Abstract
The primary purpose of the present study was to compare the microbial profiles of the tongue dorsa of healthy subjects and subjects with halitosis by using culture-independent molecular methods. Our overall goal was to determine the bacterial diversity on the surface of the tongue dorsum as part of our ongoing efforts to identify all cultivable and not-yet-cultivated species of the oral cavity. Tongue dorsum scrapings were analyzed from healthy subjects with no complaints of halitosis and subjects with halitosis, defined as an organoleptic score of 2 or more and volatile sulfur compound levels greater than 200 ppb. 16S rRNA genes from DNA isolated from tongue dorsum scrapings were amplified by PCR with universally conserved bacterial primers and cloned into Escherichia coli. Typically, 50 to 100 clones were analyzed from each subject. Fifty-one strains isolated from the tongue dorsa of healthy subjects were also analyzed. Partial sequences of approximately 500 bases of cloned inserts from the 16S rRNA genes of isolates were compared with sequences of known species or phylotypes to determine species identity or closest relatives. Nearly complete sequences of about 1,500 bases were obtained for potentially novel species or phylotypes. In an analysis of approximately 750 clones, 92 different bacterial species were identified. About half of the clones were identified as phylotypes, of which 29 were novel to the tongue microbiota. Fifty-one of the 92 species or phylotypes were detected in more than one subject. Those species most associated with healthy subjects were Streptococcus salivarius, Rothia mucilaginosa, and an uncharacterized species of Eubacterium (strain FTB41). Streptococcus salivarius was the predominant species in healthy subjects, as it represented 12 to 40% of the total clones analyzed from each healthy subject. Overall, the predominant microbiota on the tongue dorsa of healthy subjects was different from that on the tongue dorsa of subjects with halitosis. Those species most associated with halitosis were Atopobium parvulum, a phylotype (clone BS095) of Dialister, Eubacterium sulci, a phylotype (clone DR034) of the uncultivated phylum TM7, Solobacterium moorei, and a phylotype (clone BW009) of Streptococcus. On the basis of our ongoing efforts to obtain full 16S rRNA sequences for all cultivable and not-yet-cultivated species that colonize the oral cavity, there are now over 600 species.
Halitosis, or oral malodor, is a common complaint of up to one-third of the general population and a large concern for the many individuals whom it affects (6, 19). Halitosis can arise from a variety of sources including the sinuses, gastrointestinal tract, ingested food, lungs, and, most frequently, the oral cavity. Oral production of malodorous substances is most commonly associated with by-products of bacterial metabolic degradation and occurs on oral surfaces, in periodontal pockets, and especially on the dorsal tongue surface. These products result from microbial fermentation of proteins, peptides, and mucins found in saliva, blood, gingival crevicular fluid, lysed neutrophils, desquamated epithelial cells, and any residual food retained on the oral surfaces (16). The most conspicuous malodorous compounds are termed volatile sulfur compounds (VSCs), with hydrogen sulfide, methyl mercaptan, and dimethyl sulfide accounting for roughly 90% of the VSCs (32). Many oral bacteria, especially gram-negative anaerobic species found in the subgingival plaque, produce a diverse array of malodorous compounds as by-products of their metabolism including VSCs and short-chain organic acids such as valeric acid, butyric acid, putrescine, and skatole (16). Species that produce such malodorous compounds include Treponema denticola, Porphyromonas gingivalis, Prevotella intermedia, Tannerella forsythensis, Porphyromonas endodontalis, and Eubacterium species (25, 26).
Halitosis has been correlated with the presence and severity of periodontal disease and by the amount of coating on the tongue (3, 20, 21). Various methods of detecting and quantifying oral odor have been proposed, including organoleptic odor rating schemes (smelling the breath) (27, 29, 35) and analytical techniques involving gas chromatography, mass spectrometry, and cryo-osmoscopy. Rosenberg and colleagues (28) have reported on the use of a portable sulfide monitor called a Halimeter (Interscan, Chatsworth, Calif.) to quantitate the levels of VSCs in mouth breath and have shown that these levels significantly correlate with the measurements made by organoleptic odor rating schemes. In individuals with oral malodor, tongue-coating samples have been shown to hydrolyze N-benzoyl-dl-arginine-2-naphthylamide (BANA) (2, 4, 11, 21). Since the BANA test detects an arginine hydrolase produced by proteolytic bacteria, this test provides additional information on the bacterial flora associated with malodor.
