Abstract
The receptor for the pituitary glycoprotein hormone FSH (FSHR) and the nuclear hormone receptor steroidogenic factor 1 (SF-1) play important roles in control of the hypothalamic-pituitary-gonadal axis. FSHR is essential for integrating the pituitary FSH signal to gonadal response, while SF-1 is an important transcriptional regulator of many genes that function within this axis and is essential for the development of gonads and adrenal glands. Given the critical role of SF-1 in regulation of the gonads and the coexpression of FSHR and SF-1 in Sertoli and granulosa cells, we examined the ability of SF-1 to regulate transcription of the FSHR gene. We found that SF-1 stimulated rat FSHR promoter activity in a dose-dependent and promoter-specific manner. Examination of various promoter deletion mutants indicated that SF-1 acts through the proximal promoter region and upstream promoter sequences. An E box element within the proximal promoter is essential for activation of the FSHR promoter by SF-1. This element binds the transcriptional regulators USF1 and USF2 (upstream stimulatory factors 1 and 2) but not SF-1, as shown by electrophoretic mobility shift assays. In addition, functional studies identified a requirement for the USF proteins in SF-1 activation of FSHR and mapped an important regulatory domain within exons 4 and 5 of USF2. Co-transfection studies revealed that activation of protein kinase A leads to inhibition of SF-1-stimulated transcription of FSHR, while it synergized with SF-1 to activate the equine LH β-promoter (eβ). Thus, stimulation of the cAMP pathway differentially regulates SF-1 activation of the FSHR and eβ-promoters.
INTRODUCTION
FSH is an important pituitary glycoprotein that serves to regulate gonadal function (1). This hormone elicits its effects by binding a cell surface receptor found only in the gonads, where hormone binding results in a number of cellular changes that assist germ cell development (2–13). The cDNA and gene for the FSH receptor (FSHR) have been cloned and characterized from several different species (14–19). FSHR expression studies have revealed that transcription of this gene occurs only within Sertoli cells of the testis and granulosa cells of the ovary (3, 20–22). Thus, elucidation of the transcriptional mechanisms that regulate the FSHR gene will provide insight into both cell-specific transcriptional events and mechanisms that control the response of the gonads to FSH.
Transient transfection analysis of various deletion and block replacement mutants of the FSHR 5′-flanking region showed that basal transcriptional activity is controlled predominantly by elements located within the first 100-bp of the promoter (23, 24). Within this region, a single E box element, located approximately 30 bp upstream of the transcriptional start site, is an important control element for FSHR transcription (23–25). Additional studies have shown that the basic helix-loop-helix (bHLH)-ZIP proteins USF1 and USF2 (upstream stimulatory factors 1 and 2) bind and activate FSHR transcription through this proximal E box (23, 24, 26). Also within the promoter region, elements 3′ to the transcriptional start site have been identified as important for full promoter function (23, 27). Although these studies have contributed significant insight into FSHR gene regulation, it is important to recognize that our current understanding of FSHR transcription is primarily limited to the regulatory events represented by the conditions of experimental cell culture systems. Thus, functionally important transcription factors not present or active within the cultured cells remain unrecognized. Directly testing the regulatory effects of relevant transcription factors on FSHR promoter function therefore provides an important complementary means to identify proteins that regulate FSHR gene activity.
The transcription factor steroidogenic factor 1 (SF-1), also known as adrenal 4-binding protein (Ad4-bp), is a key regulator of endocrine function and sex determination (reviewed in Ref. 28). Its expression is limited primarily to cells of the gonads, adrenal, pituitary, and ventral medial hypothalamus, where it is thought to contribute to cell-specific properties of genes expressed within these tissues (29–34). SF-1 is a member of the nuclear hormone receptor family that was originally recognized for its role in endocrine regulation, as it bound to an important regulatory element (AGGTCA) common to promoters of several key steroidogenic enzymes (35–40). However, SF-1 not only regulates genes encoding steroidogenic enzymes but a number of genes, including the gonadotropin α- and β-subunits, the GnRH receptor, and the inhibin α-subunit, that are important for production of the gonadotropin hormones FSH and LH (39, 41–49). In addition, gene ablation studies in mice revealed that SF-1 is essential for adrenal and gonadal development and for proper function of the hypothalamic-pituitary-gonadal axis (50–53).
During development, SF-1 expression is first observed in the urogenital ridge of the embryo and is later found in discrete populations of cells that give rise to adrenocortical and gonadal cells (29). Shortly after induction of the testis, SF-1 expression is observed in the interstitial region and within the seminiferous cords, indicating that both Leydig (interstitial) and Sertoli (cords) cells express this protein (29, 30). In the developing ovary, SF-1 expression is significantly lower than what is observed in the testis but appears to increase just before the time of birth (29, 31). SF-1 expression continues after birth in the adrenal gland, testis, and ovary, where it has been detected in adrenocortical cells, testicular Leydig and Sertoli cells, and ovarian theca and granulosa cells (29, 32, 54).
In male rats, FSHR expression is first detected just after the testis begins to form in the embryo [embryonic day 14.5 (e14.5)], while in females, ovarian FSHR expression is delayed until just before birth (e20.5) (2). Although FSHR expression is not observed in the early SF-1-expressing cells of the urogenital ridge, its co-expression with SF-1 in Sertoli and granulosa cells as gonadogenesis proceeds suggests that SF-1 may influence FSHR expression. Furthermore, SF-1’s known role in the regulation of the hypothalamic-pituitary-gonadal axis suggests that its functions may extend beyond hormone production to that of hormone response through regulation of receptor levels. Studies presented herein demonstrate that SF-1 regulates transcription of the FSHR gene via a mechanism that involves regions of the FSHR 5′-flanking region and requires USF1 and USF2 binding to the proximal E box element.
RESULTS
SF-1 Regulates Expression of the Rat FSHR Promoter
To determine whether SF-1 regulates FSHR, the rat FSHR promoter (−2,700/+123) driving expression of a luciferase reporter gene was cotransfected into the placental cell line JEG3 together with an expression vector for SF-1 (RSV-SF1). JEG3 cells were chosen to examine SF-1 regulation of FSHR, as these cells have been shown to lack SF-1 and to activate known SF-1 target response elements via transient transfection (55, 56). Cotransfection with increasing amounts of the SF-1 expression vector showed a dose-dependent increase in FSHR promoter activity, as measured by the production of luciferase (Fig. 1A). RSV-CAT (Rous sarcoma virus-chloramphenicol acetyl transferase) was used as a control to equalize the amount of co-transfected DNA in each sample. In addition, the effects of SF-1 were promoter specific, as luciferase production from either the pGL3-Control (SV40 promoter) or pGL3-Basic (promoterless) vectors was only modestly effected by cotransfection with SF-1 expression vector (Fig. 1B). The stimulatory effect of SF-1 on rat FSHR promoter suggests that this transcription factor is important for proper regulation of the FSHR gene.
Fig. 1. SF-1 Regulates Transcription of the Rat FSHR Promoter.
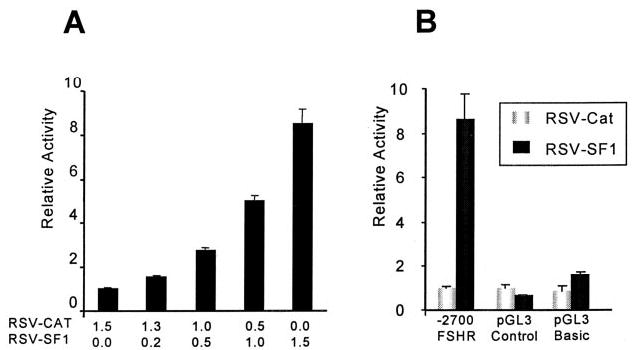
A, One microgram of FSHR(−2,700/+123)Luc vector was transiently transfected into JEG3 cells together with 200 ng RSV-βgalactosidase (a control for transfection efficiency) and increasing amounts of RSV-SF1. The total amount of cotransfected expression vector was kept constant by adjusting to 1.5 μg with RSV-CAT expression vector, which was used as a negative control. B, FSHR(−2,700/+123)Luc, pGL3-Control, or pGL3-Basic (0.5 μg) was cotransfected into JEG3 cells with either 1.5 μg RSV-CAT or RSV-SF1. Each sample also received 200 ng RSV-β-galactosidase. The relative activity represents the luciferase/β-galactosidase activity of each sample normalized to the luciferase/β-galactosidase activity of the promoter transfected with RSV-CAT alone. Error bars represent the sem.
Multiple Regions of the FSHR Promoter Are Important for Regulation by SF-1
To help delineate regions of the promoter important for response to SF-1, various deletion mutants of the FSHR promoter were tested for response to cotransfection with the SF-1 expression vector (RSV-SF1). Deletion of the −2,700-bp promoter to either −1,300 bp or −743 bp resulted in a sequential decrease in stimulation by SF-1 (Fig. 2). Deletion to −743 reduced stimulation to approximately 3-fold while further deletion to −220 had no further impact. Furthermore, deletion of the 3′-region of the promoter from +123 to +79, a region that contains an SF-1-like element (5′-TGgCCTTG-3′), had no further impact on SF-1 induction (Fig. 2; compare −220/+123 and −220/+79). Thus, several regions within the FSHR promoter (−2,700 to −1,300, −1,300 to −743, and −220 to +79) are needed for full response to SF-1, suggesting that multiple elements are required for full response to SF-1.
