Abstract
Routine analysis of Listeria monocytogenes by serotyping using traditional agglutination methods is limited in use because of the expense and limited availability of commercially prepared antisera and intra- and interlaboratory discrepancies arising from differences in antiserum preparation and visual determination of agglutination. We have adapted a commercially available set of L. monocytogenes antisera to an enzyme-linked immunosorbent assay (ELISA) format for high-throughput, low-cost serotype determination. Rather than subjective visualization of agglutination, positive antigen and antiserum reactions were scored by a quantitative, colorimetric reaction. ELISA serotyping of 89 of 101 L. monocytogenes isolates agreed with slide agglutination serotyping data, and 100 previously uncharacterized isolates were serotyped unambiguously by the ELISA method. In addition, mixed-serotype cultures of L. monocytogenes were identified by a colony immunoblot procedure, in which serogroup 1/2 and serogroup 4 colonies were discriminated by differential staining.
Listeria monocytogenes is a gram-positive, food-borne bacterial pathogen that causes listeriosis in susceptible individuals (4). In addition, L. monocytogenes is a widespread, saprophytic bacterium that can be found not only in association with soil, plants, and animal waste (5, 8, 23, 24) but also as a persistent organism in food and dairy processing environments (3, 14). All of these environments are potential sources for contamination of fresh and prepared foods with L. monocytogenes, which, in turn, poses a significant public health risk in terms of the potential for listeriosis outbreaks. In order for these risks to be minimized, subtyping of L. monocytogenes isolates has been undertaken in several laboratories in recent years to begin to identify type-specific factors contributing to virulence, persistence, and/or transmissibility of the bacterium relative to its outbreak potential (7, 10-13, 20, 22).
Current methods to subtype L. monocytogenes include DNA fingerprinting using specific (7) or random (10, 12) PCR primers, ribotyping (10, 12, 13), and pulsed-field gel electrophoresis (1). However, groupings based on these methods are still often compared to groupings based on serotype, using an agglutination method and subgrouping scheme developed by Seeliger and Höhne (18). This method differentiates L. monocytogenes into 12 different serotypes based on the reactions of somatic (O) and flagellar (H) antigens with a series of polyvalent and monovalent antisera. Serotyping of L. monocytogenes isolates is not routinely performed outside of public health reference laboratories due to the limited availability and high cost of commercially produced antisera and to the inconvenience and reliability issues associated with producing one's own antisera using specific reference strains of L. monocytogenes. Conversely, using a reference laboratory for serotyping large numbers of L. monocytogenes isolates may also be prohibitively costly or time-consuming for individual laboratories. We sought to adapt a commercial serotyping kit to a format that would be cost-effective, reliable, and of sufficiently high throughput to facilitate serotype determination of L. monocytogenes. Using a 96-well enzyme-linked immunosorbent assay (ELISA) format instead of agglutination as a means to score reactions with each antiserum, this method provides a semiquantitative measurement of positive and negative reactions and requires only a fraction of the antisera used in the agglutination assay. In addition, we used antisera from the same serotyping kit in colony immunoblot experiments to identify mixed-serotype L. monocytogenes cultures, as an initial method to resolve ambiguous intra- and interlaboratory serotyping results.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
L. monocytogenes strains used in this study are listed in Table 1. Isolates were confirmed as L. monocytogenes by production of turquoise colonies on BCM L. monocytogenes plating medium (Biosynth International, Naperville, Ill.) (15) and by PCR amplification of an iap gene fragment using L. monocytogenes-specific primers (2). Stock cultures of all strains were stored at −80°C in Bacto tryptic soy broth without dextrose (Difco) containing 0.6% (wt/vol) yeast extract (Difco) (TSYE) and 1 M glycerol. Working cultures were maintained on TSYE agar and grown at 30°C. Prior to serotype determination, single colonies of each strain were inoculated onto brain heart infusion (BHI) (Difco) motility plates containing 0.3% (wt/vol) agar and grown for 24 h at 30°C. Bacteria from the edges of the motility plate-grown colonies were then inoculated into 5 ml of BHI broth and incubated overnight at 30°C.
TABLE 1.