Effective treatments of oral malodor consist of reducing the bacterial load on the tongue and teeth through twice-daily tooth brushing with fluoride toothpaste and daily tongue debridement with a toothbrush or other mechanical device, alone or in combination with the use of antimicrobial mouth rinses such as chlorhexidine (4, 11, 33, 36).
Although the bacteria of the tongue have been implicated as a major source of odor production in subjects with halitosis, the bacterial composition of the tongue is still not well characterized. Studies of cultivable tongue microbiota have been limited by the difficulties of in vitro growth techniques, the low percentage of recovery of total organisms, and the inadequacy of microbial identification (7, 10). For example, Kazor et al. (10) were able to recover only up to 30% of the viable microbial count using a growth medium supplemented with human blood and saliva. These findings suggest that much of the tongue microbiota has not yet been cultivated, necessitating the use of molecular approaches to better characterize the tongue microflora.
Using culture-independent molecular methods, we had previously detected over 500 species or phylotypes in the subgingival plaque of healthy subjects and subjects with periodontal diseases (22), dental plaque in children with rampant caries (1), and noma (24). Other investigators have used similar techniques to determine the bacterial diversity of saliva (31), dentoalveolar abscesses (5), and subgingival plaque of a subject with gingivitis (12). The purpose of this study was to determine the bacterial diversity on the tongue dorsum and to compare the predominant bacteria (including not-yet-cultivated species) that are present on the surface of the tongue dorsa of subjects with and without oral malodor.
MATERIALS AND METHODS
Subject population.
Halitosis in healthy adult subjects with self-reported halitosis (n = 6) was confirmed by an organoleptic rating of 2 or more and VSC measurements greater than 200 ppb (Table 1). BANA hydrolysis (BANA Test; OraTec Corp, Manassas, Va.) was also determined (Table 1), since positive scores by the BANA test have previously been associated with a component of oral malodor (2, 4, 11, 21). Five healthy subjects without evidence of halitosis were used as controls.
TABLE 1.
Clinical parameters of study population
| Odor status | Subject clone | Organoleptic scorea | Volatile sulfur compound concn (ppb)b | Tongue coating | BANA test result for tongue |
|---|---|---|---|---|---|
| Malodor | M1 | 3 | 350 | + | +/− |
| M2 | 4 | 411 | + | − | |
| M3 | 3 | 452 | + | +/− | |
| M4 | 4 | 642 | + | +/− | |
| M5 | 4 | 346 | + | − | |
| M6 | 4 | 749 | + | + | |
| Healthy | H1 | 1 | 87 | − | − |
| H2 | 2 | 160 | + | +/− | |
| H3 | 2 | 144 | + | − | |
| H4 | 2 | 113 | − | − | |
| H5 | 2 | 132 | − | +/− |
A score of ≥2 is associated with malodor.
A level of >200 ppb is associated with malodor (Halimeter; Interscan Corporation).
Sample collection.
Samples were collected by scraping the tongue surface from the vallate papilla area to the anterior tongue border with a sterile wooden tongue depressor. All tongue scrapings were used directly for clonal analysis as described below. Strains were isolated from some samples on either ETSA medium (enriched Trypticase soy agar to which hemin, menadione, nitrate, and lactate were added to support growth of oral species) or MM10 medium (1/10 dilution of ETSA medium) (17).
Sample lysis.
Pure cultures or tongue scrapings were directly suspended in 50 μl of 50 mM Tris buffer (pH 7.6)-1 mM EDTA (pH 8)-0.5% Tween 20. Proteinase K (200 μg/ml) was added. The samples were then heated at 55°C for 2 h. Proteinase K was inactivated by heating at 95°C for 5 min.
Amplification of 16S rRNA cistrons by PCR and purification of PCR products.