Fig. 2. SF-1 Stimulation Maps to Several Regions of the FSHR Promoter.
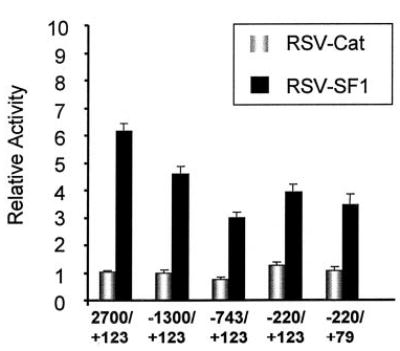
One microgram of the indicated FSHR promoter deletion mutant was cotransfected into JEG3 cells with either 0.5 μg RSV-CAT or RSV-SF1. Each sample also received 200 ng RSV-β-galactosidase. The relative activity represents the luciferase/β-galactosidase activity of each sample normalized to the luciferase/β-galactosidase activity of FSHR(−2,700/+123)Luc transfected with RSV-CAT. Error bars represent the sem.
The E Box Is Required for Stimulation by SF-1
Although SF-1 stimulation required regions of the promoter upstream of −743, an approximate 3-fold induction of the −220/+79 promoter suggested that a SF-1- responsive element(s) reside within this region. Additional studies further refined this region to between −31 bp and +79, as a promoter truncated to −31 bp still responded to SF-1 stimulation (data not shown). Using various block replacement mutants in the context of the −220/+123 FSHR promoter, we evaluated regions within the proximal promoter (−31 to +79) that are necessary for response to SF-1. Although several mutants had somewhat diminished responses, mutation μ9.2 had the most severe effect on SF-1 transactivation (Fig. 3A). Interestingly, the μ9.2 mutation also eliminated SF-1 response when placed in the context of the −2,700/+123 promoter that contains the upstream SF-1-responsive regions (Fig. 3B).
Fig. 3. An E box in the Proximal Promoter Is Required for SF-1 Regulation of the Promoter.
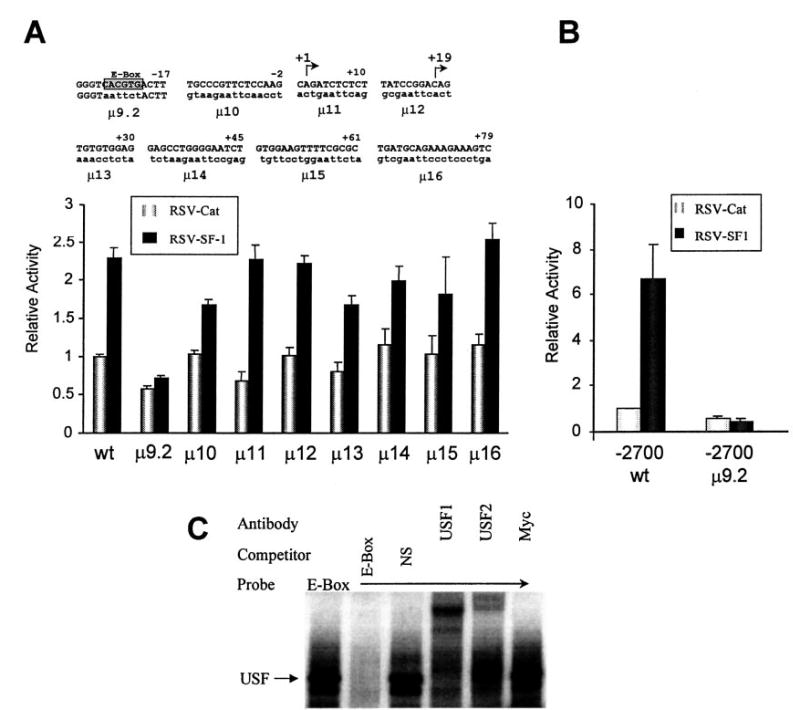
A (top), Sequence of the FSHR-proximal promoter region (top strand) and various mutations (bottom strand) examined for SF-1 responsiveness. One microgram of FSHR(−220/+123) Luc wild-type (wt) promoter or various mutants in the same context (−220/+123) were cotransfected into JEG3 cells with 200 ng RSV-β-galactosidase and 0.5 μg of either RSV-CAT or RSV-SF1. The relative activity represents the luciferase/β-galactosidase activity of each sample normalized to the luciferase/β-galactosidase activity of FSHR(−220/+123)Luc transfected with RSV-CAT. B, 0.5 μg of either FSHR(−2,700/+123)Luc or FSHR(−2,700/+123)μ 9.2Luc, which contain a mutation through the proximal E box, was cotransfected into JEG3 cells with 1.5 μg of either RSV-CAT or RSV-SF1. The relative activity represents the luciferase/β-galactosidase activity of each sample normalized to the luciferase/β-galactosidase activity of FSHR(−2,700/+123)Luc transfected with RSV-CAT. Error bars represent the sem. C, EMSA. A radiolabeled probe corresponding to the FSHR E box (5′-TCTTGGTGGGTCACGTGACTTTGCCCGT-3′) was used in EMSA with nuclear extracts from JEG3 cells. Radiolabeled probe (25 fmol) was incubated with approximately 8 μg of nuclear extract and resolved on a 4% polyacrylamide gel as described in Materials and Methods. Where indicated, unlabeled homologous (WT) or nonspecific (NS; 5′-CTAGAGTCGACCTGCAGGCATGCAAGCTTGGCATTC-3′) DNAs were added to the reaction at a concentration equal to 100× that of the probe. Where shown, antibodies specific for the bHLH proteins USF1, USF2, or c-Myc were added to the reactions before the addition of the probe. The major specific USF complexes are marked by an arrow.
The μ9.2 mutation disrupts an E box element that has been shown to be important for FSHR promoter function (23–25). As has been observed in other cell types, the E box bound the transcription factors USF1 and USF2 in JEG3 cells (Fig. 3C). Although SF-1 has not previously been shown to bind this element, careful inspection of this region revealed the presence of an SF-1-like element (gGGTCA) that includes part of the E box sequence (underlined, 5′-GGGTCACGT-GACTT-3′). To determine whether SF-1 regulates the FSHR promoter by directly binding the region containing the proximal E box, DNA/protein binding studies were performed. A known SF-1 binding site from the human α-subunit promoter (hαGSE) (46) was used as a probe to determine whether the FSHR E box could compete for SF-1 binding. As a source of SF-1 protein, we used nuclei isolated from the pituitary gonadotrope cell line αT3 (57). An electrophoretic mobility shift assay (EMSA) identified several specific complexes bound to the hαGSE (Fig. 4A). One of these complexes cross-reacted with an antibody generated to SF-1, indicating that SF-1 was present in the bound proteins (Fig. 4A). In addition, the SF-1 band was competed with homologous sequence [gonadotrope-specific element (GSE)] but not with as much as a 400 molar excess of the E box, indicating that the E box does not bind SF-1. Interestingly, several of the slower migrating complexes did compete with the E box. The inability of the FSHR E box to bind SF-1 was also indicated by studies in which the E box was used as a probe. In this case, several specific complexes were observed, but none of the bands cross-reacted with the SF-1 antibody (Fig. 4A). However, antibodies generated to USF1 and USF2 were able to supershift most of the bound complexes. SF-1 binding to the GSE was also observed when Sertoli cell nuclear extracts were used and similarly revealed that the E box was unable to compete for SF-1 binding (Fig. 4B). To assure that other complexes did not obscure SF-1 binding to the E box, in vitro transcribed/translated (TNT) SF-1 was used in an EMSA together with the E box probe. In these studies, no SF-1 binding was observed with as much as 10 μl of the SF-1 TNT mixture (Fig. 4C). However, SF-1 binding was clearly observable with the GSE probe. Thus, while the E box is essential for SF-1 regulation of the FSHR promoter, these studies indicate that regulation does not require the direct binding of SF-1 to the element. SF-1 regulation may, therefore, occur via a mechanism involving interactions with the USF proteins.
Fig. 4. SF-1 Does Not Interact with the FSHR E Box.
