Abstract
We have previously reported the cloning and characterization of the MP1 gene in Penicillium marneffei and the AFMP1 gene in Aspergillus fumigatus and their use for serodiagnosis of penicilliosis and aspergilloma and invasive aspergillosis, respectively. In this study, we describe the cloning of the AFLMP1 gene, which encodes the homologous antigenic cell wall protein in Aspergillus flavus, the most common Aspergillus species associated with human disease in our locality and in other Asian countries and the second most common Aspergillus species associated with human disease in Western countries. AFLMP1 codes for a protein, Aflmp1p, of 273 amino acid residues, with a few sequence features that are present in Mp1p and Afmp1p, the homologous antigenic cell wall proteins in P. marneffei and A. fumigatus, respectively, as well as several other cell wall proteins of Saccharomyces cerevisiae and Candida albicans. It contains a serine- and threonine-rich region for O glycosylation, a signal peptide, and a putative glycosylphosphatidylinositol attachment signal sequence. Specific anti-Aflmp1p antibody was generated with recombinant Aflmp1p protein purified from Escherichia coli to allow further characterization of Aflmp1p. Indirect immunofluorescence analysis indicated that Aflmp1p is present on the surface of the hyphae of A. flavus. Finally, it was observed that patients with aspergilloma and invasive aspergillosis due to A. flavus develop a specific antibody response against Aflmp1p. This suggested that the recombinant protein and its antibody may be useful for serodiagnosis in patients with aspergilloma or invasive aspergillosis, and the protein may represent a good cell surface target for host humoral immunity.
Since the last decade, Aspergillus species have been gaining prominence as opportunistic pathogens. In immunocompetent hosts, Aspergillus species rarely causes serious illnesses, except for aspergilloma in patients with preexisting chronic lung diseases. On the other hand, invasive aspergillosis is one of the most important infectious causes of mortality in patients with hematological malignancies and bone marrow transplant (BMT) recipients, with an incidence of 6% in a recent study on 230 BMT recipients (25). Furthermore, up to 2.5% of solid organ transplant recipients, 12% of patients with AIDS, and 40% of patients with chronic granulomatous disease could be affected by this infection (10). The mortality rate in patients with invasive aspergillosis with pulmonary involvement and persistent neutropenia was 95% (8). Of all the known Aspergillus species, Aspergillus flavus is the most common one associated with human disease in our locality and in other Asian countries, and is the second most common species associated with human disease in Western countries (10, 25).
The successful management of invasive aspergillosis is hampered by difficulties in establishing diagnosis. The “gold standard” for making a diagnosis is to obtain a positive culture of Aspergillus and to demonstrate histological evidence of mycelial invasion from tissue biopsy. Due to the very sick nature of these patients and often the presence of bleeding diathesis, tissue biopsy is often not possible or not acceptable to patients. For serological diagnosis of invasive aspergillosis, although commercial kits for antigen detection assays with monoclonal antibody against the galactomannan antigen extract are available for clinical use, no antigen detection kit based on recombinant antigens of Aspergillus is presently available. Due to less cross-reaction, recombinant antibody and antigen detection tests may offer a higher specificity and reproducibility. Moreover, recombinant antigens and generated antibodies are easy to standardize.
We have previously described the cloning and characterization of a highly antigenic cell wall mannoprotein in Penicillium marneffei (Mp1p) and its homologue in Aspergillus fumigatus (Afmp1p) (2, 26). We have also shown that enzyme-linked immunosorbent assays based on recombinant Mp1p and Afmp1p are very useful for serodiagnosis of penicilliosis and A. fumigatus aspergillosis, respectively (3, 4, 7, 24). Since there is no recombinant antigen-based kit for serodiagnosis of A. flavus infections, it would be logical to search for the homologue of Mp1p and Afmp1p in A. flavus and examine its potential for serodiagnostic purposes.