The 16S rRNA genes (rDNAs) were amplified under standard conditions with a universal primer set (forward primer, 5′-GAG AGT TTG ATY MTG GCT CAG; reverse primer, 5′- GAA GGA GGT GWT CCA RCC GCA) (22). Primers were synthesized commercially (Operon Technologies, Alameda, Calif.). PCR was performed in thin-walled tubes with a Perkin-Elmer 9700 thermocycler. One microliter of the DNA template was added to a reaction mixture (final volume, 50 μl) containing 20 pmol of each primer, 40 nmol of deoxynucleoside triphosphates, and 1 U of Taq 2000 polymerase (Stratagene, La Jolla, Calif.) in buffer containing Taqstart antibody (Sigma Chemical Co.). In a hot-start protocol, the samples were preheated at 95°C for 8 min, followed by amplification under the following conditions: denaturation at 95°C for 45 s, annealing at 60°C 45 s, and elongation for 1.5 min, with an additional 5 s for each cycle. A total of 30 cycles were performed; this was then followed by a final elongation step at 72°C for 10 min. The results of PCR amplification were examined by electrophoresis in a 1% agarose gel. DNA was stained with ethidium bromide and visualized under short-wavelength UV light.
Cloning procedures.
Cloning of PCR-amplified DNA was performed with the TOPO TA cloning kit (Invitrogen, San Diego, Calif.) according to the instructions of the manufacturer. Transformation was done with competent Escherichia coli TOP10 cells provided by the manufacturer. The transformed cells were then plated onto Luria-Bertani agar plates supplemented with kanamycin, and the plates were incubated overnight at 37°C. The colonies were then placed into 40 μl of 10 mM Tris. One microliter was used as the template to determine the correct sizes of the inserts in a PCR with an M13 (−40) forward primer and an M13 reverse primer. The sizes of the inserts (approximately 1,500 bp) were determined by PCR with flanking vector primers, followed by electrophoresis on a 1% agarose gel. Prior to sequencing of the fragments, the PCR-amplified 16S rDNA fragments were purified and concentrated with Microcon 100 (Amicon), followed by use of the QIAquick PCR purification kit (Qiagen).
16S rRNA sequencing.
Purified DNA from the PCR was sequenced with an ABI Prism cycle sequencing kit (BigDye Terminator Cycle Sequencing kit with AmpliTaq DNA Polymerase FS; Perkin-Elmer). The primers used for sequencing have been reported previously (22). Quarter dye chemistry was used with 80 μM primers and 1.5 μl of PCR product in a final volume of 20 μl. Cycle sequencing was performed with an ABI 9700 instrument, with 25 cycles of denaturation 96°C for 10 s, annealing, and extension at 60°C for 4 min. The sequencing reactions were run on an ABI 377 DNA sequencer.
16S rRNA sequencing and data analysis of unrecognized inserts.
A total of 741 clones with the insert of the correct size of approximately 1,500 bases were analyzed (typically, 50 to 100 per subject). In addition, 51 strains from healthy subjects without halitosis were analyzed. A sequence of approximately 500 bases was obtained first to determine identity or approximate phylogenetic position. Full sequences of about 1,500 bases were obtained by using five to six additional sequencing primers (22) for those species deemed novel. For identification of closest relatives, the sequences of the unrecognized inserts were compared to the 16S rRNA gene sequences of over 4,000 microorganisms in our database and over 50,000 sequences in the Ribosomal Database Project (18) and GenBank databases. Programs for data entry, editing, sequence alignment, secondary structure comparison, similarity matrix generation, and phylogenetic tree construction were written by F. E. Dewhirst (23). The similarity matrices were corrected for multiple base changes at single positions by the method of Jukes and Cantor (9). Similarity matrices were constructed from the aligned sequences by using only those sequence positions for which data were available for 90% of the strains tested. Phylogenetic trees were constructed by the neighbor-joining method of Saitou and Nei (30). TREECON, a software package for the Microsoft Windows environment, was used for the construction and drawing of evolutionary trees (34).
We are aware of the potential creation of 16S rDNA chimera molecules during the PCR (14). The percentage of chimeric inserts in 16S rRNA libraries ranged from 1 to 15%. Chimeric sequences were identified by using the Chimera check program of the Ribosomal Database Project, treeing analysis, or base signature analysis. Species identifications of the chimeras were obtained, but the partial sequences were not included in the phylogenetic analysis for tree construction.
Nucleotide sequence accession numbers.