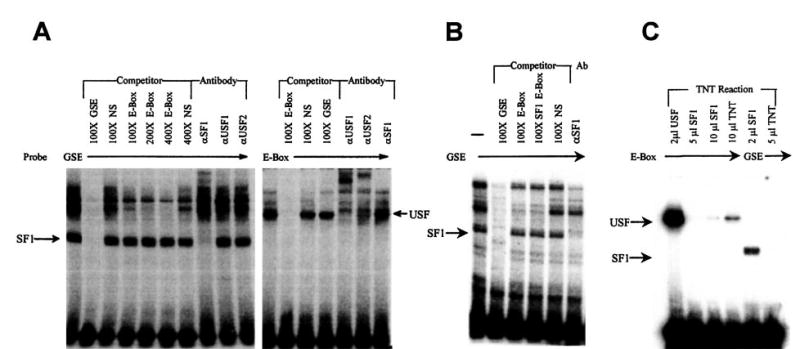
Radiolabeled probe (25 fmol) corresponding to either the human α-subunit GSE (5′-TTTCATGGGCTGACCTTGTCGTCAC-CATCAC-3′) or the FSHR E box (as in legend of Fig. 3) was used in EMSA with nuclei isolated from pituitary gonadotrope cell line αT3 (A) or primary cultures of Sertoli cells (B). EMSAs were performed with either the hαGSE or FSHR E box probe in the presence of in vitro transcribed/translated USF1, SF-1, or no DNA control (TNT) (C). Unlabeled competitors were added at the indicated amounts relative to labeled probe. Where indicated, antibodies specific for USF1, USF2, or SF-1 were added to the reactions. Arrows mark specific complexes for USF and SF-1.
The USF Proteins Are Needed for Activation of the FSHR Promoter by SF-1
To determine the role of the USF proteins in SF-1 regulation of FSHR, several mutant USF proteins that lack different protein domains were employed. The mutant USF proteins, U1ΔN and U2ΔN (Fig. 5A), efficiently bind DNA but are unable to transactivate due to the absence of the amino-terminal transactivation domains, while the U1ΔB and U2ΔB mutants lack a functional DNA binding domain (58, 59). Both sets of mutants inhibit transactivation by USF1 and USF2. The ΔN mutants have been shown to inhibit FSHR transcription by binding to the FSHR E box and interfering with the ability of endogenous USF to transactivate (26). The ΔB mutants inhibit USF transactivation by dimerizing with endogenous USF proteins and preventing interactions with responsive elements (59). Cotransfection of either U1ΔN or U2ΔN effectively blocked transactivation of the FSHR promoter by SF-1 (Fig. 5B). In addition, both ΔB mutants were able to significantly reduce SF-1 transactivation (Fig. 5B). Thus, binding of USF proteins lacking the transactivation domain to the E box or sequestering endogenous USF proteins significantly inhibited SF-1 activation of the FSHR promoter. Further analysis of the regions within the transactivation domain of USF2 revealed the requirement for amino acids encoded in exons 4 and 5 in SF-1 regulation (Fig. 5, A and C). Thus, deletion of the first 123 amino acids (Δ7–123), which are encoded for in the first four exons, significantly diminished SF-1 regulation. Extending the deleted region to include exon 5 (α1–99) further diminished SF-1 response. Additionally, deletion of the amino acids encoded by either exon 4 or 5 alone significantly reduced SF-1 activation. In contrast, cotransfection with either wild-type USF2 or a mutant in which the USR, a highly conserved USF-specific region, was deleted had little impact on SF-1’s ability to transactivate the FSHR promoter (Fig. 5C). These studies indicate that the amino acids encoded by exons 4 and 5 but not the USR are important for the regulation by SF-1. Importantly, several of these mutants only slightly diminished promoter activity by themselves, suggesting that USF requirements for SF-1 stimulation differ from the requirements for basal promoter function (Fig. 4 and Ref. 26).
Fig. 5. SF-1 Stimulation of the FSHR Promoter Requires Functional USF Proteins.
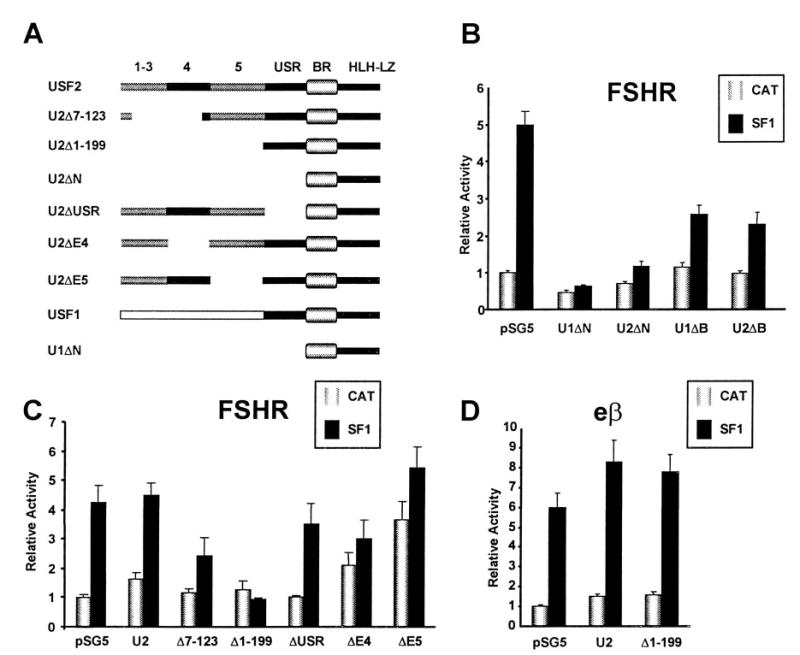
Schematic diagram of the USF proteins with the different domains or exons listed at the top (A). FSHR(−2,700/+123)Luc (0.5 μg) was cotransfected with either 0.5 μg empty expression vector (pSG5) or expression vectors for the USF mutants U1ΔN, U1ΔB, U2ΔN, and U2ΔB and 0.5 μg of either RSV-CAT or RSV-SF1 (B). Transfections were done similar to that in panel B except using expression vectors for USF2 and mutants U2Δ7–123, U2Δ1–199, U2ΔUSR, U2ΔE4, and U2ΔE5 (C). eβ (0.5 μg) was cotransfected as described in panel B with wild-type USF2 and the U2Δ1–199 mutant (D). The relative activity represents the luciferase/β-galactosidase activity of each sample normalized to the luciferase/β-galactosidase activity of FSHR(−2,700/1123)Luc or eβLuc transfected with pSG5 and RSV-CAT. Error bars represent the sem.
To determine whether the requirement for the USF proteins in SF-1 transactivation was specific to the FSHR promoter, we examined the effects of wild-type USF2 and the Δ1–199 mutant on the equine LH β-subunit (eβ) promoter. Similar to previous reports, we observed a significant transcriptional increase from the eβ promoter in response to cotransfected SF-1 (Fig. 5D and Ref. 42). However, in contrast to our observations on the FSHR promoter, the USF2 Δ1–199 mutant did not affect the ability of SF-1 to transactivate the eβ promoter (Fig. 5D). Thus, the requirement for the USF proteins in SF-1 transactivation is specific to the FSHR promoter and suggests that the mechanism by which SF-1 activates genes is dependent on promoter context.
Stimulation of the cAMP Pathway Differentially Affects SF-1 Activation of the FSHR and eβ Promoters
Previous studies on the α-inhibin promoter indicated that stimulation of the cAMP pathway enhanced the actions of SF-1 (41). While activation of this pathway is known to stimulate inhibin production, evidence for its regulation of FSHR suggests both positive and negative regulatory effects on expression (60–65). To determine whether the cAMP pathway influences SF-1 activation of FSHR, we transfected cells with the FSHR promoter and expression vectors for SF-1 (RSV-SF1) and the catalytic subunit of protein kinase A (RSV-PKA), which acts as a constitutive activator of this pathway. Cotransfection with RSV-PKA alone had little impact on FSHR promoter activity, while in the presence of RSV-SF-1, RSV-PKA blocked the ability of SF-1 to induce FSHR transcription (Fig. 6). We also examined the effects of cotransfections with RSV-PKA and RSV-SF-1 on the promoters for the equine α (eα) and β (eβ) subunits of LH (Fig. 6). PKA activated both the eβ and eα promoters, while only the eβ promoter was induced by SF-1. Interestingly, cotransfection with RSV-SF-1 and RSV-PKA resulted in a synergistic effect on the activity of the eβ promoter, while RSV-SF-1 reduced the inductive effects of PKA on the eα promoter (Fig. 6). Treatment with the cAMP analog, 8-bromo-cAMP, had similar effects to those observed with the RSV-PKA (data not shown). Thus, activation of the cAMP pathway resulted in differential effects on SF-1- stimulated promoter activity, as it blocked SF-1-activated transcription from the FSHR promoter and stimulated it on the eβ promoter.
Fig. 6. PKA Differentially Regulates SF-1 Stimulation of the FSHR and eβ Promoters.
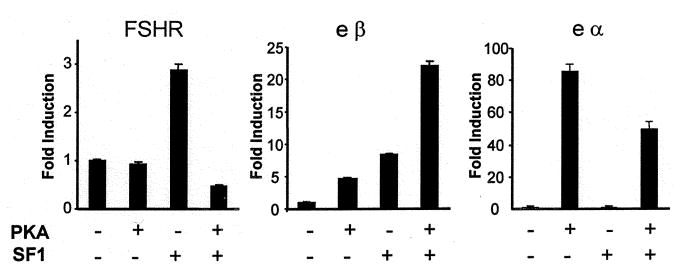
Rreporter vector (0.5 μg) containing either the promoter for FSHR, eβ, or eα was cotransfected in the presence or absence of RSV-PKA-ver2 (an expression vector for the catalytic subunit of PKA) and with or without RSV-SF1. RSV-CAT was used to normalize for the amount of transfected DNA. Fold induction represents the activity of the promoter in each transfected sample relative to the activity of the promoter transfected in the presence of RSV-CAT alone. No induction of the eα promoter was observed in the presence of SF-1. Error bars represent the sem.