In this study, we report the cloning of the AFLMP1 gene, which encodes an antigenic cell wall protein of A. flavus (Aflmp1p). Sequence analysis reveals that Aflmp1p is homologous to Mp1p and Afmp1p. Indirect immunofluorescent microscopic study indicates that Aflmp1p is specifically located in the cell walls of A. flavus. Finally, our results show that patients with invasive A. flavus infections develop high levels of specific antibody against Aflmp1p, suggesting that Aflmp1p may represent a good cell surface target of host humoral immunity.
The A. flavus strain isolated from a BMT recipient was used throughout the study. A 1-ml suspension of conidia obtained by flushing the surface of A. flavus colonies grown on Sabouraud agar at 37°C for 4 days was used to inoculate 25 ml of brain heart infusion medium (Oxoid, Hampshire, United Kingdom) in a 500-ml conical flask at 37°C in a gyratory shaker. A 2-day-old culture was harvested for DNA or RNA extraction.
A. flavus genomic DNA extraction was performed by using the DNeasy Plant Maxi kit in accordance with the manufacturer's instructions (Qiagen, Hilden, Germany). A 240-bp fragment of the putative AFLMP1 gene was amplified by using degenerate PCR primers LPW151 (5′-ANCTCATCTCCAAGAAGGAC-3′) and LPW153 (5′-GGCGTCNANACCCTTCTG-3′) (Gibco BRL), which were designed by pairwise alignment of the AFMP1 gene of A. fumigatus and the homologous gene in Aspergillus nidulans (obtained from a BLAST search of the A. nidulans EST database at www.genome.ou.edu and by assembly of contigs r5e08a1.r1 and z4 h04a1.r1). The PCR mixture (50 μl) contained A. flavus DNA, PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, and 2 mM MgCl2), 200 μM each dNTP, and 1.25 U of AmpliTaq Gold (Applied Biosystem). The mixtures were amplified in 40 cycles of 94°C for 1 min, 45°C for 1 min, and 72°C for 1 min in an automated thermal cycler (Perkin-Elmer Cetus, Gouda, The Netherlands). Distilled water was used as the negative control. Ten microliters of each amplified product was electrophoresed in 1.0% (wt/vol) agarose gel, with a molecular size marker (ΦX-174 DNA HaeIII digest; Roche) being electrophoresed in parallel. Electrophoresis in Tris-borate-EDTA buffer was performed at 100 V for 1.5 h. The gel was stained with ethidium bromide (0.5 μg/ml) for 15 min, rinsed, and photographed under UV light illumination.
The 240-bp PCR product was gel purified by using the QIAquick PCR purification kit (Qiagen, Hilden, Germany). Both strands of the PCR product were sequenced twice by using the PCR primers LPW151 and LPW153 with an ABI 377 automated sequencer used in accordance with the manufacturer's instructions (Perkin-Elmer, Foster City, Calif.).
The sequences upstream and downstream of the 240-bp fragment were obtained by rapid amplification of cDNA ends (RACE) by using the 5′/3′ RACE kit, with LPW168 (5′-GGAGATGGCCCAGAGCCTCTCCGCTGGT-3′) and LPW171 (5′-ACCAGCGGAGAGGCTCTGGGCAATCTCC-3′) being used as gene-specific primers in accordance with the manufacturer's instructions (Roche), and by subsequent DNA sequencing by using LPW168, LPW171, LPW441 (5′-ATGAGATTCTCCGCTATC-3′), and LPW442 (5′-TTAGACAGCGACGGCAAT-3′) as the sequencing primers.
The phylogenetic relationships of Mp1p (2), Afmp1p (26), Aflmp1p, and the homologous region in A. nidulans were determined by using the neighbor-joining method with Clustal X (20). A total of 93 amino acid positions were included in the analysis.