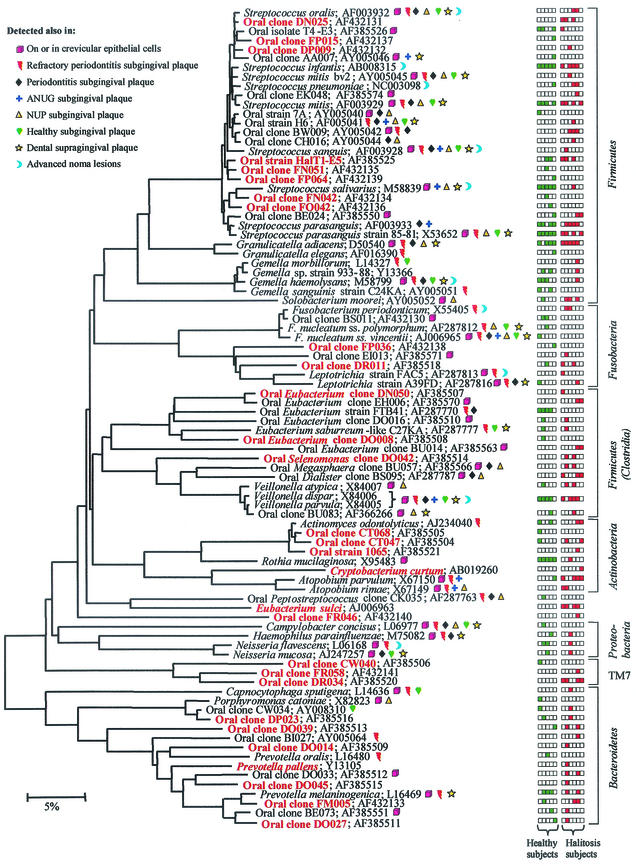
The complete 16S rDNA sequences of clones representing novel phylotypes defined in this study, the sequences of known species not reported previously, and published sequences are available for electronic retrieval from the EMBL, GenBank, and DDBJ nucleotide sequence databases under the accession numbers shown in Fig. 1.
FIG.1.
Phylogenetic tree of bacterial species or phylotypes of six phyla identified from the tongue dorsa of healthy subjects and subjects with halitosis. The information presented includes bacterial species or phylotype clone and sequence accession numbers. Novel phylotypes are definedas those taxa that are <98.5 to 99% similar in sequence comparisons to their closest relatives. Species or phylotypes detected only on the tongue dorsum and not at any other oral site are highlighted in red. Color-coordinated characters indicate health status category or some other site at which each species was identified. Bar, 5% difference in nucleotide sequence. ANUG, acute necrotizing ulcerative gingivitis; NUP, necrotizing ulcerative periodontitis.
RESULTS AND DISCUSSION
A phylogenetic tree of the prevalent species and phylotypes detected on the surface of the tongue dorsum for each of the subjects is shown in Fig. 1. Collectively, the overall bacterial diversity of the tongue dorsum is striking: 92 different bacterial taxa or phylotypes belonging to six bacterial phyla. Only 38, or about 40%, of the total number were identified as known species. Consequently, about 60% of the total were identified as phylotypes. As shown in Fig. 1, 29 of these phylotypes were unique to the tongue dorsum, in that they were not found from the sequence analysis of over 6,000 clones from other oral sites, including the subgingival plaque of healthy subjects and subjects with, periodontitis, acute necrotizing ulcerative gingivitis, and refractory periodontitis (22); the supragingival plaque of children with rampant caries (1); or advanced noma lesions (24); nor were they found on or in crevicular epithelial cells of healthy subjects and subjects with periodontitis (13). In our ongoing studies, we have detected over 300 novel phylotypes and 200 known species in oral sites. At present, we estimate that over 700 bacterial species are present in the oral cavity, over half of which we cannot presently cultivate. At the time of this publication, a list of 630 species or phylotypes of the oral cavity was compiled. The creation of a website is in progress; however, an updated version of this list can be obtained from the corresponding author.
The number of species or phylotypes that were detected in each subject ranged from 12 to 29, with 16 to 21 species or phylotypes in tongue samples from subjects without malodor and from 12 to 29 species or phylotypes in tongue samples from subjects with malodor (Table 2). The bacterial profiles for each of these subjects are depicted in the colored columns of boxes in Fig. 1. Those species most associated with health were Streptococcus salivarius, Rothia mucilaginosa (Stomatococcus mucilaginosus), and an uncharacterized, cultivable species of Eubacterium (strain FTB41) (Fig. 1 and Table 2). The 15 most prevalent species or phylotypes are listed in Table 2, where they comprise 60 to 85% of the total clones in subjects without malodor and 20 to 88% of the total clones in subjects with malodor. A prevalent species is defined as a species that was detected in at least three subjects. It is noteworthy that S. salivarius was by far the most predominant species detected in healthy subjects: in one subject (subject H1), S. salivarius represented more than 40% of the detectable species. In contrast, S. salivarius was detected in only one of the subjects with halitosis and was detected at very low levels.
