Abstract
A 5′ nuclease TaqMan PCR was developed for the quantitative detection of the periodontopathic bacteria Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. The relative numbers of bacteria were measured by the comparative threshold cycle method. This simplified method is a way of obtaining the relative quantities of these organisms from specimens and of monitoring the effect of therapy.
Periodontitis is an inflammation of the supporting tissues of the teeth (4). It is generally accepted that periodontal diseases are infectious diseases caused by oral bacteria (5). Actinobacillus actinomycetemcomitans is a nonmotile, gram-negative, capnophilic, fermentative coccobacillus that has been implicated in the etiology of localized juvenile periodontitis (12, 16, 18), while Porphyromonas gingivalis is a gram-negative, black-pigmented anaerobe that is strongly implicated as a major pathogen in adult periodontitis (14, 19). Several PCR-based systems that use oral specimens for the detection of oral bacterial infections, especially periodontitis, have been reported (1, 2, 13, 15). Most previous diagnostic systems are qualitative and are therefore unsuitable for the evaluation of treatment, as quantitative analysis is essential for monitoring the effect of therapy in treatment trials.
The TaqMan assay based on the 5′-3′ exonuclease activity of Taq polymerase has been developed for quantitative detection of DNA (9). Briefly, an oligonucleotide probe that has a reporter fluorescent dye attached to its 5′ end and a quencher dye attached to its 3′ end is used for the assay. When the probe hybridizes to its target template, the reporter dye is cleaved by the 5′ nuclease activity of Taq polymerase and becomes capable of emitting a fluorescent signal, since it is no longer suppressed by the quencher dye (8).
This report describes a simple, rapid method for the relative quantification of major periodontopathic bacteria, including A. actinomycetemcomitans and P. gingivalis, in saliva and subgingival plaque. The method uses a TaqMan PCR assay and the comparative threshold cycle (ΔΔCt) method. This is the first reported TaqMan method developed for the detection of A. actinomycetemcomitans.
The bacterial strains used in this study are listed in Table 1. The strains of A. actinomycetemcomitans and P. gingivalis were cultured as described previously (15, 17). Subgingival plaque and saliva samples from patients with periodontitis were prepared as described previously (15).
TABLE 1.
Strains and amplification results
| Strain | Source | Amplification with the following primersh:
|
||
|---|---|---|---|---|
| Aa | Pg | Universal | ||
| A. actinomycetemcomitans | ||||
| ATCC 29523 | ATCCa | + | − | + |
| Y4 | Socranskyb | + | − | + |
| NCTC 9710 | NCTCc | + | − | + |
| IDH 781 | Asikainend | + | − | + |
| IDH 1705 | Asikainen | + | − | + |
| P. gingivalis | ||||
| W83 | KUe | − | + | + |
| W50 | KU | − | + | + |
| 381 | KU | − | + | + |
| ATCC 33277 | KU | − | + | + |
| ATCC 49417 | KU | − | + | + |
| Treponema denticola | ||||
| ATCC 35404 | Ishiharaf | − | − | + |
| ATCC 35405 | Ishihara | − | − | + |
| Bacteroides forsythus ATCC 43037 | Honmag | − | − | + |
| Fusobacterium nucleatum ATCC 10953 | KU | − | − | + |
| Prevotella intermedia ATCC 25611 | ATCC | − | − | + |
| Haemophilus aphrophilus NCTC 5908 | KU | − | − | + |
| Eikenella corrodens 1085 | KU | − | − | + |
| Streptococcus anginosus FW73 | KU | − | − | + |
| Streptococcus sobrinus 6715 | KU | − | − | + |
| Streptococcus gordonii DL1 | KU | − | − | + |
| Streptococcus mutans Xc | KU | − | − | + |
| Streptococcus salivarius HT9R | KU | − | − | + |
| Escherichia coli DH5α | GIBCO BRL | − | − | + |
ATCC, American Type Culture Collection, Manassas, Va.
S. S. Socransky, Forsyth Dental Center, Boston, Mass.
NCTC, National Collection of Type Cultures, London, England.
S. Asikainen, University of Helsinki, Helsinki, Finland.
KU, culture collection in the Department of Preventive Dentistry, Kyushu University Faculty of Dental Science, Fukuoka, Japan.
K. Ishihara, Tokyo Dental College, Chiba, Japan.
K. Honma, Tokyo Dental College, Chiba, Japan.
Aa and Pg, Aa-specific and Pg-specific primer pairs, respectively.