To produce a fusion plasmid for protein purification, primers LPW230 (5′-GGAATTCCACCCCCCTCGTTGAG-3′) and LPW231 (5′-CCGCTCGAGTTAGACAGCGACGGC-3′) were used to amplify the AFLMP1 gene from cDNA. The sequence coding for amino acid residues 18 to 273 of Aflmp1p was amplified and cloned into the BamHI and EcoRI sites of expression vector pGEX-5X-3 in frame and downstream of the glutathione S-transferase (GST) coding sequence. The GST-Aflmp1p fusion protein was expressed and Aflmp1p was purified with a GST gene fusion system (Pharmacia) in accordance with the manufacturer's instructions.
To produce polyclonal guinea pig antibody, 10 ml of mycelial sediment from a 2-day-old culture was washed three times in phosphate-buffered saline (PBS) (13.7 mM sodium chloride, 0.27 mM potassium chloride, 1 mM phosphate buffer [pH 7.4]) and suspended in PBS with 0.05% phenol at a turbidity of McFarland standard 3. An equal volume of complete Freund's adjuvant was mixed with 500 μl of mycelial suspension and injected intramuscularly into a guinea pig's thigh. Incomplete Freund's adjuvant was used in subsequent immunizations, and a total of three inoculations were completed in 2 months.
To prepare guinea antibody specific against Aflmp1p, 70 μg of purified Aflmp1p recombinant protein was mixed with an equal part of complete Freund's adjuvant and injected intramuscularly into a guinea pig's thigh. Incomplete Freund's adjuvant was used in subsequent injections. Serum samples were taken 2 weeks after the third injection.
Aflmp1p samples were run on a sodium dodecyl sulfate (SDS)-10% polyacrylamide gel and electroblotted onto a nitrocellulose membrane (Bio-Rad, Hercules, Calif.). The blot was cut into strips, and the strips were incubated with preimmunized guinea pig serum, serum of a guinea pig immunized with A. flavus, sera from two patients with computed tomography- and culture-documented A. flavus aspergilloma, sera of two acute myeloid leukemia patients with culture- and histology-documented A. flavus invasive aspergillosis, sera of two patients with candidemia, sera of two patients with P. marneffei infection, or sera from six normal blood donors. All guinea pig sera were diluted at 1:4,000, and human sera were diluted at 1:500. Antigen-antibody interaction was detected with the ECL fluorescence kit (Amersham Life Science, Buckinghamshire, United Kingdom).
Whole-cell A. flavus lysate was run on an SDS-10% polyacrylamide gel and electroblotted onto a nitrocellulose membrane (Bio-Rad). The blot was cut into strips, and the strips were incubated with preimmunized guinea pig serum or serum of a guinea pig immunized with Aflmp1p diluted at 1:4,000. Antigen-antibody interaction was detected with the ECL fluorescence kit (Amersham Life Science).
In the indirect immunofluorescent assay, A. flavus germinating spores were harvested and washed twice in PBS. Cells were deposited on Teflon-coated slides, air dried, and fixed in cold acetone for 10 min. Guinea pig serum with antibodies specific against Aflmp1p were added to the fixed cells and incubated in a humidity chamber at 37°C for 60 min. A guinea pig serum with antibodies against whole-cell A. flavus and a preimmune guinea serum were used as the positive and negative controls, respectively. The cells were then washed with PBS, air dried, and incubated with affinity-purified fluorescein isothiocyanate-conjugated anti-guinea pig immunoglobulin G (Zymed) at 37°C for 60 min. The cells were mounted and observed under UV light. A positive signal of antigen-antibody interaction was indicated by the presence of apple green fluorescence.