DISCUSSION
FSHR and SF-1 are both integral components of the hypothalamic-pituitary-gonadal axis. SF-1 regulates transcription of many genes within this axis, while FSHR is essential for integrating the pituitary FSH signal to gonadal response. The important role of SF-1 in endocrine regulation is evident from its involvement in the transcriptional control of a number of genes important for either the biosynthesis or regulation of steroid hormones (66). Thus, SF-1 has been shown to regulate genes encoding steroid hormone biosynthetic enzymes and genes needed for LH and FSH production. With respect to the latter, SF-1 has been shown to regulate the genes for the gonadotropin β-subunit, the GnRH receptor, and the α-subunit for the FSH-regulatory protein inhibin (39, 41–49). Here, we show that SF-1 acts at yet another level of the axis to modulate FSH response in the gonads through transcriptional regulation of the FSHR gene.
The studies presented in this report support our current hypothesis that SF-1 stimulates FSHR transcription by binding several low-affinity binding sites within the upstream portion of the FSHR promoter and interacting with the USF proteins bound to the E box (Fig. 7). Cotransfection studies revealed that SF-1 stimulated FSHR promoter activity and that stimulation required multiple sites within the 5′-flanking region. Optimal SF-1 response was observed with the largest FSHR promoter construct (−2,700/+123) and SF-1 stimulation decreased sequentially as larger regions of the promoter were deleted from the 5′-end. Furthermore, characterization of additional deletion mutants (−2,343, −2,049, and −1,023) resulted in similarly modest changes in SF-1 response (data not shown). The moderate impact of these deletions on the full stimulation suggested that multiple weak SF-1 binding sites upstream of −734 are needed for full response of the FSHR promoter to SF-1. Response within the proximal promoter region (−220/+79) mapped to an E box element previously shown to be regulated by the transcription factors USF1 and USF2 (23, 24, 26). Interestingly, mutation of the E box in the context of the −2,700/+123 promoter eliminated all response to SF-1, demonstrating the absolute requirement for this element in SF-1 regulation of FSHR. In the absence of SF-1 binding to this element, the data suggest a mechanism in which SF-1 interacts with the USF proteins, either directly or indirectly via a secondary protein, to stimulate FSHR transcription (Fig. 7, options 1 and 2, respectively). In support of this hypothesis, basal promoter activity and SF-1-stimulated promoter activity had differential requirements for the USF proteins.
Fig. 7. Model for SF-1 Regulation of the FSHR Promoter.
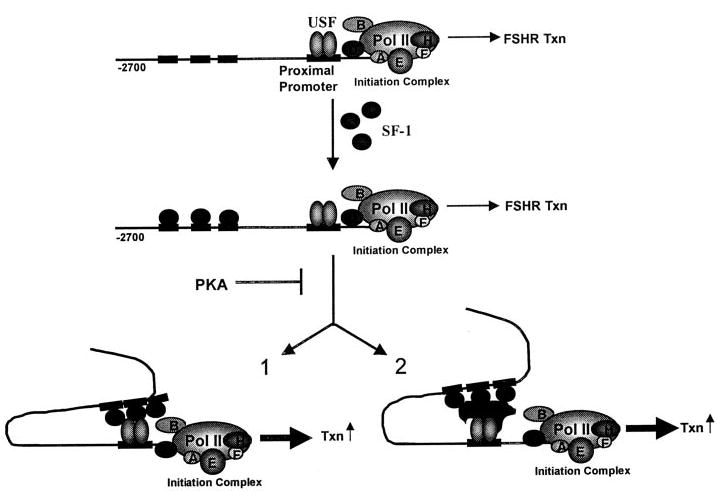
As postulated, SF-1 induces FSHR transcription by binding several sites within the FSHR promoter that span the region from −2,700 to −743. After binding to these upstream sites, SF-1 interacts either directly with the USF proteins (1) or with an intermediate regulatory protein (2). Interaction with the E box-bound complex results in a stimulation of the initiation complex and induction of transcription. While not shown in the model, the data also suggest that in the absence of these upstream sites (−2,700 to −743), SF-1 binds sufficiently to the E box-bound complex to stimulate transcription. PKA interferes with the ability of SF-1 to transactivate the FSHR promoter at a point likely to be downstream of SF1 binding.
Further confirmation of a role for SF-1 in FSHR gene regulation was recently provided by a study on the mouse FSHR promoter. In this report, the investigators showed that SF-1 activated the mouse FSHR promoter through two SF-1 binding sites located between −1,171 and −1,369 bp relative to the translations start site(67). Examination of the rat promoter sequence within this region identified two potential SF-1 binding sites similar to those described for the mouse. The elements reside at positions −1,213 (5′-TTTCCTTGG-3′) and −1,252 (5′-TTTCCTTGA-3′) relative to the major transcriptional start site. Interestingly, removal of both of these sites with the −1,300 to −743 deletion (Fig. 2) resulted in a decrease in promoter response to SF-1. However, in addition to a role for these elements, our studies further suggest that other sites important for SF-1 response reside upstream of −1,300 and that the USF proteins are important for the overall response to SF-1.
The involvement of the USF proteins in SF-1 activation of FSHR was further substantiated by studies in which plasmids expressing various USF mutants were cotransfected with the FSHR promoter and an expression vector for SF-1. These studies revealed that the amino-terminal transactivation domains of USF1 and USF2 are required for SF-1 stimulation and implicated the amino acids encoded by both exons 4 and 5 of USF2 as needed for activation by SF-1. Importantly, the effects of the USF mutants on SF-1 transactivation were specific to the FSHR promoter, as neither wild-type USF2 nor the Δ1–199 mutant influenced SF-1 activation of the eβ promoter. Interestingly, many recent reports have revealed that SF-1 commonly acts in concert with other transcription factors to regulate gene activity. Thus, SF-1 has been shown to synergize with the transcriptional regulators RAR (retinoic acid receptor), GATA-4 (a member of the GATA family of zinc proteins), Egr-1 (early growth response protein 1), Ptx-1 (pituitary homeobox 1), and Wt1 (Wilms tumor suppressor) (40, 43, 45, 55, 68–70). Therefore, a common mechanism by which SF-1 regulates transcription is one involving cooperation with other transcription factors, of which USF1 and USF2 can now be included. Although a role for USF proteins in SF-1 transcriptional activation has not been previously reported, it is of interest to note that USF and SF-1 are implicated in the regulation of both the human α-subunit and SF-1 genes, suggesting that a similar mechanism may be shared by them as well (46, 71–75).
Although the mechanisms by which SF-1 and USF cooperate to regulate FSHR are not fully understood, the finding that these transcription factors act in concert to activate transcription provides important insights into FSHR gene regulation, transactivation function of SF-1, and the expanding physiological roles of SF-1 in the hypothalamic-pituitary-gonadal axis. Within this axis, SF-1 appears to regulate both FSH production and response. Thus, through its regulation of the α-subunit of the FSH-regulatory protein inhibin, SF-1 can directly influence FSH production in the pituitary, and through its regulation of FSHR, SF-1 can influence FSH response. Interestingly, activation of the inhibin α subunit by SF-1 has been shown to dramatically increase in the presence of activated PKA, the major downstream regulator of FSH action in the gonads (41). Thus, rising levels of FSH in the presence of SF-1 should dramatically stimulate inhibin levels produced by the gonads, which would then feed back on the pituitary to specifically decrease FSH production. Remarkably, our studies revealed that activated PKA had the opposite impact on SF-1-stimulated transcription of the FSHR promoter. Thus, with SF-1 in position to regulate these two genes, FSH stimulation can lead to opposite transcriptional effects resulting in a rise in α inhibin and a fall in FSHR, which would result in a dramatic decrease in FSH signaling in the gonads. Together these observations provide a compelling molecular argument for SF-1 as a central regulator in the integrated control of FSH action in the reproductive system.
In support of our observations, several studies have demonstrated inhibitory effects of cAMP on the expression of FSHR. In the ovary, treatment with hormones such as PMSG or human CG (hCG), which stimulate the cAMP pathway, can result in either an increase or decrease in FSHR expression (65, 76). Treatment with PMSG to stimulate follicle growth results in an increase in FSHR mRNA, whereas, treatment with hCG to induce ovulation and luteinization results in a marked decrease in FSHR expression. It is therefore tempting to speculate that the differential effects of cAMP on FSHR expression in the ovary may reflect the expression level of SF-1 in granulosa cells at the time of hormone stimulation. In the testis, FSHR mRNA in Sertoli cells is significantly reduced in response to cAMP induction (62, 63). Thus, our studies suggest that SF-1 in granulosa and Sertoli cells is an important modulator of FSHR regulation by the gonadotropin hormones FSH and LH.