L. monocytogenes strains used in this study
| Isolate | Strain no. | Origin | Serotype by:
|
Source | |
|---|---|---|---|---|---|
| Agglutination | ELISA | ||||
| 10403 | RM2194 | Human, clinical | 1/2a | 1/2a | D. Portnoy |
| 32490G | RM2985 | Bulk milk | 1/2a | 1/2a | USDAa |
| 35568A | RM2989 | Bulk milk | 1/2a | 1/2a | USDA |
| 10867C | RM2990 | Bulk milk | 1/2a | 1/2a | USDA |
| 51772 | RM3015 | Cheese | 1/2a | 1/2a | ATCCb |
| 19111 | RM3023 | Poultry | 1/2a | 1/2a | ATCC |
| J0098 | RM3029 | Food | 1/2a | 1/2a | CDCc |
| 12443 | RM3102 | Monkey, clinical | 1/2a | 1/2a | R. Rabourne |
| FSL-J2-020 | RM3152 | Cow | 1/2a | 1/2a | M. Wiedmann |
| FSL-C1-056 | RM3160 | Human, sporadic | 1/2a | 1/2a | M. Wiedmann |
| FSL-J2-054 | RM3161 | Sheep | 1/2a | 1/2a | M. Wiedmann |
| FSL-J2-031 | RM3163 | Cow | 1/2a | 1/2a | M. Wiedmann |
| FSL-J2-066 | RM3164 | Sheep | 1/2a | 1/2a | M. Wiedmann |
| FSL-J2-063 | RM3165 | Sheep | 1/2a | 1/2a | M. Wiedmann |
| FSL-J1-101 | RM3175 | Human, sporadic | 1/2a | 1/2a | M. Wiedmann |
| FSL-N3-031 | RM3184 | Food, sporadic | 1/2a | 1/2a | M. Wiedmann |
| FSL-R2-499 | RM3185 | Human, epidemic | 1/2a | 1/2a | M. Wiedmann |
| 750f | RM3370 | Environmental | 1/2a | 1/2a | WADOHd |
| 841f | RM3372 | Environmental | 1/2a | 1/2a | WADOH |
| 1155f | RM3373 | Human | 1/2a | 1/2a | WADOH |
| 1157f | RM3374 | Human | 1/2a | 1/2a | WADOH |
| 1160f | RM3376 | Human | 1/2a | 1/2a | WADOH |
| 1162f | RM3378 | Human | 1/2a | 1/2a | WADOH |
| 1163f | RM3379 | Environmental | 1/2a | 1/2a | WADOH |
| 1165f | RM3381 | Environmental | 1/2a | 1/2a | WADOH |
| 1166f | RM3382 | Human | 1/2a | 1/2a | WADOH |
| 1445f | RM3385 | Blood vessel | 1/2a | 1/2a | WADOH |
| 17209 | RM2991 | Sheep brain | 1/2b | 1/2b | I. Wesley |
| 16888 | RM2995 | Cow brain | 1/2b | 4b | I. Wesley |
| G848 | RM3024 | Unknown | 1/2b | 1/2b | CDC |
| FSL-J2-064 | RM3155 | Cow | 1/2b | 1/2b | M. Wiedmann |
| FSL-J1-177 | RM3156 | Human, sporadic | 1/2b | 1/2b | M. Wiedmann |
| FSL-J2-035 | RM3157 | Goat | 1/2b | 1/2b | M. Wiedmann |
| 9900101f | RM3368 | Environmental | 1/2b | 1/2b | WADOH |
| 9900104f | RM3369 | Human | 1/2b | 1/2b | WADOH |
| 842f | RM3371 | Blood vessel | 1/2b | 1/2b | WADOH |
| 1159f | RM3375 | Human | 1/2b | 1/2b | WADOH |
| 1164f | RM3380 | Human | 1/2b | 1/2b | WADOH |
| G-3321 | RM3014 | Human | 1/2c | 1/2c | CDC |
| H9666 | RM3017 | Blood | 1/2c | 1/2c | CDC |
| H9333 | RM3018 | Blood | 1/2c | 1/2c | CDC |
| H9066 | RM3019 | Mushrooms | 1/2c | 1/2c | CDC |
| H9067 | RM3020 | Cheese | 1/2c | 1/2c | CDC |
| H7973 | RM3021 | Blood | 1/2c | 1/2c | CDC |
| FSL-J1-094 | RM3166 | Human, sporadic | 1/2c | 1/2, nonmotile | M. Wiedmann |
| 9900096f | RM3367 | Environmental | 1/2c | 1/2c | WADOH |
| J0095 | RM3026 | Pie | 3a | 3a | CDC |
| FSL-C1-115 | RM3167 | Human, sporadic | 3a | 1/2a | M. Wiedmann |
| FSL-J1-169 | RM3158 | Human, sporadic | 3b | 1/2b | M. Wiedmann |
| J0096 | RM3027 | Chicken | 3c | 3c | CDC |
| FSL-J1-049 | RM3159 | Human, sporadic | 3c | 3c | M. Wiedmann |
| FSL-J1-031 | RM3168 | Human, sporadic | 4a | 4c | M. Wiedmann |
| FSL-J1-168 | RM3169 | Human, sporadic | 4a | 4c | M. Wiedmann |
| FSL-X1-010 | RM3171 | Unknown | 4a | 4a | M. Wiedmann |
| F2379 | RM2199 | Cheese, outbreak | 4b | 4b | D. Portnoy |
| 8807-X2 | RM2983 | Sheep brain | 4b | 4b | WADDLe |
| 013668A | RM2984 | Cow brain | 4b | 4b | WADDL |
| 35584A | RM2986 | Bulk milk | 4b | 4b | USDA |
| 2149 | RM2987 | Human, clinical | 4b | 4b | I. Wesley |
| 2219 | RM2988 | Coleslaw | 4b | 4b | I. Wesley |
| 2223 | RM2992 | Cucumber | 4b | 4d/4e | I. Wesley |
| 13565A | RM2996 | Bulk milk | 4b | 4b | USDA |
| 11056A | RM2997 | Bulk milk | 4b | 4b | USDA |
| 2207 | RM2998 | Human, stillbirth | 4b | 4b | I. Wesley |
| 19115 | RM3013 | Human | 4b | 4b | ATCC |
| J0097 | RM3028 | Human | 4b | 4b | CDC/PICK> |
| G3990 | RM3098 | Cheese | 4b | 4b | R. Rabourne |
| G3982 | RM3099 | Cheese | 4b | 4b | R. Rabourne |
| H7550 | RM3100 | Hot dog | 4b | 4b | R. Rabourne |
| ScottA | RM3101 | Human, clinical | 4b | 4b | R. Rabourne |
| 12375 | RM3103 | Monkey, clinical | 4b | 4b | R. Rabourne |
| FSL-J1-225 | RM3150 | Human, clinical | 4b | 4b | M. Wiedmann |
| FSL-N1-225 | RM3151 | Human, epidemic | 4b | 4b | M. Wiedmann |
| FSL-J1-110 | RM3153 | Food, epidemic | 4b | 4b | M. Wiedmann |
| FSL-C1-122 | RM3154 | Human, sporadic | 4b | 4b | M. Wiedmann |
| FSL-J1-158 | RM3173 | Goat | 4b | 4b | M. Wiedmann |
| FSL-J1-108 | RM3176 | Human, epidemic | 4b | 4b | M. Wiedmann |
| FSL-J1-116 | RM3177 | Human, epidemic | 4b | 4b | M. Wiedmann |
| FSL-J1-119 | RM3178 | Human, epidemic | 4b | 4b | M. Wiedmann |