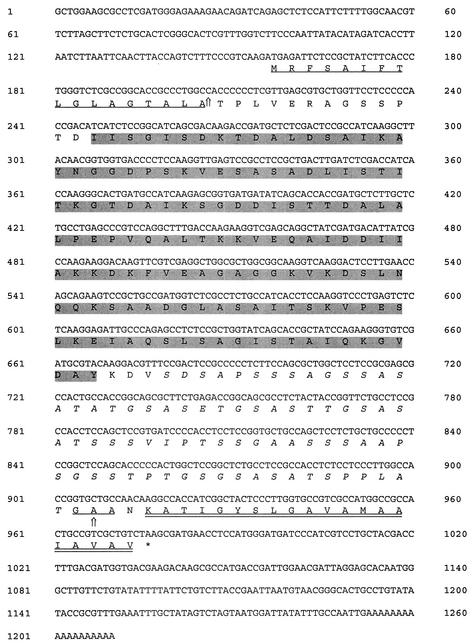
After a 240-bp fragment was amplified and sequenced by using degenerate PCR primers designed by multiple alignment of the AFMP1 gene of A. fumigatus and the homologous gene in A. nidulans (encoding the putative protein Anmp1p), the complete sequence of the gene was obtained by rapid amplification of cDNA ends. Bidirectional DNA sequencing revealed that the cDNA contained a single open reading frame encoding 273 amino acid residues with a predicted molecular mass of 26.3 kDa. The DNA and predicted protein sequences are shown in Fig. 1.
FIG. 1.
DNA and amino acid sequences of Aflmp1p. AFLMP1 cDNA contains a single open reading frame encoding 273 amino acid residues with a predicted molecular mass of 26.3 kDa. The N-terminal cleavable signal peptide of 17 amino acids is underlined. The C-terminal cleavable GPI signal peptide of 20 amino acids is double underlined. A 75-amino-acid serine- and threonine-rich region is in italics, indicating that this protein may have many O glycosylation sites. The 141-amino-acid region CR4, showing homology to CR1 and CR2 of Mp1p in P. marneffei, CR3 of Afmp1p in A. fumigatus, and CR5 of the homologous protein in A. nidulans, is shaded in gray.
Protein sequence analysis revealed that this protein has several features that are similar to the Mp1p of P. marneffei, Afmp1p of A. fumigatus, and several other fungal cell wall proteins (1, 11, 12, 15-19, 22) (Fig. 1 and 2A). Similar to Mp1p and Afmp1p, this protein has a putative N-terminal signal peptide found in most secretory proteins (18, 23). It also has a putative C-terminal glycosylphosphatidylinositol (GPI) membrane attachment signal sequence that is commonly used for cytoplasmic membrane attachment (5, 21) and has been implicated in fungal cell wall assembly (9). After processing, this protein should have 256 amino acid residues with a predicted molecular mass of 24.6 kDa as a polypeptide. The gene and protein were named AFLMP1 (for Aspergillus flavus mannoprotein 1) and Aflmp1p, respectively. Aflmp1p is expected to be a glycosylated protein, since it has potential O glycosylation sites. Aflmp1p contains a 75-amino-acid serine- and threonine-rich region in its C-terminal half as a site for O glycosylation, similar to Mp1p, Afmp1p, and other yeast cell wall proteins of S. cerevisiae and C. albicans (1). However, unlike Mp1p, which contains two potential N glycosylation sites, Aflmp1p does not contain any potential N glycosylation sites.
FIG. 2.
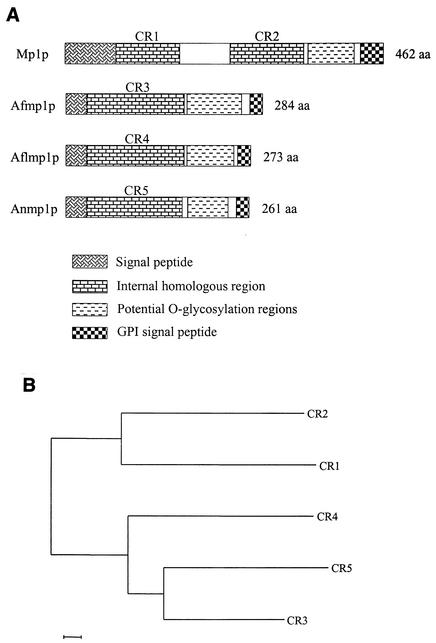
(A) Diagrammatic representation of Mp1p of P. marneffei, Afmp1p of A. fumigatus, Aflmp1p of A. flavus, and the homologous protein of A. nidulans, showing the regions representing the signal peptide, internal homologous regions (CR1 to CR5), potential O glycosylation regions, and GPI signal peptide. (B) Phylogenetic tree based on the amino acid sequences of CR1 and CR2 in P. marneffei, CR3 in A. fumigatus, CR4 in A. flavus, and CR5 in A. nidulans. The tree was inferred from amino acid data by the neighbor-joining method. The scale bar indicates the estimated number of substitutions per 50 amino acids by using the Jukes-Cantor correction.
