Abstract
We analyzed 62 clinical isolates of streptogramin A-resistant (SGAr) Staphylococcus aureus collected between 1981 and 2001 in 14 hospitals located in seven French cities. These isolates, including five with decreased susceptibility to glycopeptides, were distributed into 45 antibiotypes and 38 SmaI genotypes. Each of these genotypes included between 1 and 11 isolates, the SmaI patterns of which differed by no more than three bands. Although numerous clones were identified, we observed the spread of monoclonal isolates either within the same hospital or within hospitals in distinct cities and at large time intervals. Hybridization with probes directed against 10 SGAr genes (vatA, vatB, vatC, vatD, vatE, vgaA, vgaB, vgaAv, vgbA, and vgbB) revealed six patterns: vgaAv (21 isolates), vatA-vgbA (24 isolates), vgaAv-vatB-vgaB (14 isolates), vgaAv-vatA-vgbA (1 isolate), vgaAv-vatA-vgbA-vatB-vgaB (1 isolate), and vgaA (1 isolate). We detected at least one SGAr determinant in all of the tested isolates. vgaAv, which is part of the recently characterized transposon Tn5406, was found in 59.7% of the tested isolates. Of the 16 streptogramin B-susceptible isolates, 14 carried vgaAv alone and were susceptible to the mixtures of streptogramins, whereas the 2 isolates carrying vgaAv-vatB-vgaB were resistant to these mixtures. vatA-vgbA was found on plasmids of the same apparent size in 26 (42%) of the tested clinical isolates from 18 unrelated SmaI genotypes. The possible dissemination of some of the multiple clones characterized in the present study with an expected increased selective pressure of streptogramins following the recent licensing of Synercid (quinupristin-dalfopristin) must be carefully monitored.
Methicillin-resistant Staphylococcus aureus (MRSA) has become a major nosocomial pathogen worldwide. Glycopeptides have been the reference drugs for the treatment of MRSA infections (16, 21, 30). After the emergence of clones with decreased susceptibility to these antibiotics, first reported as sporadic cases in several countries and more recently also as outbreaks, alternative treatments such as quinupristin-dalfopristin (Synercid) have been promoted (17, 21, 33). Quinupristin and dalfopristin are derivatives of pristinamycins IA and IIA, respectively (10). Quinupristin-dalfopristin is an injectable, semisynthetic, mixture with a synergistic activity against most gram-positive pathogens. Since 1999, quinupristin-dalfopristin has been available for use in hospitals for the treatment of infections caused by gram-positive cocci that are resistant to other antibiotics.
Quinupristin-dalfopristin and the natural antibiotics produced by streptomycetes, such as streptogramin, pristinamycin, synergistin, mikamycin, and virginiamycin, are mixtures of two classes of compounds, A and B, with distinct primary structures (10, 14). The A compounds are polyunsaturated cyclic macrolactones, and the B compounds are cyclic hexadepsipeptides. Both types of compounds bind different targets in the peptidyltransferase domain of the 23S ribosomal subunit and inhibit protein elongation at different steps. Compounds A and B are bacteriostatic when used separately but act synergistically when combined, such that in some cases they are bactericidal, mainly against gram-positive bacteria.
In countries such as France, where natural mixtures (pristinamycin and synergistin) have been used orally and topically since 1960, the prevalence of clinical isolates of staphylococci resistant to mixtures of compounds A and B (pristinamycin MICs of >2 mg liter−1) varies from 0 to 44% in hospitals (22). Virginiamycin was long used in animal feed as a growth promoter in both Europe and the United States but was banned in Europe in 1999. The first staphylococcal clinical isolates resistant to the mixtures were reported in France in 1975 (14). Staphylococcal resistance to synergistic mixtures is always associated with resistance to A compounds (pristinamycin IIA MICs of ≥8 mg liter−1) but is not necessarily associated with resistance to B compounds (1).
In staphylococci, resistance to B compounds is mediated by (i) the methylation of 23S rRNA, which confers resistance to macrolide-lincosamide-streptogramin B (MLSB) when the erm genes are expressed constitutively (32); (ii) the hydrolysis of the drug by lyases (24) encoded by the vgbA (7) and vgbB (5) genes, which are always located on plasmids conferring resistance to A compounds; (iii) probable efflux ABC proteins encoded by the msr genes, which confers resistance to streptogramin B (SGB) and C14-C15 macrolides (26, 27); or (iv) a mutation in the L22 ribosomal protein of S. aureus, which is responsible for cross-resistance to SGB and erythromycin (23). Resistance to A compounds may be conferred by two categories of genes: (i) vatABCDE (4, 5, 8, 18, 25, 35), encoding related acetyltransferases that inactivate the drug and are found on staphylococcal and/or Enterococcus faecium plasmids, and (ii) vgaAB (2, 6) and lsa (28), encoding related ATP-binding proteins, the resistance mechanism of which has not been elucidated. vga-type genes are found in staphylococcal plasmids but not in E. faecium strains, whereas lsa conferring resistance to SGA and to clindamycin is present in the chromosome of all tested E. faecalis strains. A variant of vgaA (vgaAv) (19) carried by a transposon found in staphylococci, Tn5406 (20), which is located on plasmids and/or the chromosome, was recently characterized.