The mechanisms by which PKA influences SF-1-activated transcription are not fully understood. Our data, and data with the α inhibin promoter, demonstrate the importance of promoter context in the integration of SF-1 and cAMP transcriptional signals. Studies with the α inhibin promoter suggested that the synergistic effects of SF-1 and activated PKA were the result of direct interaction between SF-1 and the cAMP response element binding protein, CREB, which are bound to different but closely spaced response elements on the promoter (41). Our studies also demonstrate synergistic effects between activated PKA and SF-1 but on a different promoter, the eβ promoter. This is a novel finding and the mechanism has not been fully explored. While studies have demonstrated a role for SF-1 in the regulation of eβ, information regarding cAMP regulation is lacking (42). In the horse, this gene is expressed in both the placenta and pituitary, where, in the latter, it is under the control of the hypothalamic peptide GnRH. Although not extensively studied, the cAMP pathway has been implicated in GnRH stimulation of pituitary gonadotrope cells. Thus the adenylate cyclase inhibitor 2′,3′-dideoxyadenosine blocked GnRH-induced LHβ mRNA, and GnRH was shown to induce phosphorylation of CREB, a known downstream target of PKA (77, 78). In the placenta, cAMP is known to regulate the human β-subunit of CG, which is functionally similar to the equine β subunit in these cells (79). These observations, together with our findings on the effects of SF-1 and activated PKA, suggest that the cAMP pathway is an important regulator of the LH and CG β genes and SF-1 is involved in modulating the effects.
With respect to the FSHR promoter, PKA stimulation had the opposite effect on transcriptional induction by SF-1, providing a molecular explanation for the reported negative changes in FSHR mRNA in response to cAMP induction (41, 80–84). While our current understanding of how these two signals converge to regulate FSHR is limited, our favored hypothesis is that PKA activation leads to modification of SF-1 and prevents its interaction with the USF proteins (Fig. 7). Alternatively, the transcriptional repressor ICER (inducible cAMP early repressor), which is induced by FSH in testicular Sertoli cells, may bind the FSHR promoter and interfere with SF-1-activated transcription. Previous reports provide evidence for ICER regulation of FSHR (85). Further analysis of the protein interactions and domains required for regulation of transcription will provide additional insight into the mechanisms required for SF-1 activation and PKA repression of FSHR promoter activity.
MATERIALS AND METHODS
DNA Constructs
Rat FSHR promoter/luciferase constructs FSHR(−2,700/+123)Luc, FSHR(−1,300/+123)Luc, FSHR(−763/+123)Luc, FSHR(−220/+123)Luc, and FSHR(−220/+79)Luc are described elsewhere (23). For FSHR(−2,700/+123)μ9.2, the 343-bp EcoRV/XhoI fragment from −220 to +123 was removed and replaced with the analogous region generated from FSHR(−220/+123)μ9.2. Expression vectors for the USF were kindly provided by Dr. Michele Sawadogo (59). PGL3-Control and pGL3-Basic were purchased from Promega Corp. (Madison, WI). The cDNA for mouse SF-1 was generously provided by Dr. Keith Parker and subcloned downstream of the Rous Sarcoma viral promoter as described (42). RSV-CAT is described elsewhere (86). The eβ and eα promoters were a gift from Dr. Michael Wolfe and are described elsewhere (42, 87). The expression vector for the catalytic subunit of PKA (RSV-PKA-ver2) was generously provided by Dr. Richard Maurer and is described elsewhere (88). Plasmid DNAs were prepared from overnight bacterial cultures using QIAGEN DNA plasmid columns according to the supplier’s protocol (QIAGEN, Chatsworth, CA).
Transfection and Enzyme Analysis
The choriocarcinoma cell line JEG3 was cultured in DMEM supplemented with 10% FBS. For transfection, cells were seeded onto six-well plates (35 mm/well) at a density of 110,000 cells per well. Unless otherwise stated, cells were transfected with 1 μg of FSHR-luciferase construct, 0.2 μg RSV-β-galactosidase, and 0.5 μg of expression vector using 5 μl of lipofectamine. The lipofectamine/DNA complex was replaced after 16 h with JEG3 media (see above). Cells were harvested and assayed approximately 60 h after transfection according to previously described procedures (89). To control for transfection efficiency, luciferase activity generated from the FSHR promoter was normalized to the β-galactosidase activity from the cotransfected RSV-β-galactosidase vector. Data were averaged over a minimum of three independent experiments.
Electrophoretic Mobility Shift Analysis
The gonadotrope cell line αT3 was grown in culture as indicated elsewhere (57, 89) and nuclei were isolated as described (90). Nuclear extracts from primary Sertoli cells and JEG3 cells were prepared as described (73). In vitro transcription/translation reactions were done according to the manufacturer’s recommendation (Promega Corp., Madison. WI) using cDNA vectors encoding human USF1 and mouse SF-1 as indicated above. EMSAs were performed as previously described using approximately 3 × 105 αT3 nuclei or 5–10 μg of nuclear proteins from JEG3 cells and 25 fmol radiolabeled double-stranded oligodeoxynucleotide probe (91). End-labeled oligodeoxynucleotide probes were generated using T4 kinase. DNA sequences of the human α-subunit GSE and FSHR E box oligodeoxynucleotides are 5′-TTTCATGGGCTGACCTTGTCGTCACCATCAC-3′ and 5′-TCTTGGTGGGTCACGTGACTTTGCCCGT-3′, respectively. All reaction components were incubated at room temperature for 30 min in the absence of probe. Competitors were added at the indicated molar ratios, and 1 μl of antibody was used for supershift analysis. After the addition of probe, reactions were incubated an additional 15 min on ice and then resolved on a 4% nondenaturing polyacrylamide gel. Antibodies for USF1 (C-20)X and USF2 (C-20)X were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), and the SF-1 antibody was purchased from Upstate Biotechnology, Inc. (Lake Placid, NY).
Acknowledgments
I would like to thank the Center of Reproductive Sciences at the University of Kansas Medical Center for imaging, cell culture, and DNA sequencing services. I also thank Dr. Michael Wolfe for generously providing αT3 nuclei and Daren Rice and Jiang-kai Chen for their technical assistance.
Footnotes
This work was supported by NIH Grant R29HD-3521701A1 (to L.L.H.).
References
- 1.Pierce J, Parsons TF. Glycoprotein hormones: structure, function. Annu Rev Biochem. 1981;50:465–495. doi: 10.1146/annurev.bi.50.070181.002341. [DOI] [PubMed] [Google Scholar]
- 2.Rannikki AS, Zhang F-P, Huhtaniemi IT. Ontogeny of follicle-stimulating hormone receptor gene expression in the rat testis and ovary. Mol Cell Endocrinol. 1995;107:199–210. doi: 10.1016/0303-7207(94)03444-x. [DOI] [PubMed] [Google Scholar]
- 3.Camp TA, Rahal JO, Mayo KE. Cellular localization and hormonal regulation of follicle-stimulating hormone and luteinizing hormone receptor messenger RNAs in the rat ovary. Mol Endocrinol. 1991;5:1405–1417. doi: 10.1210/mend-5-10-1405. [DOI] [PubMed] [Google Scholar]
- 4.Griswold MD 1993 Actions of FSH on mammalian Sertoli cells. In: Russell LD, Griswold MD (eds) The Sertoli Cell. Cache River Press, Clearwater, MN, pp 493–508
- 5.Heckert L, Griswold MD1992 The changing functions of follicle-stimulating hormone in the testes of prenatal, newborn, immature, and adult rats. In: Hunzicker-Dunn M, Schwartz NB (eds) Follicle-Stimulating Hormone: Regulation of Secretion and Molecular Mechanisms of Action. Springer-Verlag, New York, pp 237–245
- 6.Zirkin BR, Awoniyi C, Griswold MD, Russell LD, Sharpe R. Is FSH required for adult spermatogenesis? J Androl. 1994;15:273–276. [PubMed] [Google Scholar]