Sequence analysis further revealed that the only region of Aflmp1p homologous to Mp1p, Afmp1p, and Anmp1p is a 141-amino-acid residue region (CR4) situated upstream to the N terminus of the serine- and threonine-rich region (Fig. 2A and B). This sequence shares similarity with two distinct but conserved regions in Mp1p, hereby named conserved regions 1 and 2 (CR1 and CR2). The corresponding regions in Afmp1p and Anmp1p were named CR3 and CR5, respectively. Within the CR4 141-amino-acid region, the sequence identities between Aflmp1p and CR3 of Afmp1p and CR5 of Anmp1p are 63% and 57%, respectively, which are higher than the sequence identities between Aflmp1p and CR1 and CR2 of Mp1p (41% in both cases). In comparison, CR1 and CR2 of Mp1p are 54% identical. This suggests that CR1 and CR2 probably evolved from a duplication event after the separation of Aspergillus and Penicillium.
To produce recombinant Aflmp1p protein, the AFLMP1 sequence minus the signal peptide (codon 18 to 273) was amplified by PCR and cloned in frame with GST in expression plasmid pGEX-5X-3. The GST-Aflmp1p fusion protein was expressed in E. coli, and Aflmp1p was subsequently purified. The purified Aflmp1p was separated on an SDS gel followed by Coomassie blue staining. After purification, a single band was visible on the gel at about 30 kDa, consistent with the expected molecular mass of 24.6 kDa for Aflmp1p.
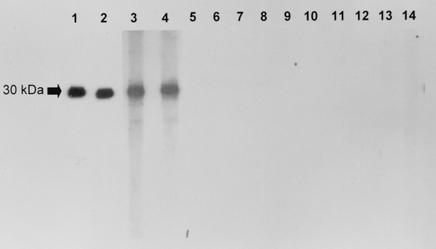
Western blot analysis showed that recombinant Aflmp1p reacted strongly with serum of a guinea pig immunized with A. flavus (Fig. 3, lane 1) or Aflmp1p (Fig. 3, lane 2) but not with preimmunized guinea pig serum (Fig. 3, lane 3), indicating the Aflmp1p is highly immunogenic in guinea pigs.
FIG. 3.
Western blot analysis of purified Aflmp1p of A. flavus by using guinea pig sera. Strong antigen-antibody interaction was detected with sera of guinea pigs immunized with A. flavus (lane 1) or Aflmp1p (lane 2), but not with preimmune guinea pig serum (lane 3).
A band of about 70 kDa was observed when sonicated cell lysate was reacted with guinea pig serum after Aflmp1p immunization (Fig. 4, lane 1). This may represent glycosylated Aflmp1p in A. flavus.
FIG. 4.
Western blot analysis of Aflmp1p in sonicated A. flavus cell lysate by using guinea pig sera. A band of about 70 kDa (arrow) was detected with the sera of guinea pigs immunized with Aflmp1p (lane 1) but not with preimmune guinea pig serum (lane 2).