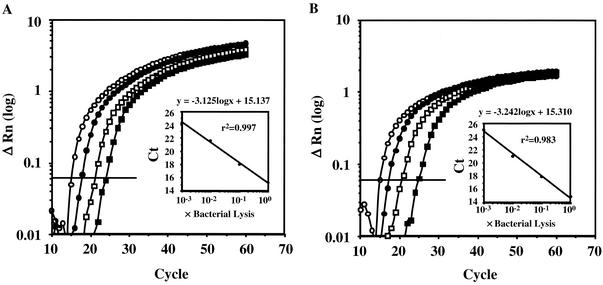
The oligonucleotide primers and probes, designed by using Primer Express (version 1.5) software (Applied Biosystems, Foster City, Calif.), are listed in Table 2. The sequences of the universal primers and a probe for a broad range of bacteria are complementary to highly conserved regions within the 16S rRNA gene (7). The A. actinomycetemcomitans- and P. gingivalis-specific primers and probes were designed from the lktA (6) and 16S rRNA genes, respectively. The specificities of the primers and probes were confirmed by conventional PCR (Table 1) and dot blot analysis with digoxigenin-labeled probes (data not shown), respectively. Conventional PCR with universal primers amplified a DNA fragment of a similar size (68 bp) from all the strains listed in Table 1. The fluorescent probes were dually labeled with a reporter dye (6-carboxyfluorescein [FAM]) covalently attached at the 5′ end and a quencher dye (6-carboxytetramethylrhodamine [TAMRA]) covalently attached at the 3′ end. The primers used for real-time PCR were also used for conventional PCR (Table 2). The conventional PCR assays used to confirm the specificities and universalities of the primers were performed as follows: 94°C for 5 min, followed by 25 cycles of 94°C for 15 s, 55°C for 30 s, and 72°C for 1 min. For each real-time PCR, 20 μl of a mixture containing 1 μl of lysed cells, 1× TaqMan Universal PCR master mixture (Applied Biosystems), each sense and antisense primer at a concentration of 200 nM, and 250 nM TaqMan probe was placed in each well of a 96-well plate. Amplification and detection were performed with the ABI PRISM 7700 sequence detection system (Applied Biosystems) with the following cycle profile: 50°C for 2 min, 95°C for 10 min, and 60 cycles at 95°C for 15 s and 58°C for 1 min. Optimal AmpErase uracil-N-glycosylase enzyme activity requires a 2-min step at 50°C (10). Ct is defined as the cycle at which the fluorescence becomes detectable above the background fluorescence and is inversely proportional to the logarithm of the initial number of template molecules. A standard curve was plotted for each primer-probe set by using the Ct values obtained from amplification of known quantities of DNA. To check the linearity of the detection system, solutions of lysed A. actinomycetemcomitans or P. gingivalis were amplified in successive 10-fold dilutions in a series of real-time PCRs so that a correlation coefficient could be calculated from the standard curve of Ct values. Detection and quantification were linear over the range of DNA concentrations examined. The quantity of DNA was linear over the range from 30 pg to 3 μg per reaction mixture for both species (Fig. 1A and 1B). The number of A. actinomycetemcomitans or P. gingivalis DNA copies was normalized to the number of 16S rRNA gene DNA copies by the method of Biéche et al. (3), with modifications. Briefly, we measured both the species-specific (A. actinomycetemcomitans and P. gingivalis) and the control-specific (16S rRNA gene) fluorescence for each specimen. In addition, we measured both types of fluorescence in four serial 10-fold dilutions of sample lysate. Then, we constructed standard curves for both the targets and the control for each sample. The results, expressed as the fold difference (N) in the number of target gene copies relative to the number of 16S rRNA gene copies, were determined as follows: N = 2ΔΔCt = 2(ΔCt target − ΔCt 16S rRNA), where ΔΔCt is ΔCt target minus ΔCt 16S rRNA and ΔCt is the difference in threshold cycles for target and reference. The ΔCt values for the sample and 16S rRNA were determined by subtracting the average Ct value for the target gene from the average Ct value for the 16S rRNA gene. This study determined the numbers of A. actinomycetemcomitans and P. gingivalis bacteria in saliva and subgingival plaque samples from 10 patients with periodontitis (Table 3). Furthermore, conventional PCR was performed for comparison of the sensitivities of both the conventional and the real-time PCR analyses. The conventional PCR analysis was performed under the same conditions used for the real-time PCR, and the sensitivities were compared (Table 3). The real-time PCR analysis was more sensitive than the conventional PCR analysis in this assay.
TABLE 2.