| FSL-J1-126 | RM3179 | Human, epidemic | 4b | 4b | M. Wiedmann |
| FSL-N1-227 | RM3180 | Food, epidemic | 4b | 4b | M. Wiedmann |
| FSL-N3-008 | RM3181 | Food, epidemic | 4b | 4b | M. Wiedmann |
| FSL-N3-013 | RM3182 | Food, epidemic | 4b | 4b | M. Wiedmann |
| FSL-N3-022 | RM3183 | Food, epidemic | 4b | 4b | M. Wiedmann |
| 9900094f | RM3366 | Human, spinal fluid | 4b | 4b | WADOH |
| 1161f | RM3377 | Human | 4b | 4b | WADOH |
| 1167f | RM3383 | Human | 4b | 4b | WADOH |
| 1329f | RM3384 | Human | 4b | 4b | WADOH |
| 2140f | RM3386 | Human, spinal fluid | 4b | 4b | WADOH |
| 2150f | RM3387 | Tissue | 4b | 4b | WADOH |
| 2172f | RM3388 | Stool | 4b | 4b | WADOH |
| F-4565f | RM3390 | Human | 4b | 4b | CDC |
| G-1092f | RM3391 | Human | 4b | 4b | CDC |
| 19116 | RM3022 | Chicken | 4c | 4c | ATCC |
| J0099 | RM3030 | Bull | 4c | 4c | CDC |
| FSL-X1-009 | RM3170 | Unknown | 4c | 4b | M. Wiedmann |
| FSL-X1-008 | RM3172 | Unknown | 4c | 4b | M. Wiedmann |
| 36467Cf | RM3389 | Bulk milk | 4c | 4c | USDA |
| J0094 | RM3025 | Human | 4d | 4d | CDC |
| 19118 | RM3016 | Chicken | 4e | 4b | ATCC |
| FSL-M1-004 | RM3162 | Human, sporadic | N/A | 3a | M. Wiedmann |
USDA, U.S. Department of Agriculture.
ATCC, American Type Culture Collection, Manassas, Va.
CDC, Centers for Disease Control and Prevention, Atlanta, Ga.
WADOH, Washington State Department of Health, Olympia.
WADDL, Washington Animal Disease Diagnostic Laboratory, Pullman.
Tested by three different lots of serotyping reagents.
Serotyping.
One hundred one L. monocytogenes isolates were serotyped by both the agglutination method and the ELISA method. The two methods were performed independently by two different laboratories. Twenty-six strains were tested three times using three different lots of serotyping reagents (Table 1).
(i) Agglutination method.
Serotyping was performed by a slide agglutination assay using commercially prepared antisera (Listeria antiserum Seiken kit; Denka Seiken Co., Tokyo, Japan) according to the manufacturer's instructions.
(ii). ELISA method.
Cultures of L. monocytogenes grown as described were pelleted by centrifugation (14,000 × g for 5 min) and resuspended in an equal volume of 0.2% (wt/vol) NaCl. Cells to be used for O-antigen determination were autoclaved for 30 min at 121°C, allowed to cool to room temperature, centrifuged and resuspended in 0.2% NaCl to an optical density at 600 nm of 0.3 to 0.4. Cells to be used for H-antigen determination were centrifuged, resuspended in an equal volume of 4% (wt/vol) formaldehyde-0.2% NaCl, and incubated at room temperature for 1 h. Cells were then washed, pelleted by centrifugation, and resuspended in 0.2% NaCl to an optical density at 600 nm of 0.3 to 0.4. Prepared cells were added (70 μl/well) to ELISA well strips (MaxiSorp flat-bottom well strips; Nalge Nunc International, Rochester, N.Y.) and allowed to dry at 40°C overnight. All subsequent steps were performed at room temperature. After rinsing the wells with distilled, deionized water (ddH2O), nonspecific surfaces were blocked with 200 μl/well of casein blocking solution (0.5% casein, 30 mM NaN3, 150 mM NaCl, 10 mM Tris-HCl [pH 7.5]) for 1 h. After removing blocking solution and rinsing with ddH2O, individual antisera (Listeria antiserum Seiken kit) were diluted in dilution buffer (1% bovine serum albumin, 0.1% Tween 20, 2.7 mM KCl, 15 mM NaN3, 150 mM NaCl, 10 mM Tris-HCl [pH 7.5]), and 100 μl was added to each well and incubated for 1 h. Wells were washed twice with wash buffer (0.1% Tween 20, 150 mM NaCl, 10 mM Tris-HCl [pH 7.5]) and twice with ddH2O. Alkaline phosphatase-conjugated goat anti-rabbit antibody (Zymed Laboratories, South San Francisco, Calif.) was diluted 1:1,000 in dilution buffer, and 100 μl was added to each well and incubated for 1 h. After washing the wells as before, 100 μl of 1-mg/ml p-nitrophenyl phosphate in 1 M diethanolamine-0.5 mM MgCl2 · 6H2O, pH 9.8, was added to each well and incubated for 30 min. p-Nitrophenyl phosphate hydrolysis was measured as A405 using a SpectraMax 340 microplate reader (Molecular Devices Corp., Sunnyvale, Calif.). Average values of duplicate reactions of each strain for each O-factor antiserum and H-factor antiserum were calculated relative to the maximum O-factor antiserum and H-factor antiserum reactions, respectively, for that strain. For each strain-antiserum combination, duplicate wells containing cell suspensions incubated without primary antisera and antisera incubated without cell suspensions were included as negative controls. Serotypes were assigned according to the scheme described by Seeliger and Höhne (18).