TABLE 2.
Percentage of prevalent species or phylotypes on tongue dorsum
| Species or phylotype | % Clones from:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Healthy subjects (no. of clones analyzed)
|
Halitosis subjects (no. of clones analyzed)
|
||||||||||
| H1 (102) | H2 (51) | H3 (68) | H4 (65) | H5 (76) | M1 (50) | M2 (56) | M3 (46) | M4 (81) | M5 (78) | M6 (47) | |
| Atopobium parvulum | 3 | 6 | 4 | 8 | 51a | ||||||
| Cryptobacterium curtum | 19 | ||||||||||
| Dialister sp. clone BS095 | 4 | 8 | 2 | ||||||||
| Eubacterium sulci | 2 | 4 | 4 | 6 | |||||||
| Fusobacterium periodonticum | 5 | 24 | |||||||||
| Granulicatella adiacens | 8 | 21 | 5 | 14 | 4 | 2 | 7 | 7 | 4 | ||
| Neisseria flavescens | 4 | 7 | 24 | ||||||||
| Rothia mucilaginosa | 6 | 4 | 3 | 10 | 5 | 4 | |||||
| Streptococcus infantis | 1 | 2 | 9 | 6 | 5 | 4 | 4 | 20 | 1 | ||
| Streptococcus parasanguis | 27 | 18 | 7 | 32 | 8 | 18 | 9 | 2 | 7 | 9 | 4 |
| Streptococcus pneumoniae | 1 | 10 | 3 | ||||||||
| Streptococcus salivarius | 41 | 24 | 26 | 12 | 12 | 5 | 6 | ||||
| Streptococcus sp. clone BW009 | 4 | 1 | 19 | ||||||||
| Streptococcus strain HalT4-E3 | 1 | 2 | 24 | 8 | 6 | ||||||
| Veillonella parvula/V. dispar | 3 | 6 | 3 | 11 | 22 | 2 | 10 | 1 | |||
| No. of species detected | 21 | 22 | 18 | 16 | 16 | 16 | 29 | 15 | 26 | 28 | 12 |
| % of total detected | 85 | 60 | 76 | 76 | 69 | 68 | 20 | 71 | 68 | 59 | 88 |
Values in boldface indicate that the species comprised at least 10% of the tongue microbiota.
Those species most associated with halitosis were Atopobium parvulum, Eubacterium sulci, Fusobacterium periodonticum, a phylotype (clone BS095) of Dialister, a phylotype (clone BW009) of Streptococcus, a phylotype (clone DR034) of the uncultivated phylum TM7 (8), and Solobacterium moorei (Fig. 1 and Table 2). Note that in most of the samples, several species or phylotypes represented a significant proportion of the total (Table 2). Although some species were not detected in all subjects, they were the predominant species in one or more samples. For example, Cryptobacterium curtum was detected in only one of the samples from a subject with halitosis, but it represented about 20% of the clones analyzed in that subject. Other species, such as Granulicatella (Abiotrophia) adiacens, Streptococcus parasanguis, Streptococcus infantis, and Veillonella spp., were commonly detected in most of the samples (Table 2).
The tongue dorsum harbors a highly diverse, yet characteristic, bacterial population. In healthy subjects, S. salivarius was by far the predominant species. In contrast, S. salivarius was typically absent from subjects with halitosis. Although bacteria other than S. salivarius appeared to be associated with halitosis, it is not known if they are directly involved in oral malodor. Cultural studies have associated R. mucilaginosa with malodor (7) and S. salivarius and Veillonella parvula or Veillonella dispar as common healthy tongue organisms (C. E. Kazor and W. J. Loesche, unpublished data).
On the basis of our sequence analyses, the tongue dorsum possesses a unique microbiota: about one-third of the bacterial population was found only on the tongue and not in or on the surfaces of other oral sites. However, a sample of sufficient size (e.g., in a large clinical trial) is necessary to provide the power to detect differences in microbial compositions to identify more precisely those species that are associated with halitosis and health. Such studies will be accomplished by using 16S rRNA-based oligonucleotide probes in checkerboard DNA-DNA hybridization assays (1) or eventually by using oligonucleotide microarrays.