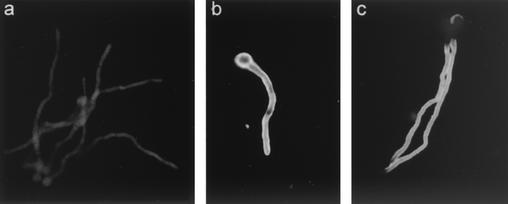
To examine the distribution of Aflmp1p in A. flavus, fixed sections of A. flavus cells were examined by indirect immunofluorescence light microscopy with guinea pig anti-Aflmp1p antibody. Indirect immunofluorescence showed that Aflmp1p was specifically located on the surface of A. flavus (Fig. 5c). A negative control with preimmune guinea pig serum showed no staining (Fig. 5a).
FIG. 5.
Indirect immunofluorescence staining of A. flavus hyphae with preimmune guinea pig serum (a), guinea pig anti-A. flavus whole-cell antibody (b), and guinea pig anti-Afmp1p antibody (c). Apple green fluorescence indicates a positive signal for antigen-antibody interaction.
Western blot analysis showed that recombinant Aflmp1p reacted specifically with sera from the four patients with aspergilloma or invasive aspergillosis (Fig. 6, lanes 1 to 4). No specific reaction was seen between Aflmp1p and sera from either healthy blood donors (Fig. 6, lanes 5 to 10), the two patients with documented C. albicans fungemia (Fig. 6, lanes 11 and 12), or the two patients with documented P. marneffei infections (Fig. 6, lanes 13 and 14).
FIG. 6.
Western blot analysis of purified Aflmp1p of A. flavus by using human sera. Strong antigen-antibody interaction was detected with the sera of two patients with aspergilloma (lanes 1 and 2) and two patients with invasive aspergillosis (lanes 3 and 4). Controls were sera from healthy blood donors (lanes 5 to 10) and from patients infected with C. albicans (lanes 11 and 12) and P. marneffei (lanes 13 and 14).
In this study, we describe the cloning of the complete open reading frame of the AFLMP1 gene of A. flavus. DNA sequence analysis of AFLMP1 revealed that it encodes a protein (Aflmp1p) of 273 amino acid residues, of which the 141-amino-acid region CR4 shows homology to CR1 and CR2 of Mp1p in P. marneffei, CR3 of Afmp1p in A. fumigatus, and CR5 of the homologous protein in A. nidulans. Examination of the Aflmp1p sequence identified several features that are common to Mp1p, Afmp1p, and cell wall proteins in other fungi (1, 11, 15 to 19, 22). These include an N-terminal signal peptide (23), a serine- and threonine-rich region in the C-terminal half that acts as a site for O glycosylation (1), and a C-terminal GPI membrane attachment signal, which is utilized by many proteins to anchor themselves to eukaryotic cell membranes (5, 21). Unlike Mp1p, Aflmp1p does not possess any potential N glycosylation sites, but it has a GAA site for cleavage by phospholipase. Like other surface proteins, on release from the cell membrane into the cell wall, Aflmp1p may fulfill many important physiological functions, such as cell-cell recognition, cell adhesion, receptor functions, and transport of ion and nutrients.
Indirect immunofluorescence study with anti-Aflmp1p antibody reveals that Aflmp1p is specifically located in the cell wall of A. flavus. The similarity in both the anatomical localization and functional motifs among Aflmp1p, Afmp1p, Mp1p, and other fungal cell wall proteins may imply that similar cell wall proteins may be present in other fungi. Since it has been shown previously that Mp1p and Afmp1p—and in this study, Aflmp1p—are important antigenic proteins in P. marneffei, A. fumigatus, and A. flavus and are useful for the diagnosis of the corresponding infections, the cloning of similar genes in other pathogenic fungi may be very rewarding in helping the serological diagnosis of other fungal infections.
The cloning of AFLMP1 would have direct implications for the laboratory diagnosis of A. flavus infections. Since we have shown in the present study that patients with culture-documented A. flavus aspergilloma and some with A. flavus invasive aspergillosis, but not normal blood donors or patients with C. albicans or P. marneffei infections, possess antibodies against Aflmp1p, an enzyme-linked immunosorbent assay with purified Aflmp1p or antibody against Aflmp1p may enhance the sensitivity and specificity of the serological tests for antibody and antigen detection, respectively, in patients with A. flavus infections.