Colony immunoblotting.
Colonies of L. monocytogenes grown on BHI plates at 30°C were transferred by colony lift to nitrocellulose filters (pore size, 0.45 μm; Schleicher and Schuell, Keene, N.H.) for 10 min. Filters were washed (twice for 10 min each) in wash buffer to remove cell debris and incubated in 20 ml of casein blocking solution for 1 h. As the primary antibody, L. monocytogenes O-factor antiserum I/II (from the Listeria antiserum Seiken kit), which is specific for all serotype 1/2 and 3 strains, was diluted 1:2,000 in 10 ml of dilution buffer and incubated with blocked filters for 30 min. Filters were then washed (twice for 10 min each) with 20 ml of wash buffer and incubated for 30 min with 10 ml of a 1:2,000 dilution of alkaline phosphatase-conjugated goat anti-rabbit antibody. After washing (twice for 10 min each) with 20 ml of wash buffer, colonies positively reacting with antiserum I/II were detected using 10 ml of naphthol-AS-phosphate (0.2 mg/ml)-Fast Red TR (0.1 mg/ml; multicolor detection set Red; Roche Molecular Biochemicals, Indianapolis, Ind.). After color development, alkaline phosphatase was inactivated by incubating the filters for 10 min in 50 mM EDTA, pH 8.0, at 80°C. The filters were then blocked as before, and the same procedure was performed, except that L. monocytogenes O-factor antiserum V/VI (from the Listeria antiserum Seiken kit), which is specific for all serotype 4 strains, was used as the primary antibody. Alkaline phosphatase activity specific to positive reactions with antiserum V/VI were detected using 10 ml of naphthol-AS-GR-phosphate (0.2 mg/ml)-Fast Blue B (0.35 mg/ml; multicolor detection set Green; Roche).
RESULTS
Development of ELISA-format serotyping method.
The ELISA protocol to serotype L. monocytogenes isolates was adapted from standard immunoassay methods (9), using the slide agglutination protocol of the Listeria antiserum Seiken serotyping kit as a guide for sample preparation. Strains J0094, J0095, J0096, J0097, J0098, and J0099 (serotypes 4d, 3a, 3c, 4b, 1/2a, and 4c, respectively) were used as serotype reference strains (Table 1) in initial experiments to determine the proper dilution level for discrimination between positive and negative reactions for all antisera. We determined that the O-factor and H-factor antisera gave suitable results at dilutions of 1:1,000 and 1:500, respectively (data not shown). All subsequent ELISA serotype determinations were performed using antisera diluted to these levels.
Using the serotype reference strains, cell preparation methods were compared. Cell suspensions that were autoclaved for 30 min, boiled for 1 h, incubated at 65°C for 1 h, or not treated reacted similarly to the appropriate positive O-factor antisera. However, autoclaved cells tended to react less to negative O-factor antisera relative to cells treated by the other methods; that is, lower background reactions were observed. In addition, formaldehyde-treated cells were prone to give false-positive reactions to O antiserum IX (data not shown). Since the ELISA method was to be compared directly to the antiserum kit slide agglutination method, which recommended using autoclaved cells, all subsequent O-antigen determinations were performed with autoclaved cells. Similarly, to directly compare H-antigen determination by the ELISA method to that by the slide agglutination method, isolates initially were subcultured three times on BHI motility agar, as recommended in the slide agglutination kit protocol, to increase the proportion of fully flagellated cells and thereby render a robust reaction with the appropriate H-factor antisera. Subsequent experiments showed that a single 24-h passage on BHI motility agar was sufficient in the ELISA protocol to yield accurate reactions with H-factor antisera. Treatment of the cell suspensions by autoclaving was less effective than treatment with formaldehyde in preserving H-antigens for serotyping. Therefore, formaldehyde-treated cell suspensions and autoclaved cell suspensions were considered optimal for determining H-antigens and O-antigens, respectively.
Interpretation of ELISA reactions.