Oligonucleotide primers and probes
| Designation | Sequence | Amplicon size (bp) | Target | Source or reference |
|---|---|---|---|---|
| Primers | ||||
| Aa1956-F | 5′-CAGCATCTGCGATCCCTGTA-3′ | 147 | lktA | JP2 |
| Aa2102-R | 5′-TCAGCCCTTTGTCTTTCCTAGGT-3′ | |||
| Pg1198-F | 5′-TACCCATCGTCGCCTTGGT-3′ | 126 | 16S rRNA | W83 |
| Pg1323-R | 5′-CGGACTAAAACCGCATACACTTG-3′ | |||
| Uni152-F | 5′-CGCTAGTAATCGTGGATCAGAATG-3′ | 69 | 16S rRNA | 8 |
| Uni220-R | 5′-TGTGACGGGCGGTGTGTA-3′ | |||
| Fluorescent probes | ||||
| Aa2034T | 5′-FAM-TCGAGTATTCCTCAAGCATTCTCGCACG-TAMRA-3′ | lktA | JP2 | |
| Pg1238T | 5′-FAM-GCTAATGGGACGCATGCCTATCTTACAGCT-TAMRA-3′ | 16S rRNA | W83 | |
| Uni177T | 5′-FAM-CACGGTGAATACGTTCCCGGGC-TAMRA-3′ | 16S rRNA | 8 |
FIG. 1.
Amplification of genomic DNA from lysed cells. Serial dilutions of genomic DNA were from A. actinomycetemcomitans (A) or P. gingivalis (B). The relative fluorescence (ΔRn) was monitored as the increase in the intensity of the reporter dye relative to the intensity of the passive internal reference dye. The threshold fluorescence, or the level at which the threshold cycle was determined, is shown. The standard curves were generated from the amplification plots in the insets (correlation coefficients, 0.997 for A. actinomycetemcomitans and 0.983 for P. gingivalis). Ct is the cycle number at which the threshold fluorescence is reached.
TABLE 3.
Comparison of conventional PCR and real-time PCR
| Sample and patient no. | Conventional PCR result
|
Real-time PCR resultc
|
||
|---|---|---|---|---|
| Aaa | Pgb | % of Aa | % of Pg | |
| Saliva | ||||
| 1 | − | + | NDd | 0.20 ± 0.02 |
| 2 | + | − | 0.19 ± 0.04 | ND |
| 3 | + | + | 0.22 ± 0.04 | 1.56 ± 0.06 |
| 4 | + | − | 0.20 ± 0.03 | 5.11 × 10−4 ± 1.16 × 10−4 |
| 5 | − | + | ND | 0.64 ± 0.02 |
| 6 | − | + | ND | 1.34 ± 0.13 |
| 7 | + | + | 0.11 ± 0.01 | 0.08 ± 0.00 |
| 8 | − | + | ND | 0.81 ± 0.06 |
| 9 | − | − | ND | ND |
| 10 | − | − | ND | ND |
| Subgingival plaque | ||||
| 1 | − | + | ND | 1.33 ± 0.20 |
| 2 | − | − | 7.16 × 10−4 ± 0.47 × 10−4 | ND |
| 3 | − | − | 4.69 × 10−3 ± 0.45 × 10−3 | ND |
| 4 | + | − | 0.11 ± 0.03 | ND |
| 5 | − | + | ND | 0.38 ± 0.04 |
| 6 | − | + | ND | 6.40 ± 0.43 |
| 7 | + | + | 0.02 ± 0.00 | 0.20 ± 0.02 |
| 8 | − | + | ND | 1.10 ± 0.16 |
| 9 | − | − | ND | ND |
| 10 | − | − | ND | ND |
Aa, A. actinomycetemcomitans.
Pg, P. gingivalis.
Data are expressed as means ± standard deviation (n = 3).
ND, not detected.
One way to quantify bacteria is to use absolute quantification, which requires very precise sample collection. Another way is to use relative quantification by the ΔΔCt method. From a clinical perspective, analysis of the percentage of specific bacteria in a region is often required to evaluate treatment. Lyons et al. (11) pointed out the importance of relative quantification rather than determination of the absolute number of a single species in a mixed sample.
The percentages of A. actinomycetemcomitans and P. gingivalis bacteria in each subgingival plaque sample varied by a few orders of magnitude (Table 3). Our results for P. gingivalis are consistent with those from a previous report (11). The proportion of A. actinomycetemcomitans bacteria ranged from 0 to 0.11%. Similar results were obtained with the saliva samples, in which the proportion of P. gingivalis bacteria ranged from 0 to 1.56% and that of A. actinomycetemcomitans bacteria ranged from 0 to 0.22%.
The simplified ΔΔCt method is accurate and useful for the relative quantification of periodontopathic bacteria. Further studies of this real-time PCR-based quantitative detection system might be useful both for the evaluation of treatment and for elucidation of the etiologic role of unculturable oral bacteria in periodontitis.
Acknowledgments
This investigation was supported by Grant-in-Aid for the Encouragement of Young Scientists 13771265 (to Y.N.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; a research grant from the Takeda Science Foundation (to Y.N.); and research fellowships from the Japan Society for the Promotion of Science for Young Scientists (to N.S.).