Besides laboratory diagnosis, we speculate that Aflmp1p may have the potential to be used as a vaccine in patients at high risk of developing invasive A. flavus infections. From our results, Aflmp1p was shown to be closely associated with humoral immunity, and antibodies have been suggested to be important against certain extracellular opportunistic fungi (6, 13, 14); immunization could be administered through the mucosal route to stimulate the production of secretory immunoglobulin A. Furthermore, elevation of the antibody response might lead to lysis of the mold by stimulating the complement pathway and might facilitate phagocytosis of the mold by opsonization.
Nucleotide sequence accession number.
The nucleotide sequence of the AFLMP1 gene has been deposited with GenBank under accession no. AF461762.
Acknowledgments
This work was partly supported by the Research Grant Council Grant, AIDS Trust Fund, University Development Fund, and Committee of Research and Conference Grant, The University of Hong Kong.
REFERENCES
- 1.Bailey, D. A., P. J. F. Feldmann, M. Bovey, N. A. R. Gow, and A. J. P. Brown. 1996. The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J. Bacteriol. 178:5353-5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao, L., C. M. Chan, C. Lee, S. S. Wong, and K. Y. Yuen. 1998. MP1 encodes an abundant and highly antigenic cell wall mannoprotein in the pathogenic fungus Penicillium marneffei. Infect. Immun. 66:966-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao, L., D. L. Chen, C. Lee, C. M. Chan, K. M. Chan, N. Vanittanakom, D. N. Tsang, and K. Y. Yuen. 1998. Detection of specific antibodies to an antigenic mannoprotein for diagnosis of Penicillium marneffei penicilliosis. J. Clin. Microbiol. 36:3028-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao, L., K. M. Chan, D. Chen, N. Vanittanakom, C. Lee, C. M. Chan, T. Sirisanthana, D. N. Tsang, and K. Y. Yuen. 1999. Detection of cell wall mannoprotein Mp1p in culture supernatants of Penicillium marneffei and in sera of penicilliosis patients. J. Clin. Microbiol. 37:981-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caras, I. W., and G. N. Weddell. 1989. Signal peptide for protein secretion directing glycophospholipid membrane anchor attachment. Science 243:1196-1198. [DOI] [PubMed] [Google Scholar]
- 6.Cassone, A., S. Conti, F. De Bernardis, and L. Polonelli. 1997. Antibodies, killer toxins and antifungal immunoprotection: a lesson from nature? Immunol. Today 18:164-169. [DOI] [PubMed] [Google Scholar]
- 7.Chan, C. M., P. C. Woo, A. S. Leung, X. Y. Che, and L. Cao. 2002. Detection of specific antibodies to an antigenic mannoprotein for serodiagnosis of Aspergillus fumigatus aspergillosis. J. Clin. Microbiol. 40:2041-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denning, D. W., and D. A. Stevens. 1990. Antifungal and surgical treatment of invasive aspergillosis: review of 2,121 published cases. Rev. Infect. Dis. 12:1147-1201. [DOI] [PubMed] [Google Scholar]
- 9.De Nobel, H., and P. N. Lipke. 1994. Is there a role for GPIs in yeast cell-wall assembly? Trends Cell Biol. 4:42-45. [DOI] [PubMed] [Google Scholar]