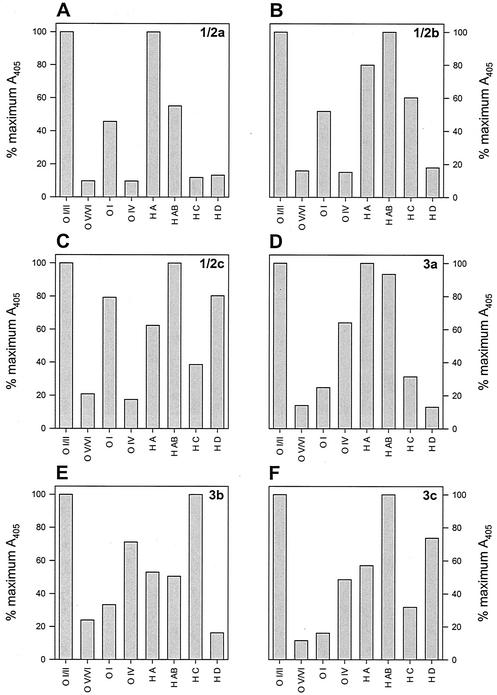
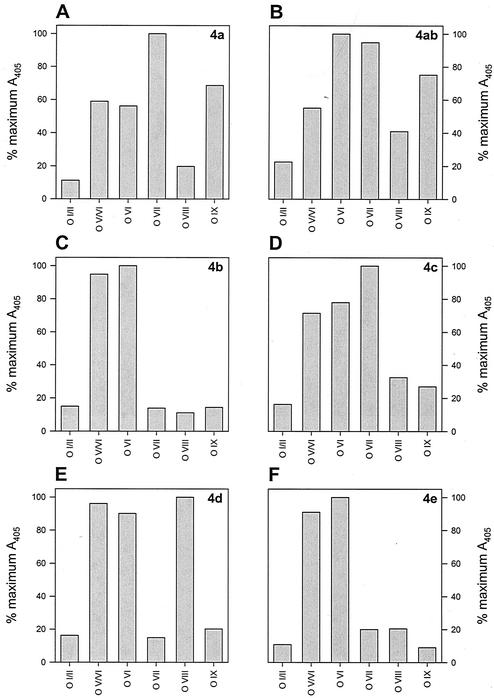
As with the slide agglutination method, serotype assignment using the ELISA method is a three-step process. First, the reactions of O-factor antisera I/II and V/VI are compared; strains that react positively to antiserum I/II will react negatively to antiserum V/VI, and vice versa. Strains reacting positively with antiserum I/II are differentiated into serogroup 1/2 by a positive reaction to O-factor antiserum I and serogroup 3 by a positive reaction to O-factor antiserum IV. These serogroups are further differentiated into serotypes by their specific reaction to H-factor antisera (Table 2). Strains reacting positively with antiserum V/VI (i.e., serogroup 4) are differentiated into serotypes 4a, 4ab, 4b, 4c, 4d, and 4e by their reactions to O-factor antisera VI, VII, VIII, and IX; all serogroup 4 strains react identically to H-factor antisera (Table 2). Positive and negative reactions were scored relative to the maximal positive reaction of each strain to all O-factor and H-factor antisera, with negative reactions typically at levels less than 25% of the maximal positive reaction for O-antigens (Fig. 1 and 2). Reactions to H-factor antisera were generally weaker than those to O-factor antisera, and so interpretation of results was more subjective, especially for 1/2c and 3c strains (Fig. 1C and E). Nevertheless, graphic interpretations of data obtained using the ELISA method were less ambiguous than visual positive-negative interpretations obtained using the slide agglutination method.
TABLE 2.
Antigen components of each L. monocytogenes serotype
| Serotype | O-antigensa | H-antigens |
|---|---|---|
| 1/2a | I, II | A, B |
| 1/2b | I, II | A, B, C |
| 1/2c | I, II | B, D |
| 3a | II, IV | A, B |
| 3b | II, IV | A, B, C |
| 3c | II, IV | B, D |
| 4a | (V), VII, IX | A, B, C |
| 4ab | V, VI, VII, IX | A, B, C |
| 4b | V, VI | A, B, C |
| 4c | V, VII | A, B, C |
| 4d | (V), VI, VIII | A, B, C |
| 4e | V, VI, (VIII), (IX) | A, B, C |
Antigens in parentheses may not be present in all isolates.
FIG. 1.
ELISA O-antigen and H-antigen reactions of serogroup 1/2 and 3 L. monocytogenes strains. (A) Strain J0098, serotype 1/2a; (B) strain G848, serotype 1/2b; (C) strain G-3321, serotype 1/2c; (D) strain J0095, serotype 3a; (E) strain cpp81, serotype 3b; (F) strain J0096, serotype 3c.
FIG. 2.
ELISA O-antigen reactions of serogroup 4 L. monocytogenes strains. (A) Strain FSL-X1-010, serotype 4a; (B) strain JL2-10, serotype 4ab; (C) strain J0097, serotype 4b; (D) strain J0099, serotype 4c; (E) strain J0094, serotype 4d; (F) strain 19118, serotype 4e. Note that serotype 4e is indistinguishable from 4b in this case.
Comparison of ELISA and slide agglutination to assign serotypes.
The ELISA serotyping method was compared to the slide agglutination method using 101 different L. monocytogenes isolates (Table 1). Of these strains, results obtained by the ELISA method matched those obtained by slide agglutination for 89 of the 101 isolates (88%). For isolates of the clinically important serotypes 1/2a, 1/2b, and 4b, the ELISA method agreed with the slide agglutination method for 27 of 27 strains (100%), 10 of 11 strains (91%), and 38 of 39 strains (97%), respectively. Conversely, one isolate (19118) that was designated serotype 4e by slide agglutination was identified as serotype 4b by ELISA due to a negative reaction with O-factor antiserum IX, and one isolate that was designated serotype 4b by slide agglutination was identified by ELISA as serotype 4d/4e due to a positive reaction with O-factor antiserum VIII. It is noteworthy that in the serotyping scheme presented by Seeliger and Höhne (18) (Table 2), not all serotype 4e strains necessarily contain O-antigens VIII and IX. Such strains would be indistinguishable from serotype 4b. Also, the scheme cannot distinguish between serotypes 4e and 4d solely by the presence of O-antigen VIII (Table 2). During testing, two strains (FSL-J1-094 and G3990) were nonmotile on motility agar, and a third strain (FSL-J1-116) was delayed in motility, requiring a second passage on motility agar. Because of this defect, strain FSL-J1-094 (serogroup 1/2) could not be typed fully due to the lack of H-factor antiserum-reactive flagella; strains G3990 and FSL-J1-116 were both typed as 4b, so H-antigen determinations were not necessary.