- 7.Aittomaki K, Lucena JLD, Pakarinen P, Sistonen P, Tapanainenkila J, Gromoll J, Kaskikari R, Sankila E-M, Lehvaslaiho H, Engel AR, Nieschlag E, Huhtaniemi I, de la Chapell A. Mutation in the follicle-stimulating hormone receptor gene causes hereditary hypergonadotropic ovarian failure. Cell. 1995;82:959–968. doi: 10.1016/0092-8674(95)90275-9. [DOI] [PubMed] [Google Scholar]
- 8.DeManno DA, Hunzicker-Dunn M 1992 FSH Regulation of cAMP-dependent protein kinase regulatory subunits in rat and porcine ovarian tissues. In: Hunzicker-Dunn M, Schwartz NB (eds) Follicle-Stimulating Hormone: Regulation of Secretion and Molecular Mechanisms of Action. Springer-Verlag, New York, pp 156–166
- 9.Dorrington JH, Bendell JJ, Khan SA. Interactions between FSH, estradiol-17 β and transforming growth factor-β regulate growth and differentiation in the rat gonad. J Steroid Biochem Mol Biol. 1993;44:441–447. doi: 10.1016/0960-0760(93)90248-u. [DOI] [PubMed] [Google Scholar]
- 10.Kangasniemi M, Kaipia A, Pekka M, Toppari J, Huhtaniemi I, Parvinen M. Cellular regulation of follicle-stimulating hormone (FSH) binding in rat seminiferous tubules. J Androl. 1990;11:336–343. [PubMed] [Google Scholar]
- 11.Kanzaki M, Hattori M, Kojima I. Growth of differentiation: determination by FSH of the action of insulin-like growth factor-I in cultured rat granulosa cells. Endocr J. 1996;43:15–23. doi: 10.1507/endocrj.43.15. [DOI] [PubMed] [Google Scholar]
- 12.Luderer U, Schwartz NB 1992 An overview of FSH regulation and action. In: Hunzicker-Dunn M, Schwartz NB (eds) Follicle-Stimulating Hormone: Regulation of Secretion and Molecular Mechanisms of Action. Springer-Verlag, New York, pp 1–28
- 13.Matthews CG, Borgato S, Beck-Peccoz P, Adams M, Tone Y, Gambino G, Casagrande S, Tedeschini G, Benedetti A, Chatterjee VKK. Primary amenorrhoea and infertility due to a mutation in the β subunit of follicle stimulating hormone. Nature. 1993;5:83–86. doi: 10.1038/ng0993-83. [DOI] [PubMed] [Google Scholar]
- 14.Gromoll J, Pekel E, Nieschlag E. The structure and organization of the human follicle-stimulating hormone receptor gene. Genomics. 1996;35:308–311. doi: 10.1006/geno.1996.0361. [DOI] [PubMed] [Google Scholar]
- 15.Heckert LL, Daley IJ, Griswold MD. Structural organization of the follicle-stimulating hormone receptor gene. Mol Endocrinol. 1992;6:70–80. doi: 10.1210/mend.6.1.1738373. [DOI] [PubMed] [Google Scholar]
- 16.Huhtaniemi I, Eskola V, Pakarinen P, Matikainen T, Sprengel R. The murine luteinizing hormone and follicle-stimulating hormone receptor genes: transcriptional initiation sites, putative promoter sequences, and promoter activity. Mol Cell Endocrinol. 1992;88:55–66. doi: 10.1016/0303-7207(92)90009-u. [DOI] [PubMed] [Google Scholar]
- 17.Sprengel R, Braun T, Nikolics K, Segaloff DL, Seeburg PH. The testicular receptor for follicle stimulating hormone: structure and functional expression of cloned cDNA. Mol Endocrinol. 1990;4:525–530. doi: 10.1210/mend-4-4-525. [DOI] [PubMed] [Google Scholar]
- 18.Tena-Sempere M, Manna PR, Huhtaniemi I. Molecular cloning of the mouse follicle-stimulating hormone receptor complementary deoxyribonucleic acid: functional expression of alternatively spliced variants and receptor inactivation by a C566T transition in exon 7 of the coding sequence. Biol Reprod. 1999;60:1515–1527. doi: 10.1095/biolreprod60.6.1515. [DOI] [PubMed] [Google Scholar]
- 19.Remy JJ, Lahbib-Mansais Y, Yerle M, Bozon V, Couture L, Pajot E, Greber D, Salesse R. The porcine follitropin receptor: cDNA cloning, functional expression and chromosomal localization of the gene. Gene. 1995;163:257–261. doi: 10.1016/0378-1119(95)00385-j. [DOI] [PubMed] [Google Scholar]
- 20.Dankbar B, Brinkworth MH, Schlatt S, Weinbauer GF, Nieschlag E, Gromoll J. Ubiquitous expression of the androgen receptor and testis-specific expression of the FSH receptor in the cynomolgus monkey (Macaca fascicularis) revealed by a ribonuclease protection assay. J Steroid Biochem Mol Biol. 1995;55:35–41. doi: 10.1016/0960-0760(95)00148-s. [DOI] [PubMed] [Google Scholar]
- 21.Heckert LL, Griswold MD. Expression of follicle-stimulating hormone receptor mRNA in rat testes and Sertoli cells. Mol Endocrinol. 1991;5:670–677. doi: 10.1210/mend-5-5-670. [DOI] [PubMed] [Google Scholar]
- 22.Means AR. Specific interaction of 3H-FSH with rat testis binding sites. Adv Exp Med Biol. 1973;36:431–448. doi: 10.1007/978-1-4684-3237-4_20. [DOI] [PubMed] [Google Scholar]
- 23.Heckert LL, Daggett MA, Chen J. Multiple promoter elements contribute to activity of the follicle-stimulating hormone receptor (FSHR) gene in testicular Sertoli cells. Mol Endocrinol. 1998;12:1499–1512. doi: 10.1210/mend.12.10.0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goetz TL, Lloyd TL, Griswold MD. Role of E box and initiatory region in the expression of the rat follicle-stimulating hormone receptor. J Biol Chem. 1996;271:33317–33324. doi: 10.1074/jbc.271.52.33317. [DOI] [PubMed] [Google Scholar]
- 25.Linder CC, Heckert LL, Goetz TL, Griswold MD. Follicle-stimulating hormone receptor gene promoter activity. Endocrine. 1994;2:957–966. [Google Scholar]
- 26.Heckert LL, Sawadogo M, Daggett MA, Chen J. The USF proteins regulate transcription of the follicle-stimulating hormone receptor but are insufficient for cell-specific expression. Mol Endocrinol. 2000;14:1836–1848. doi: 10.1210/mend.14.11.0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gromoll J, Dankbar B, Gudermann T. Characterization of the 5′ flanking region of the human follicle-stimulating hormone receptor gene. Mol Cell Endocrinol. 1994;102:93–102. doi: 10.1016/0303-7207(94)90102-3. [DOI] [PubMed] [Google Scholar]
- 28.Lala DS, Rice DA, Parker KL. Steroidogenic factor 1, a key regulator of steroidogenic enzyme expression, is the mouse homolog of fushi tarazu-factor 1. Mol Endocrinol. 1992;6:1249–1258. doi: 10.1210/mend.6.8.1406703. [DOI] [PubMed] [Google Scholar]
- 29.Ikeda Y, Shen W-H, Ingraham HA, Parker KL. Developmental expression of mouse steroidogenic factor-1, an essential regulator of the steroid hydroxylases. Mol Endocrinol. 1994;8:654–662. doi: 10.1210/mend.8.5.8058073. [DOI] [PubMed] [Google Scholar]
- 30.Hatano O, Takayama K, Imai T, Waterman MR, Takakusu A, Omura T, Morohashi K. Sex-dependent expression of a transcription factor, Ad4BP, regulating steroidogenic P-450 genes in the gonads during prenatal and postnatal rat development. Development. 1994;120:2787–2797. doi: 10.1242/dev.120.10.2787. [DOI] [PubMed] [Google Scholar]
- 31.Shen W-H, Moore CD, Ikeda Y, Parker KL, Ingraham HA. Nuclear receptor steroidogenic factor 1 regulates the Mullerian inhibiting substance gene: a link to the sex determination cascade. Cell. 1994;77:651–661. doi: 10.1016/0092-8674(94)90050-7. [DOI] [PubMed] [Google Scholar]
- 32.Morohashi K, Hatano O, Nomura M, Takayama K, Hara M, Yoshii H, Takakusu A, Omura T. Function and distribution of a steroidogenic cell-specific transcription factor, Ad4BP. J Steroid Biochem Mol Biol. 1995;53:81–88. doi: 10.1016/0960-0760(95)00041-w. [DOI] [PubMed] [Google Scholar]
- 33.Ingraham HA, Lala S, Ikeda Y, Luo X, Shen W-H, Nachtigal MW, Abbud R, Nilson JH, Parker KL. The nuclear receptor steroidogenic factor 1 acts at multiple levels of the reproductive axis. Genes Dev. 1994;8:2303–2312. doi: 10.1101/gad.8.19.2302. [DOI] [PubMed] [Google Scholar]
- 34.Roselli CE, Jorgensen EZ, Doyle MW, Ronnekleiv OK. Expression of the orphan receptor steroidogenic factor-1 mRNA in the rat medial basal hypothalamus. Brain Res Mol Brain Res. 1997;44:66–72. doi: 10.1016/s0169-328x(96)00187-8. [DOI] [PubMed] [Google Scholar]
- 35.Rice DA, Mouw AR, Bogerd AM, Parker KL. A shared promoter element regulates the expression of three steroidogenic enzymes. Mol Endocrinol. 1991;5:1552–1561. doi: 10.1210/mend-5-10-1552. [DOI] [PubMed] [Google Scholar]