- 10.Latge, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lipke, P. N., D. Wojciechowicz, and J. Kurjan. 1989. AG alpha 1 is the structural gene for the Saccharomyces cerevisiae alpha-agglutinin, a cell surface glycoprotein involved in cell-cell interactions during mating. Mol. Cell. Biol. 9:3155-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maretens, J., J. Verhaegen, H. Demuynch, P. Brock, G. Verhoef, P. Vandenberghe, J. Van Eldere, L. Verbist, and M. Boogaerts. 1999. Autopsy-controlled prospective evaluation of serial screening for circulating galactomannan by a sandwich enzyme-linked immunosorbent assay for hematological patients at risk for invasive aspergillosis. J. Clin. Microbiol. 37:3223-3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matthews, R., S. Hodgetts, and J. Burnie. 1995. Preliminary assessment of a human recombinant antibody fragment to hsp90 in murine invasive candidiasis. J. Infect. Dis. 171:1668-1671. [DOI] [PubMed] [Google Scholar]
- 14.Matthews, R. C., J. P. Burnie, D. Howat, T. Rowland, and F. Walton. 1991. Autoantibody to heat-shock protein 90 can mediate protection against systemic candidosis. Immunology 74:20-24. [PMC free article] [PubMed] [Google Scholar]
- 15.Moukadiri, I., J. Armero, A. Abad, R. Sentandreu, and J. Zueco. 1997. Identification of a mannoprotein present in the inner layer of the cell wall of Saccharomyces cerevisiae. J. Bacteriol. 179:2154-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nuoffer, C., P. Jeno, A. Conzelmann, and H. Riezman. 1991. Determinants for glycophospholipid anchoring of the Saccharomyces cerevisiae GAS1 protein to the plasma membrane. Mol. Cell. Biol. 11:27-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roy, A., C. F. Lu, D. L. Marykwas, P. N. Lipke, and J. Kurjan. 1991. The AGA1 product is involved in cell surface attachment of the Saccharomyces cerevisiae cell adhesion glycoprotein a-agglutinin. Mol. Cell. Biol. 11:4196-4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saporito-Irwin, S. M., C. E. Birse, P. S. Sypherd, and W. A. Fonzi. 1995. PHR1, a pH-regulated gene of Candida albicans, is required for morphogenesis. Mol. Cell. Biol. 15:601-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teunissen, A. W., E. Holub, J. van der Hucht, J. A. van den Berg, and H. Y. Steensma. 1993. Sequence of the open reading frame of the FLO1 gene from Saccharomyces cerevisiae. Yeast 9:423-427. [DOI] [PubMed] [Google Scholar]
- 20.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Udenfriend, S., and K. Kodukula. 1995. How glycosylphosphatidylinositol-anchored membrane proteins are made. Annu. Rev. Biochem. 64:563-591. [DOI] [PubMed] [Google Scholar]
- 22.van der Vaart, J. M., L. H. Caro, J. W. Chapman, F. M. Klis, and C. T. Verrips. 1995. Identification of three mannoproteins in the cell wall of Saccharomyces cerevisiae. J. Bacteriol. 177:3104-3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Heijne, G. 1986. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 14:4683-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woo, P. C., C. M. Chan, A. S. Leung, S. K. Lau, X. Y. Che, S. S. Wong, L. Cao, and K. Y. Yuen. 2002. Detection of cell wall galactomannoprotein Afmp1p in culture supernatants of Aspergillus fumigatus and in sera of aspergillosis patients. J. Clin. Microbiol. 40:4382-4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuen, K. Y., P. C. Woo, M. S. Ip, R. H. Liang, E. K. Chiu, H. Siau, P. L. Ho, F. F. Chen, and T. K. Chan. 1997. Stage-specific manifestation of mold infections in bone marrow transplant recipients: risk factors and clinical significance of positive concentrated smears. Clin. Infect. Dis. 25:37-42. [DOI] [PubMed] [Google Scholar]
- 26.Yuen, K. Y., C. M. Chan, K. M. Chan, P. C. Woo, X. Y. Che, A. S. Leung, and L. Cao. 2001. Characterization of AFMP1: a novel target for serodiagnosis of aspergillosis. J. Clin. Microbiol. 39:3830-3837. [DOI] [PMC free article] [PubMed] [Google Scholar]