In addition to those strains serotyped by both methods, 97 L. monocytogenes strains that previously were not serotyped were characterized by the ELISA protocol. These strains were isolated from various sources, including produce, meats and dairy products, food processing environments, animals and humans, soils, and environmental samples. The majority of these isolates (86%) were serotyped unambiguously as 1/2a (17 strains), 1/2b (39 strains), or 4b (27 strains) by the ELISA protocol. The remaining isolates were serotyped as 1/2c (5 strains), 3b (3 strains), 4a (2 strains), 3a (1 strain), 4ab, (1 strain), 4c (1 strain), and 4d (1 strain).
Identification of mixed-serotype cultures.
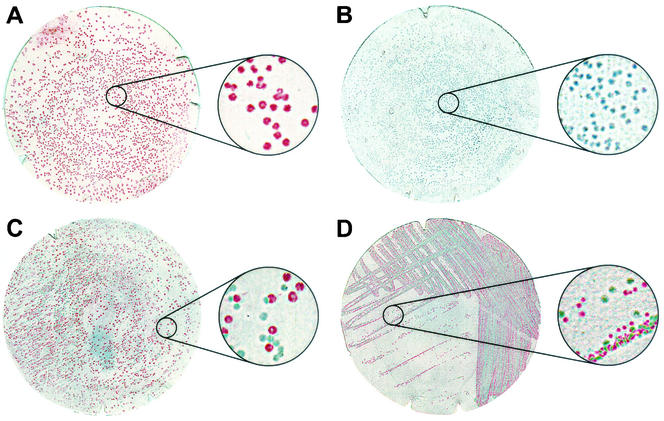
Initial comparisons of the serotyping methods using L. monocytogenes strain 19118 (Table 1) were problematic in that slide agglutination resulted in serotype 4e, while independent ELISA tests on cultures from two separate colony picks identified the strain as serotype 4b and 1/2c. Since the initial discrimination between positive and negative reactions to O-factor antisera I/II and V/VI generally is very robust, we hypothesized that the stock culture of strain 19118 contained a mixture of these two serotypes rather than hypothesizing that the discrepancy resulted from inconsistent performance of the ELISA. To test this hypothesis, a colony immunoblot assay was developed using the O-factor antisera I/II and V/VI to discriminate between serogroups 1/2 and 4. Colony lifts on nitrocellulose were probed sequentially using each O-factor antiserum as the primary antibody and a unique color alkaline phosphatase substrate to correspond to each primary antibody. Using this method, colonies of serotype 1/2a reference strain J0098 and serotype 4b reference strain J0097 could be distinguished on a plate containing a mixture of both strains (Fig. 3C). No false-positive colonies resulting from cross-reaction of O-factor antiserum I/II with strain J0097 or O-factor antiserum V/VI with strain J0098 were observed (Fig. 3A and B). Performing this colony immunoblot procedure on a plate streaked for individual colonies of strain 19118 confirmed the presence of serogroups 1/2 and 4 (Fig. 3D), consistent with the initial, seemingly conflicting ELISA serotyping results, and demonstrated that this was, in fact, a mixed culture of serotype 4b strain 19118 contaminated with a serotype 1/2c strain.
FIG. 3.
Colony immunoblotting of single- and mixed-serotype cultures of L. monocytogenes. Blots of serotype 1/2a strain J0098 (A), serotype 4b strain J0097 (B), or a mixture of strains J0098 and J0097 (C) were probed sequentially with O-factor antiserum I/II (stained red) and O-factor antiserum V/VI (stained green). (D) A streak plate of strain 19118 was blotted and probed sequentially with O-factor antiserum V/VI (stained red) and O-factor antiserum I/II (stained green). Insets are magnifications (×5) of the indicated region of each filter to show detail.