We have become aware of the vatA-vgbA combination (33; data not shown), which was not found in the 52 unrelated staphylococci tested previously (1). Indeed, vatA-vgbA was always associated with vgaA. Related plasmids (15 to 45 kb) carrying this combination have been detected in strains belonging to various staphylococcal species and isolated prior to 1997 (3).
The aim of the present study was to type and analyze the distribution of SGA resistance (SGAr) genes, because of their uncommon phenotype, in a collection of 62 SGAr clinical isolates of S. aureus sent to us from 1981 to 2001 by French hospital microbiologists.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The relevant characteristics of the 62 S. aureus SGAr clinical isolates used are reported in Table 1. These isolates were collected from different patients between 1981 and 2001 in 14 hospitals located in seven French cities. Thirty-five patients were infected (56.5%), whereas seventeen (27.4%) were colonized. No information was available for the other 10 patients. S. aureus strains BM3093 (3), BM3318 (19), and BM10692 (3), S. cohnii subsp. cohnii BM10711 (5), and E. faecium strain K14 (18) were used as positive controls for the detection of streptogramin resistance genes. S. aureus strain NCTC 8325, the SmaI pattern of which served as a size standard in pulsed-field gel electrophoresis, was also used. The following recombinant plasmids were used as probes in hybridization experiments: pIP1652-vatA (8), pIP1692-vatB (4), pIP1740-vatC (5), pIP1795-vatD (25), pIP1802-vatE (18), pIP1653-vgaA (6), pIP1799-vgaAv (19), pIP1705-vgaB (2), pIP1654-vgbA (7), pIP1741-vgbB (5), and pIP1644-IS257tnp (12).
TABLE 1.
Relevant characteristics of 62 S. aureus clinical isolates resistant to SGA
| SmaI geno- typea | Yr | Source (city or city-hospital) | Antibiotic resistance markersb | Presence or absence of streptogramin resistance genec:
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| vgaAv | vgaA | vatA | vatB | vgaB | vgbA | ||||
| 1A | 1999 | Paris-A | CHL PRI PIB PIIA PEF | − | − | + (9.5) | − | − | + (9.5) |
| 2A | 1997 | Grenoble | PEN STR KAN TOB GEN MLSBi PRI PIIA LIN CLI SUL TMP | + (0.6 + 1.1 + 1.3) | − | − | + (5.5) | + (5.5) | − |
| 3 | 2001 | Cahors | PRI PIB PIIA | − | − | + (9.5) | − | − | + (9.5) |
| 4 | 1999 | Villiers-St Denis | PEN MET PRI PIB PIIA TECI | − | − | + (8.0) | − | − | + (8.0) |
| 5 | 1999 | Paris-A | PEN TET MIN PRI PIB PIIA SUL FUC | − | − | + (9.5) | − | − | + (9.5) |
| 6 | 1999 | Paris-A | PEN MET STR SPT KAN NEO TOB GEN TET MLSBc PRI PIIA SUL PEF RIF | − | − | + (9.5) | − | − | + (9.5) |
| 7 | 2000 | Remiremont | PEN STR SPT KAN TOB GEN TET MLSBc PRI PIIA SUL PEF RIF TECI | + (0.6 + 1.1) | − | − | + (5.0) | + (5.0) | − |
| 8a | 1998 | Paris-A | PEN MET KAN NEO TOB PRI PIB PIIA PEF | − | − | + (9.5) | − | − | + (9.5) |
| 8b | 1998 | Paris-A | PEN PRI PIB PIIA | − | − | + (9.5) | − | − | + (9.5) |