It has been suggested that the majority of cases of oral malodor are due to bacterial proteolytic activity in the mouth, such as might be measured by the BANA test (16). Since known BANA test-positive oral species typically found in subgingival plaque, i.e., P. gingivalis, T. denticola, T. forsythensis, and various Capnocytophaga species, were not detected (15), other tongue bacterial species are likely responsible for the BANA reaction of the tongue coating. Now that we have identified additional predominant bacterial species present on the tongue dorsa of individuals with halitosis, it would be of interest to examine the ability of these other cultivable species to hydrolyze the BANA substrate and to produce VSCs and other by-products that may contribute to the clinical presentation of malodor.
Acknowledgments
This study was supported by NIH grants DE12465 and DE11443 from the National Institute of Dental and Craniofacial Research.
REFERENCES
- 1.Becker, M. R., B. J. Paster, E. J. Leys, M. L. Moeschberger, S. G. Kenyon, J. L. Galvin, S. K. Boches, F. E. Dewhirst, and A. L. Griffen. 2002. Molecular analysis of bacterial species associated with early childhood caries. J. Clin. Microbiol. 40:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosy, A., G. V. Kulkarni, M. Rosenberg, and C. A. McCulloch. 1994. Relationship of oral malodor to periodontitis: evidence of independence in discrete subpopulations. J. Periodontol. 65:37-46. [DOI] [PubMed] [Google Scholar]
- 3.De Boever, E. H., M. DeUzeda, and W. J. Loesche. 1994. Relationship between volatile sulfur compounds, BANA-hydrolyzing bacteria and gingival health in patients with and without complaints of oral malodor. J. Clin. Dent. 4:114-119. [PubMed] [Google Scholar]
- 4.De Boever, E. H., and W. J. Loesche. 1995. Assessing the contribution of anaerobic microflora of the tongue to oral malodor J. Am. Dent. Assoc. 126:1384-1393. [DOI] [PubMed] [Google Scholar]
- 5.Dymock, D., A. J. Weightman, C. Scully, and W. G. Wade. 1996. Molecular analysis of microflora associated with dentoalveolar abscess. J. Clin. Microbiol. 34:537-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frexinos, J., P. Denis, H. Allemand, S. Allouche, F. Los, and G. Bonnelye. 1998. Descriptive study of digestive functional symptoms in the French general population. Gastroenterol. Clin. Biol. 22:785-791. [PubMed]
- 7.Hartley, M. G., M. A. El-Maaytah, C. McKenzie, and J. Greenman. 1996. The tongue microbiota of low odour and malodorous individuals. Microb. Ecol. Health Dis. 9:215-223. [Google Scholar]
- 8.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism, vol. 3. Academic Press, Inc., New York, N.Y. [Google Scholar]
- 10.Kazor, C. E., J. R. Flowers, J. Stoll, and W. J. Loesche. 1999. Oral malodor: defining the normal tongue flora. J. Dent. Res. 78:421. [Google Scholar]
- 11.Kozlovsky, A., D. Gordon, I. Gelernter, W. J. Loesche, and M. Rosenberg. 1994. Correlation between the BANA test and oral malodor parameters. J. Dent. Res. 73:1036-1042. [DOI] [PubMed] [Google Scholar]
- 12.Kroes, I., P. W. Lepp, and D. A. Relman. 1999. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. USA 96:14547-14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levin, I. M., C. N. Lau, S. S. Socransky, A. D. Haffajee, L. Martin, J. L. Galvin, S. K. Boches, B. J. Paster, and F. E. Dewhirst. 1999. Cultivable and uncultivable species on or in gingival epithelial cells. J. Dent. Res. 78:453. [Google Scholar]
- 14.Liesack, W., H. Weyland, and E. Stackebrandt. 1991. Potential risk of gene amplification by PCR as determined by 16S rDNA analysis of a mixed-culture of strict barophilic bacteria. Microb. Ecol. 21:191-198. [DOI] [PubMed] [Google Scholar]
- 15.Loesche, W. J., W. A. Bretz, D. Kerschensteiner, J. A. Stoll, S. S. Socransky, P. P. Hujoel, and D. E. Lopatin. 1990. Development of a diagnostic test for anaerobic periodontal infections based on plaque hydrolysis of benzoyl-dl-arginine naphthylamide. J. Clin. Microbiol. 28:1551-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loesche, W. J., and C. E. Kazor. 2002. Microbiology and treatment of halitosis. Periodontology 2000 28:256-279. [DOI] [PubMed] [Google Scholar]
- 17.Loesche, W. J., and S. A. Syed. 1973. The predominant cultivable flora of carious plaque and carious dentine. Caries Res. 7:201-216. [DOI] [PubMed] [Google Scholar]
- 18.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyazaki, H., S. Sakao, K. Yasuhiro, and T. Tadamichi. 1995. Oral malodor in the general population of Japan, p. 119-136. In M. Rosenberg (ed.), Bad breath research perspectives. Ramot Publishing, Tel Aviv, Israel.