DISCUSSION
Serotyping of L. monocytogenes as a primary subtyping method has been in use for decades, even though it has been noted repeatedly that serotyping is ambiguous, sometimes variable within and between laboratories, and limited in its usefulness to demonstrate a correlation of strains between outbreaks of listerial infections (16, 17, 19). The ELISA serotyping protocol described in this work is a means to attempt to rectify the ambiguities and intra- and interlaboratory variability inherent to the traditional serotyping method, while at the same time allowing laboratories to perform more analyses with significant cost and time savings. Because serotype reactions depend on the quality of the antisera used, which in turn depends on which standardized strains and antigen preparation methods are chosen, the likelihood of inaccurate or inconsistent assignment of serotype rises when laboratories prepare their own antisera, especially for nonclinical isolates (i.e., those not of serotypes 1/2a, 1/2b, or 4b) (6, 16). The standard slide agglutination method relies on visual acuity and judgement to produce accurate data; this may be a substantial source of variability when comparing data between individuals in a laboratory and between different laboratories. In addition, the agglutination assays typically use 10 to 100 μl of antiserum per reaction, depending on methodology. Using the commercially prepared antiserum kit and dilution factors described, the ELISA protocol requires 0.2 μl of each O-factor antiserum and 0.4 μl of each H-factor antiserum to determine the serotype of one isolate in duplicate, so the contents of the kit are sufficient to perform ELISA serotyping in duplicate on 10,000 strains. Since the titers of the antisera vary on a lot-to-lot basis, the dilution levels used with each kit must be optimized. The use of a commercially prepared set of antisera and a semiquantitative ELISA format greatly reduce the variability of antiserum quality as well as the inconsistencies in judgement associated with weakly agglutinating antigen-antiserum combinations. Using L. monocytogenes strains of known serotypes as references (Fig. 1 and 2), serotypes of uncharacterized isolates can be assigned easily and with little ambiguity. With the exception of a strain previously serotyped as 4e by agglutination, which was consistently serotyped as 4b by ELISA, strains representative of all serotypes were typeable by this method. The distinctions between serotypes 4b and 4e and between serotypes 4d and 4e are poorly defined in the original scheme (Table 2). Modifications to this scheme proposed by Garcia et al. (6) make the distinctions marginally clearer, in that all serotype 4e strains, and some serotype 4b strains, were proposed to contain O-antigen IX. In contrast, neither of these serotypes reacted positively to O-factor antiserum IX in the ELISA, using cells prepared as described. Also, two strains typed as 4a by agglutination were typed as 4c by ELISA, the difference resulting from negative reactions to O-factor antiserum IX in the ELISA. These data suggest that O-antigen IX may be unstable during cell preparation or intermittently expressed at sufficient levels to detect by ELISA in certain L. monocytogenes strains, and that further optimization of O-factor antiserum IX reaction conditions is necessary for consistent use in the ELISA method.
Another source of variability in serotype identification may arise when L. monocytogenes cultures contain mixtures of strains of different serotypes. This scenario arose when our laboratories assigned different serotypes, most obviously in serogroups 1/2 and 4, to apparently identical samples of particular strains (Table 1). The discrepancy was investigated by developing a serogroup-specific colony immunoblot method, which could distinguish these serogroups by differential staining (Fig. 3). L. monocytogenes isolates are routinely obtained through enrichment procedures, the result of which may contain more than one strain of a particular serotype. For example, single-colony picks from an enrichment culture from one soil sample showed that the soil contained two different serotypes of L. monocytogenes (strains TP1-1 and TP1-3; data not shown). Picking one or a small number of colonies following an enrichment protocol for serotyping may therefore not include all serotypes present in the original sample. Alternatively, the colony immunoblot method could be used on entire streak or spread plates to quickly ascertain whether they contained mixtures of, in this case, serogroup 1/2 or 3 and serogroup 4 strains. This assay could be developed further in order to distinguish between serogroups 1/2 and 3, and different serogroup 4 types.
As an epidemiological tool, serotyping is not sufficiently discriminatory to determine positive correlations between food-borne isolates and clinical isolates in cases of listeriosis outbreaks, though a negative relationship between isolates could be demonstrated by differing serotype results in these instances (19). More-discriminatory molecular methods are currently in use (7, 10, 12, 13, 16, 20, 22) and offer a greater degree of confidence in determining epidemiological relatedness of food-borne and clinical L. monocytogenes isolates. However, because serotype designation has been correlated with virulence potential (only 3 of the 14 serotypes cause a vast majority of human listeriosis cases (21), serotyping does provide relevant subtyping information. The ELISA and colony immunoblotting techniques described in this paper are useful means to quickly and economically serotype L. monocytogenes isolates and confirm the serotype uniformity of a culture before more-detailed epidemiological analyses are performed.
Acknowledgments
We thank C. Donnelly, L. Graves, J. Hu, G. Inami, M. Janda, H. Kinde, J. Luchansky, R. Meinersmann, D. Portnoy, R. Rabourne, I. Wesley, and M. Wiedmann for providing L. monocytogenes strains and serotype information; A. Bates, J. Reynolds, and W. Muraoka for technical assistance; and J. Lindsay for facilitating sharing of strains.
This work was supported by U.S. Department of Agriculture Agricultural Research Service CRIS project 5325-42000-040-00D and CWU 5348-32000-017-00D.