Footnotes
This article is respectfully dedicated to the memory of Toshihiko Koga, who died on 14 October 2001.
REFERENCES
- 1.Amano, A., M. Kuboniwa, I. Nakagawa, S. Akiyama, I. Morisaki, and S. Hamada. 2000. Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J. Dent. Res. 79:1664-1668. [DOI] [PubMed] [Google Scholar]
- 2.Ashimoto, A., C. Chen, I. Bakker, and J. Slots. 1996. Polymerase chain reaction detection of 8 putative periodontal pathogens in subgingival plaque of gingivitis and advanced periodontitis lesions. Oral Microbiol. Immunol. 11:266-273. [DOI] [PubMed] [Google Scholar]
- 3.Biéche, I., B. Parfait, V. Le Doussal, M. Olivi, M. C. Rio, R. Lidereau, and M. Vidaud. 2001. Identification of CGA as a novel estrogen receptor-responsive gene in breast cancer: an outstanding candidate marker to predict the response to endocrine therapy. Cancer Res. 61:1652-1658. [PubMed] [Google Scholar]
- 4.Christersson, L. A., C. L. Fransson, R. G. Dunford, and J. J. Zambon. 1992. Subgingival distribution of periodontal pathogenic microorganisms in adult periodontitis. J. Periodontol. 63:418-425. [DOI] [PubMed] [Google Scholar]
- 5.Genco, R. J., J. J. Zambon, and L. A. Christersson. 1988. The origin of periodontal infections. Adv. Dent. Res. 2:245-259. [DOI] [PubMed] [Google Scholar]
- 6.Goncharoff, P., D. H. Figurski, R. H. Stevens, and D. H. Fine. 1993. Identification of Actinobacillus actinomycetemcomitans: polymerase chain reaction amplification of lktA-specific sequences. Oral Microbiol. Immunol. 8:105-110. [DOI] [PubMed] [Google Scholar]
- 7.Greisen, K., M. Loeffelholz, A. Purohit, and D. Leong. 1994. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J. Clin. Microbiol. 32:335-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 9.Holland, P. M., R. D. Abramson, R. Watson, and D. H. Gelfand. 1991. Detection of specific polymerase chain reaction product by utilizing the 5′→3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88:7276-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Longo, M. C., M. S. Berninger, and J. L. Hartley. 1990. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene 93:125-128. [DOI] [PubMed] [Google Scholar]
- 11.Lyons, S. R., A. L. Griffen, and E. J. Leys. 2000. Quantitative real-time PCR for Porphyromonas gingivalis and total bacteria. J. Clin. Microbiol. 38:2362-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer, D. H., and P. M. Fives-Taylor. 1997. The role of Actinobacillus actinomycetemcomitans in the pathogenesis of periodontal disease. Trends Microbiol. 5:224-228. [DOI] [PubMed] [Google Scholar]
- 13.Oho, T., Y. Yamashita, Y. Shimazaki, M. Kushiyama, and T. Koga. 2000. Simple and rapid detection of Streptococcus mutans and Streptococcus sobrinus in human saliva by polymerase chain reaction. Oral Microbiol. Immunol. 15:258-262. [DOI] [PubMed] [Google Scholar]
- 14.Slots, J., and R. J. Genco. 1984. Black-pigmented Bacteroides species, Capnocytophaga species, and Actinobacillus actinomycetemcomitans in human periodontal disease: virulence factors in colonization, survival, and tissue destruction. J. Dent. Res. 63:412-421. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki, N., Y. Nakano, Y. Yoshida, D. Ikeda, and T. Koga. 2001. Identification of Actinobacillus actinomycetemcomitans serotypes by multiplex PCR. J. Clin. Microbiol. 39:2002-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson, M., and B. Henderson. 1995. Virulence factors of Actinobacillus actinomycetemcomitans relevant to the pathogenesis of inflammatory periodontal diseases. FEMS Microbiol. Rev. 17:365-379. [DOI] [PubMed] [Google Scholar]
- 17.Yoshimura, M., Y. Nakano, Y. Yamashita, T. Oho, T. Saito, and T. Koga. 2000. Formation of methyl mercaptan from l-methionine by Porphyromonas gingivalis. Infect. Immun. 68:6912-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zambon, J. J. 1985. Actinobacillus actinomycetemcomitans in human periodontal disease. J. Clin. Periodontol. 12:1-20. [DOI] [PubMed] [Google Scholar]
- 19.Zambon, J. J., H. S. Reynolds, and J. Slots. 1981. Black-pigmented Bacteroides spp. in the human oral cavity. Infect. Immun. 32:198-203. [DOI] [PMC free article] [PubMed] [Google Scholar]