rifampin resistance in pathogenic mycobacteria. Antimicrob. Agents Chemother. 38:2380-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams, D. L., C. W. Limbers, L. Spring, S. Jayachandra, and T. P. Gillis. 1996. PCR-heteroduplex detection of rifampin-resistant Mycobacterium tuberculosis, p. 122-129. In D. H. Persing (ed.), PCR protocols for emerging infectious diseases. A supplement to Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, D.C.
- 11.Williams, D. L., L. Spring, L. Collins, L. P. Miller, L. B. Heifets, P. R. J. Gangadharam, and T. P. Gillis. 1998. Contribution of rpoB mutations to development of rifamycin cross-resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 42:1853-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams, D. L., L. Spring, T. P. Gillis, M. Salfinger, and D. H. Persing. 1998. Evaluation of a polymerase chain reaction-based universal heteroduplex generator assay for direct detection of rifampin susceptibility of Mycobacterium tuberculosis from sputum specimens. Clin. Infect. Dis. 26:446-450. [DOI] [PubMed] [Google Scholar]
- 13.Wood, N., and J. Bidwell. 1996. Genetic screening and testing by induced heteroduplex formation. Electrophoresis 17:247-254. [DOI] [PubMed] [Google Scholar]
- 14.Yang, B., H. Koga, H. Ohno, K. Ogawa, M. Fukuda, Y. Hirata, S. Maesaki, K. Tomono, T. Tashiro, and S. Kohno. 1998. Relationship between antimycobacterial activities of rifampicin, rifabutin and KRM-1648 and rpoB mutations of Mycobacterium tuberculosis. J. Antimicrob. Chemother. 42:621-628. [DOI] [PubMed] [Google Scholar]
- 15.Yap, E. P. H., and J. O. McGee. 1994. Detection of mutations by PCR, p. 107-120. In H. G. Griffin and A. M. Griffin (ed.), PCR technology. Current innovations. CRC Press, Boca Raton, Fla.