- 20.Morita, M., and H. L. Wang. 2001. Association between oral malodor and adult periodontitis: a review. J. Clin. Periodontol. 28:813-819. [DOI] [PubMed] [Google Scholar]
- 21.Morita, M., and H. L. Wang. 2001. Relationship between sulcular sulfide levels and oral malodor in subjects with periodontal disease. J. Periodontol. 72:79-84. [DOI] [PubMed] [Google Scholar]
- 22.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paster, B. J., and F. E. Dewhirst. 1988. Phylogeny of campylobacters, wolinellas, Bacteroides gracilis, and Bacteroides ureolyticus by 16S rRNA sequencing. Int. J. Syst. Bacteriol. 38:56-62. [Google Scholar]
- 24.Paster, B. J., W. A. Falkler, Jr., C. O. Enwonwu, E. O. Idigbe, K. O. Savage, V. A. Levanos, M. A. Tamer, R. L. Ericson, C. N. Lau, and F. E. Dewhirst. 2002. Predominant bacterial species and novel phylotypes in advanced noma lesions. J. Clin. Microbiol. 40:2187-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Persson, S., R. Claesson, and J. Carlsson. 1989. The capacity of subgingival species to produce volatile sulfur compounds in human serum. Oral Microbiol. Immunol. 4:169-172. [DOI] [PubMed] [Google Scholar]
- 26.Persson, S., M. B. Edlund, R. Claesson, and J. Carlsson. 1990. The formation of hydrogen sulfide and methylmercaptan by oral bacteria. Oral Microbiol. Immunol. 5:195-201. [DOI] [PubMed] [Google Scholar]
- 27.Pitts, G., R. Pianotti, T. W. Feary, J. McGuiness, and T. Masurat. 1981. The in vivo effects of an antiseptic mouthwash on odor-producing microorganisms. J. Dent. Res. 60:1891-1896. [DOI] [PubMed] [Google Scholar]
- 28.Rosenberg, M., G. V. Kulkarni, A. Bosy, and C. A. McCulloch. 1991. Reproducibility and sensitivity of oral malodor measurements with a portable sulfide monitor. J. Dent. Res. 70:1436-1440. [DOI] [PubMed] [Google Scholar]
- 29.Rosenberg, M., I. Septon, I. Eli, R. Bar-Ness, I. Gelernter, S. Brenner, and J. Gabbay. 1991. Halitosis measurement by an industrial sulphide monitor. J. Periodontol. 62:487-489. [DOI] [PubMed] [Google Scholar]
- 30.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 31.Sakamoto, M., M. Umeda, I. Ishikawa, and Y. Benno. 2000. Comparison of the oral bacterial flora in saliva from a healthy subject and two periodontitis patients by sequence analysis of 16S rDNA libraries. Microbiol. Immunol. 44:643-652. [DOI] [PubMed] [Google Scholar]
- 32.Tonzetich, J. 1971. Direct gas chromatographic analysis of sulphur compounds in mouth air in man. Arch. Oral Biol. 16:587-597. [DOI] [PubMed] [Google Scholar]
- 33.Tonzetich, J., and S. K. Ng. 1976. Reduction of malodor by oral cleansing procedures. Oral Surg. Oral Med. Oral Pathol. 42:172-181. [DOI] [PubMed] [Google Scholar]
- 34.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 35.Yaegaki, K., and J. M. Coil. 2000. Examination, classification, and treatment of halitosis; clinical perspectives. J. Can. Dent. Assoc. 66:257-261. [PubMed] [Google Scholar]
- 36.Yaegaki, K., and K. Sanada. 1992. Biochemical and clinical factors influencing oral malodor in periodontal patients. J. Periodontol. 63:783-789. [DOI] [PubMed] [Google Scholar]