- 36.Morohashi K, Honda S, Inomata Y, Handa H, Omura T. A common trans-acting binding factor, Ad4-binding protein, to the promoters of steroidogenic P-450s. J Biol Chem. 1992;267:17913–17919. [PubMed] [Google Scholar]
- 37.Caron KM, Clark BJ, Ikeda Y, Parker KL. Steroidogenic factor 1 acts at all levels of the reproductive axis. Steroids. 1997;62:53–56. doi: 10.1016/s0039-128x(96)00159-6. [DOI] [PubMed] [Google Scholar]
- 38.Asa SL, Bamberger AM, Cao B, Wong M, Parker KL, Ezzat S. The transcription activator steroidogenic factor-1 is preferentially expressed in the human pituitary gonadotroph. J Clin Endocrinol Metab. 1996;81:2165–2170. doi: 10.1210/jcem.81.6.8964846. [DOI] [PubMed] [Google Scholar]
- 39.Duval DL, Nelson SE, Clay CM. A binding site for steroidogenic factor-1 is part of a complex enhancer that mediates expression of the murine gonadotropin-releasing hormone receptor gene. Biol Reprod. 1997;56:160–168. doi: 10.1095/biolreprod56.1.160. [DOI] [PubMed] [Google Scholar]
- 40.Halvorson LM, Ito M, Jameson JL, Chin WW. Steroidogenic factor-1 and early growth response protein 1 act through two composite DNA binding sites to regulate luteinizing hormone β-subunit gene expression. J Biol Chem. 1998;273:14712–14720. doi: 10.1074/jbc.273.24.14712. [DOI] [PubMed] [Google Scholar]
- 41.Ito M, Park Y, Weck J, Mayo KE, Jameson JL. Synergistic activation of the inhibin α-promoter by steroidogenic factor-1 and cyclic adenosine 3′,5′-monophosphate. Mol Endocrinol. 2000;14:66–81. doi: 10.1210/mend.14.1.0410. [DOI] [PubMed] [Google Scholar]
- 42.Wolfe MW. The equine luteinizing hormone β-subunit promoter contains two functional steroidogenic factor-1 response elements. Mol Endocrinol. 1999;13:1497–1510. doi: 10.1210/mend.13.9.0345. [DOI] [PubMed] [Google Scholar]
- 43.Wolfe MW, Call GB. Early growth response protein 1 binds to the luteinizing hormone-β promoter and mediates gonadotropin-releasing hormone-stimulated gene expression. Mol Endocrinol. 1999;13:752–763. doi: 10.1210/mend.13.5.0276. [DOI] [PubMed] [Google Scholar]
- 44.Keri RA, Nilson JH. A steroidogenic factor-1 binding site is required for activity of the luteinizing hormone B subunit promoter in gonaotropes of transgenic mice. J Biol Chem. 1996;271:10782–10785. doi: 10.1074/jbc.271.18.10782. [DOI] [PubMed] [Google Scholar]
- 45.Dorn C, Ou Q, Svaren J, Crawford PA, Sadovsky Y. Activation of luteinizing hormone β gene by gonadotropin-releasing hormone requires the synergy of early growth response-1 and steroidogenic factor-1. J Biol Chem. 1999;274:13870–13876. doi: 10.1074/jbc.274.20.13870. [DOI] [PubMed] [Google Scholar]
- 46.Barnhart KM, Mellon PL. The orphan nuclear receptor, steroidogenic factor-1, regulates the glycoprotein hormone α-subunit gene in pituitary gonadotropes. Mol Endocrinol. 1994;8:878–885. doi: 10.1210/mend.8.7.7527122. [DOI] [PubMed] [Google Scholar]
- 47.Ngan ES, Cheng PK, Leung PC, Chow BK. Steroidogenic factor-1 interacts with a gonadotrope-specific element within the first exon of the human gonadotropin-releasing hormone receptor gene to mediate gonadotrope-specific expression. Endocrinology. 1999;140:2452–2462. doi: 10.1210/endo.140.6.6759. [DOI] [PubMed] [Google Scholar]
- 48.Tremblay JJ, Drouin J. Egr-1 is a downstream effector of GnRH and synergizes by direct interaction with Ptx1 and SF-1 to enhance luteinizing hormone β gene transcription. Mol Cell Biol. 1999;19:2567–2576. doi: 10.1128/mcb.19.4.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Halvorson LM, Kaiser UB, Chin WW. The protein kinase C system acts through the early growth response protein 1 to increase LHβ gene expression in synergy with steroidogenic factor-1. Mol Endocrinol. 1999;13:106–116. doi: 10.1210/mend.13.1.0216. [DOI] [PubMed] [Google Scholar]
- 50.Luo X, Ikeda Y, Parker KL. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell. 1994;77:481–490. doi: 10.1016/0092-8674(94)90211-9. [DOI] [PubMed] [Google Scholar]
- 51.Luo X, Ikeda Y, Schlosser DA, Parker KL. Steroidogenic factor 1 is the essential transcript of the mouse FTZ-F1 gene. Mol Endocrinol. 1995;9:1233–1239. doi: 10.1210/mend.9.9.7491115. [DOI] [PubMed] [Google Scholar]
- 52.Sadovsky Y, Crawford PA, Woodson KG, Polish JA, Clements MA, Tourtellotte LM, Simburger K, Milbrandt J. Mice deficient in the orphan receptor steroidogenic factor 1 lack adrenal glands and gonads but express P450 side-chain-cleavage enzyme in the placenta and have normal embryonic serum levels of corticosteroids. Proc Natl Acad Sci USA. 1995;92:10939–10943. doi: 10.1073/pnas.92.24.10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peterson KR, Li QL, Clegg CH, Furukawa T, Navas PA, Norton EJ, Kimbrough TG, Stamatoyannopoulos G. Use of yeast artificial chromosomes (YACs) in studies of mammalian development: production of beta-globin locus YAC mice carrying human globin developmental mutants. Proc Natl Acad Sci USA. 1995;92:5655–5659. doi: 10.1073/pnas.92.12.5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ikeda Y, Lala DS, Luo X, Kim E, Moisan M-P, Parker KL. Characterization of the mouse FTZ-F1 gene, which encodes a key regulator of steroid hydroxylase gene expression. Mol Endocrinol. 1993;7:852–860. doi: 10.1210/mend.7.7.8413309. [DOI] [PubMed] [Google Scholar]
- 55.Nachtigal MW, Hirokawa Y, Enyeart-VanHouten DL, Flanagan JN, Hammer GD, Ingraham HA. Wilms’ tumor 1 and Dax-1 modulate the orphan nuclear receptor SF-1 in sex-specific gene expression. Cell. 1998;93:445–454. doi: 10.1016/s0092-8674(00)81172-1. [DOI] [PubMed] [Google Scholar]
- 56.Ito M, Yu R, Jameson JL. DAX-1 inhibits SF-1-mediated transactivation via a carboxy-terminal domain that is deleted in adrenal hypoplasia congenita. Mol Cell Biol. 1997;17:1476–1483. doi: 10.1128/mcb.17.3.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Windle JJ, Weiner RI, Mellon PL. Cell lines of the pituitary gonadotrope lineage derived by targeted oncogenesis in transgenic mice. Mol Endocrinol. 1990;4:597–603. doi: 10.1210/mend-4-4-597. [DOI] [PubMed] [Google Scholar]
- 58.Roy AL, Du H, Gregor PD, Novina CD, Martinez E, Roeder RG. Cloning of an inr- and E-box-binding protein, TFII-I, that interacts physically and functionally with USF1. EMBO J. 1997;16:7091–7104. doi: 10.1093/emboj/16.23.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luo X, Sawadogo M. Functional domains of the transcription factor USF2: atypical nuclear localization signals and context-dependent transcriptional activation domains. Mol Cell Biol. 1996;16:1367–1375. doi: 10.1128/mcb.16.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Minegishi T, Tano M, Nakamura K, Nakamura M, Igarashi S, Ito I, Shinozaki H, Karino S, Ibuki Y, Miyamoto K. Regulation of follicle-stimulating hormone receptor. Horm Res. 1996;46(Suppl 1):37–44. doi: 10.1159/000185180. [DOI] [PubMed] [Google Scholar]
- 61.Minegishi T, Tano M, Kishi H, Kameda T, Miyamoto K. Follicle-stimulating hormone regulation on its receptor messenger ribonucleic acid levels in cultured rat granulosa cells. Biochim Biophys Acta. 1997;1359:165–173. doi: 10.1016/s0167-4889(97)00095-5. [DOI] [PubMed] [Google Scholar]
- 62.Maguire SM, Tribley WA, Griswold MD. Follicle-stimulating hormone (FSH) regulates the expression of FSH receptor messenger ribonucleic acid in cultured Sertoli cells and in hypophysectomized rat testis. Biol Reprod. 1997;56:1106–1111. doi: 10.1095/biolreprod56.5.1106. [DOI] [PubMed] [Google Scholar]
- 63.Themmen AP, Blok LJ, Post M, Baarends WM, Hoogerbrugge JW, Parmentier M, Vassart G, Grootegoed JA. Follitropin receptor down-regulation involves a cAMP-dependent post-transcriptional decrease of receptor mRNA expression. Mol Cell Endocrinol. 1991;78:R7–R13. doi: 10.1016/0303-7207(91)90130-k. [DOI] [PubMed] [Google Scholar]
- 64.Tilly JL, LaPolt PS, Hsueh AJ. Hormonal regulation of follicle-stimulating hormone receptor messenger ribonucleic acid levels in cultured rat granulosa cells. Endocrinology. 1992;130:1296–1302. doi: 10.1210/endo.130.3.1311235. [DOI] [PubMed] [Google Scholar]