REFERENCES
- 1.Brosch, R., M. Brett, B. Catimel, J. B. Luchansky, B. Ojeniyi, and J. Rocourt. 1996. Genomic fingerprinting of 80 strains from the W. H. O. multicenter international typing study of Listeria monocytogenes via pulsed-field gel electrophoresis (PFGE). Int. J. Food Microbiol. 32:343-355. [DOI] [PubMed] [Google Scholar]
- 2.Bubert, A., I. Hein, M. Rauch, A. Lehner, B. Yoon, W. Goebel, and M. Wagner. 1999. Detection and differentiation of Listeria spp. by a single reaction based on multiplex PCR. Appl. Environ. Microbiol. 65:4688-4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chasseignaux, E., P. Gerault, M. T. Toquin, G. Salvat, P. Colin, and G. Ermel. 2002. Ecology of Listeria monocytogenes in the environment of raw poultry meat and raw pork meat processing plants. FEMS Microbiol. Lett. 210:271-275. [DOI] [PubMed] [Google Scholar]
- 4.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fenlon, D. R. 1985. Wild birds and silage as reservoirs of Listeria in the agricultural environment. J. Appl. Bacteriol. 59:537-543. [DOI] [PubMed] [Google Scholar]
- 6.Garcia, J. A., L. Dominguez, V. Briones, M. Blanco, J. F. Fernandez-Garayzabal, and G. Suarez. 1990. Revision of the antigenic structure of genus Listeria. FEMS Microbiol. Lett. 55:113-119. [DOI] [PubMed] [Google Scholar]
- 7.Jersek, B., P. Gilot, M. Gubina, N. Klun, J. Mehle, E. Tcherneva, N. Rijpens, and L. Herman. 1999. Typing of Listeria monocytogenes strains by repetitive element sequence-based PCR. J. Clin. Microbiol. 37:103-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kearney, T. E., M. J. Larkin, J. P. Frost, and P. N. Levett. 1993. Survival of pathogenic bacteria during mesophilic anaerobic digestion of animal waste. J. Appl. Bacteriol. 75:215-219. [DOI] [PubMed] [Google Scholar]
- 9.Kemeny, D. M., and S. J. Challacombe (ed.). 1988. ELISA and other solid phase assays. John Wiley & Sons, New York, N.Y.
- 10.Louie, M., P. Jayaratne, I. Luchsinger, J. Devenish, J. Yao, W. Schlech, and A. Simor. 1996. Comparison of ribotyping, arbitrarily primed PCR, and pulsed-field gel electrophoresis for molecular typing of Listeria monocytogenes. J. Clin. Microbiol. 34:15-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manzano, M., L. Cocolin, C. Cantoni, and G. Comi. 1998. A rapid method for the identification and partial serotyping of Listeria monocytogenes in food by PCR and restriction enzyme analysis. Int. J. Food. Microbiol. 42:207-212. [DOI] [PubMed] [Google Scholar]
- 12.Mereghetti, L., P. Lanotte, V. Savoye-Marczuk, N. Marquet-Van Der Mee, A. Audurier, and R. Quentin. 2002. Combined ribotyping and random multiprimer DNA analysis to probe the population structure of Listeria monocytogenes. Appl. Environ. Microbiol. 68:2849-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nadon, C. A., D. L. Woodward, C. Young, F. G. Rodgers, and M. Wiedmann. 2001. Correlations between molecular subtyping and serotyping of Listeria monocytogenes. J. Clin. Microbiol. 39:2704-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pritchard, T. J., K. J. Flanders, and C. W. Donnelly. 1995. Comparison of the incidence of Listeria on equipment versus environmental sites within dairy processing plants. Int. J. Food Microbiol. 26:375-384. [DOI] [PubMed] [Google Scholar]
- 15.Restaino, L., E. W. Frampton, R. M. Irbe, G. Schabert, and H. Spitz. 1999. Isolation and detection of Listeria monocytogenes using fluorogenic and chromogenic substrates for phosphatidylinositol-specific phospholipase C. J. Food Prot. 62:244-251. [DOI] [PubMed] [Google Scholar]
- 16.Schonberg, A., E. Bannerman, A. L. Courtieu, R. Kiss, J. McLauchlin, S. Shah, and D. Wilhelms. 1996. Serotyping of 80 strains from the W. H. O. multicentre international typing study of Listeria monocytogenes. Int. J. Food Microbiol. 32:279-287. [DOI] [PubMed] [Google Scholar]
- 17.Schuchat, A., B. Swaminathan, and C. V. Broome. 1991. Epidemiology of human listeriosis. Clin. Microbiol. Rev. 4:169-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seeliger, H. P. R., and K. Höhne. 1979. Serotyping of Listeria monocytogenes and related species. Methods Microbiol. 13:31-49. [Google Scholar]
- 19.Seeliger, H. P. R., and B. Langer. 1989. Serological analysis of the genus Listeria. Its values and limitations. Int. J. Food Microbiol. 8:245-248. [DOI] [PubMed] [Google Scholar]
- 20.Swaminathan, B., S. B. Hunter, P. M. Desmarchelier, P. Gerner-Smidt, L. M. Graves, S. Harlander, R. Hubner, C. Jacquet, B. Pedersen, K. Reineccius, A. Ridley, N. A. Saunders, and J. A. Webster. 1996. W. H. O.-sponsored international collaborative study to evaluate methods for subtyping Listeria monocytogenes: restriction fragment length polymorphism (RFLP) analysis using ribotyping and Southern hybridization with two probes derived from L. monocytogenes chromosome. Int. J. Food Microbiol. 32:263-278. [DOI] [PubMed] [Google Scholar]
- 21.Tappero, J. W., A. Schuchat, K. A. Deaver, L. Mascola, J. D. Wenger, et al. 1995. Reduction in the incidence of human listeriosis in the United States. Effectiveness of prevention efforts? JAMA 273:1118-1122. [DOI] [PubMed] [Google Scholar]
- 22.Waak, E., W. Tham, and M. L. Danielsson-Tham. 2002. Prevalence and fingerprinting of Listeria monocytogenes strains isolated from raw whole milk in farm bulk tanks and in dairy plant receiving tanks. Appl. Environ. Microbiol. 68:3366-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weis, J., and H. P. R. Seeliger. 1975. Incidence of Listeria monocytogenes in nature. Appl. Microbiol. 30:29-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welshimer, H. J. 1960. Survival of Listeria monocytogenes in soil. J. Bacteriol. 80:316-320. [DOI] [PMC free article] [PubMed] [Google Scholar]