- 65.LaPolt PS, Tilly JL, Aihara T, Nishimori K, Hsueh AJ. Gonadotropin-induced up- and down-regulation of ovarian follicle-stimulating hormone (FSH) receptor gene expression in immature rats: effects of pregnant mare’s serum gonadotropin, human chorionic gonadotropin, and recombinant FSH. Endocrinology. 1992;130:1289–1295. doi: 10.1210/endo.130.3.1537292. [DOI] [PubMed] [Google Scholar]
- 66.Parker KL, Schimmer BP. Steroidogenic factor 1: a key determinant of endocrine development and function. Endocr Rev. 1997;18:361–377. doi: 10.1210/edrv.18.3.0301. [DOI] [PubMed] [Google Scholar]
- 67.Levallet J, Koskimies P, Rahman N, Huhtaniemi I. The promoter of murine follicle-stimulating hormone receptor: functional characterization and regulation by transcription factor steroidogenic factor 1. Mol Endocrinol. 2001;15:80–92. doi: 10.1210/mend.15.1.0583. [DOI] [PubMed] [Google Scholar]
- 68.Tremblay JJ, Viger RS. Transcription factor GATA-4 enhances Mullerian inhibiting substance gene transcription through a direct interaction with the nuclear receptor SF-1. Mol Endocrinol. 1999;13:1388–1401. doi: 10.1210/mend.13.8.0330. [DOI] [PubMed] [Google Scholar]
- 69.Barnea E, Bergman Y. Synergy of SF1 and RAR in activation of Oct-3/4 promoter. J Biol Chem. 2000;275:6608–6619. doi: 10.1074/jbc.275.9.6608. [DOI] [PubMed] [Google Scholar]
- 70.Tremblay JJ, Lanctot C, Drouin J. The pan-pituitary activator of transcription, Ptx1 (pituitary homeobox 1), acts in synergy with SF-1 and Pit1 and is an upstream regulator of the Lim-homeodomain gene Lim3/Lhx3. Mol Endocrinol. 1998;12:428–441. doi: 10.1210/mend.12.3.0073. [DOI] [PubMed] [Google Scholar]
- 71.Jackson SM, Guiterrez-Hartmann A, Hoeffler JP. Upstream stimulatory factor, a basic-helix-loop-helix-zipper protein, regulates the activity of the α-glycoprotein hormone gene in pituitary cells. Mol Endocrinol. 1995;9:278–291. doi: 10.1210/mend.9.3.7539886. [DOI] [PubMed] [Google Scholar]
- 72.Nomura M, Nawata H, Morohashi K. Autoregulatory loop in the regulation of the mammalian ftz-f1 gene. J Biol Chem. 1996;271:8243–8249. doi: 10.1074/jbc.271.14.8243. [DOI] [PubMed] [Google Scholar]
- 73.Daggett MA, Rice DA, Heckert LL. Expression of steroidogenic factor 1 in the testis requires an E box and CCAAT box in its promoter proximal region. Biol Reprod. 2000;62:670–679. doi: 10.1095/biolreprod62.3.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harris AN, Mellon PL. The basic helix-loop-helix, leucine zipper transcription factor, USF (upstream stimulatory factor), is a key regulator of SF-1 (steroidogenic factor-1) gene expression in pituitary gonadotrope and steroidogenic cells. Mol Endocrinol. 1998;12:714–726. doi: 10.1210/mend.12.5.0100. [DOI] [PubMed] [Google Scholar]
- 75.Woodson KG, Crawford PA, Sadovsky Y, Milbrandt J. Characterization of the promoter of SF-1, an orphan nuclear receptor required for adrenal and gonadal development. Mol Endocrinol. 1997;11:117–126. doi: 10.1210/mend.11.2.9881. [DOI] [PubMed] [Google Scholar]
- 76.Nakamura K, Minegishi T, Takakura Y, Miyamoto K, Hasegawa Y, Ibuki Y, Igarashi M. Hormonal regulation of gonadotropin receptor mRNA in rat ovary during follicular growth and luteinization. Mol Cell Endocrinol. 1991;82:259–263. doi: 10.1016/0303-7207(91)90039-u. [DOI] [PubMed] [Google Scholar]
- 77.Kim C, Park D, Ryu K. Effect of adenylate cyclase inhibitor and protein kinase C inhibitor on GnRH-induced LH release and LH β subunit biosynthesis in rat anterior pituitary cells. Yonsei Med J. 1994;35:493–501. doi: 10.3349/ymj.1994.35.4.493. [DOI] [PubMed] [Google Scholar]
- 78.Duan WR, Shin JL, Jameson JL. Estradiol suppresses phosphorylation of cyclic adenosine 3′,5′-monophosphate response element binding protein (CREB) in the pituitary: evidence for indirect action via gonadotropin-releasing hormone. Mol Endocrinol. 1999;13:1338–1352. doi: 10.1210/mend.13.8.0322. [DOI] [PubMed] [Google Scholar]
- 79.Albanese C, Kay TW, Troccoli NM, Jameson JL. Novel cyclic adenosine 3′,5′-monophosphate response element in the human chorionic gonadotropin beta-subunit gene. Mol Endocrinol. 1991;5:693–702. doi: 10.1210/mend-5-5-693. [DOI] [PubMed] [Google Scholar]
- 80.Leers-Sucheta S, Morohashi K, Mason JI, Melner MH. Synergistic activation of the human type II 3β-hydroxysteroid dehydrogenase/δ5-δ4 isomerase promoter by the transcription factor steroidogenic factor-1/adrenal 4-binding protein and phorbol ester. J Biol Chem. 1997;272:7960–7967. doi: 10.1074/jbc.272.12.7960. [DOI] [PubMed] [Google Scholar]
- 81.Lopez D, Sandhoff TW, McLean MP. Steroidogenic factor-1 mediates cyclic 3′,5′-adenosine monophosphate regulation of the high density lipoprotein receptor. Endocrinology. 1999;140:3034–3044. doi: 10.1210/endo.140.7.6846. [DOI] [PubMed] [Google Scholar]
- 82.Naville D, Penhoat A, Durand P, Begeot M. Three steroidogenic factor-1 binding elements are required for constitutive and cAMP-regulated expression of the human adrenocorticotropin receptor gene. Biochem Biophys Res Commun. 1999;255:28–33. doi: 10.1006/bbrc.1998.9891. [DOI] [PubMed] [Google Scholar]
- 83.Sandhoff TW, Hales DB, Hales KH, McLean MP. Transcriptional regulation of the rat steroidogenic acute regulatory protein gene by steroidogenic factor 1. Endocrinology. 1998;139:4820–4831. doi: 10.1210/endo.139.12.6345. [DOI] [PubMed] [Google Scholar]
- 84.Sugawara T, Kiriakidou M, McAllister JM, Kallen CB, Strauss JF. Multiple steroidogenic factor 1 binding elements in the human steroidogenic acute regulatory protein gene 5′-flanking region are required for maximal promoter activity and cyclic AMP responsiveness. Biochemistry. 1997;36:7249–7255. doi: 10.1021/bi9628984. [DOI] [PubMed] [Google Scholar]
- 85.Monaco L, Foulkes NS, Sassone-Corsi P. Pituitary follicle-stimulating hormone (FSH) induces CREM gene expression in Sertoli cells: involvement in long-term desensitization of the FSH receptor. Proc Natl Acad Sci USA. 1995;92:10673–10677. doi: 10.1073/pnas.92.23.10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Silver BJ, Bokar J, Virgin JB, Valen E, Milsted A, Nilson JH. Cyclic AMP regulation of the human glycoprotein hormone α-gene is mediated by an 18 base pair element. Proc Natl Acad Sci USA. 1987;84:2198–2202. doi: 10.1073/pnas.84.8.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Farmerie TA, Abbud RA, Budworth PR, Clay CM, Keri RA, McDowell KJ, Wolfe MW, Nilson JH. Characterization of the equine glycoprotein hormone α-subunit gene reveals divergence in the mechanism of pituitary and placental expression. Biol Reprod. 1997;57:1104–1114. doi: 10.1095/biolreprod57.5.1104. [DOI] [PubMed] [Google Scholar]
- 88.Howard P, Day KH, Kim KE, Richardson J, Thomas J, Abraham I, Fleischmann RD, Gottesman MM, Maurer RA. Decreased catalytic subunit mRNA levels and altered catalytic subunit mRNA structure in a cAMP-resistant Chinese hamster ovary cell line. J Biol Chem. 1991;266:10189–10195. [PubMed] [Google Scholar]
- 89.Heckert LL, Schultz K, Nilson JH. Different composite regulatory elements direct expression of the human α subunit gene to pituitary and placenta. J Biol Chem. 1995;270:26497–26504. doi: 10.1074/jbc.270.44.26497. [DOI] [PubMed] [Google Scholar]
- 90.Hagenbuchle O, Wellauer PK. A rapid method for the isolation of DNA-binding proteins from purified nuclei of tissues and cells in culture. Nucleic Acids Res. 1992;20:3555–3559. doi: 10.1093/nar/20.14.3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Heckert LL, Schultz K, Nilson JH. The cAMP response elements of the human α subunit gene bind similar proteins in trophoblasts and gonadotropes but have distinct functional sequence requirements. J Biol Chem. 1996;271:31650–31656. doi: 10.1074/jbc.271.49.31650. [DOI] [PubMed] [Google Scholar]