| 9 | 2000 | Blois | PEN MET SPT MLSBc PRI PIIA TMP PEF RIF FUC FOF VANI TEC | − | + (3.2) | − | − | − | − |
| 10 | 2000 | Paris-C | MLSBc PRI PIIA PEF | + (0.6 + 1.3) | − | − | − | − | − |
| 11 | 2000 | Paris-C | PEN KAN TOB GEN MLSBc PRI PIIA FUC | + (0.6 + 1.1 + 1.3) | − | − | + (5.5) | + (5.5) | − |
| 12a | 1999 | Villiers-St Denis | PEN MET SPT KAN NEO TOB MLSBi PRI PIB PIIA PEF | − | − | + (9.5) | − | − | + (9.5) |
| 12a | 2001 | Paris-G | PEN MET SPT KAN NEO TOB MLSBi PRI PIB PIIA PEF | − | − | + (9.5) | − | − | + (9.5) |
| 12b | 2000 | Orsay | PEN MET KAN NEO TOB MLSBi PRI PIB PIIA PEF | − | − | + (9.5) | − | − | + (9.5) |
| 13 | 1981 | Paris-A | PEN MET STR SPT KAN NEO TOB GEN TET MIN MLSBc PRI PIIA | + (0.6 + 1.1) | − | − | + (7.5) | + (7.5) | − |
| 14a | 1998 | Toulouse | PEN MET KAN NEO TOB GEN PRI PIIA LIN PEF TMP | + (0.6 + 1.1 + 1.3) | − | − | + (5.5) | + (5.5) | − |
| 14b | 1999 | Toulouse | PEN MET SPT KAN NEO TOB MLSBc PRI PIIA PEF TECI | + (0.6 + 1.3) | − | − | − | − | − |
| 15a | 1991 | Paris-A | PEN MET STR SPT KAN NEO TOB GEN TET MIN MLSBc PRI PIIA PEF RIF hVANI TECI | − | − | + (9.5) | − | − | + (9.5) |
| 15b | 1999 | Paris-A | PEN MET SPT KAN NEO TOB MLSBc PRI PIIA PEF | − | − | + (9.5) | − | − | + (9.5) |
| 15c | 1993 | Paris-A | PEN MET STR SPT KAN NEO TOB GEN TET MIN MLSBc PRI PIIA SUL TMP PEF RIF FOF hVANI TEC | + (0.6 + 1.1) | − | − | + (5.0) | + (5.0) | − |
| 16aB | 1999 | Paris-A | PEN MET SPT KAN NEO TOB MLSBc PRI PIIA TMP PEF | − | − | + (8.0) | − | − | + (8.0) |
| 16bB | 1999 | Paris-A | PEN MET SPT KAN NEO TOB GEN MLSBc PRI PIIA PEF RIF | − | − | + (9.5) | − | − | + (9.5) |
| 17aB | 1998 | Paris-A | PEN MET SPT KAN NEO TOB MLSBc PRI PIIA PEF | − | − | + (8.0) | − | − | + (8.0) |
| 17bB | 1996 | Grenoble | PEN MET SPT KAN NEO TOB GEN MLSBc PRI PIIA TMP PEF FOF | + (0.6 + 1.1) | − | − | + (4.5) | + (4.5) | − |
| 18C | 1990 | Paris-A | PEN STR KAN NEO MLSBi PIIA LIN PEF | + (0.6 + 1.3) | − | − | − | − | − |
| 19C | 1999 | Paris-A | PEN MET STR KAN NEO MLSBc PRI PIIA SUL TMP FUC | − | − | + (9.5) | − | − | + (9.5) |
| 20D | 1999 | Paris-A | PEN MET SPT KAN NEO TOB MLSBc PRI PIIA PEF | − | − | + (9.5) | − | − | + (9.5) |
| 21D | 1998 | Paris-A | PEN MET KAN NEO TOB PRI PIB PIIA PEF | − | − | + (9.5) | − | − | + (9.5) |
| 22aE | 1998 | Paris-A | PEN MET SPT KAN NEO TOB MLSBc PRI PIIA PEF | − | − | + (8.0) | − | − | + (8.0) |
| 22aE | 1999 | Paris-A | PEN MET SPT KAN NEO TOB GEN MLSBc PRI PIIA PEF | + (0.6 + 1.1) | − | − | + (5.5) | + (5.5) | − |
| 22aE | 1999 | Paris-A | PEN MET SPT KAN NEO TOB GEN MLSBc PRI PIIA PEF | + (0.6 + 1.1) | − | − | + (5.5) | + (5.5) | − |
| 22aE | 1999 | Paris-A | PEN MET SPT KAN NEO TOB GEN MLSBc PRI PIIA PEF | + (0.6 + 1.1) | − | − | + (5.5) | + (5.5) | − |
| 23E | 1988 | Paris-A | PEN MET STR SPT KAN NEO TOB GEN TET MIN MLSBc PRI PIIA PEF | + (0.6 + 1.1) | − | − | + (7.5) | + (7.5) | − |
| 24aF | 1997 | Toulouse | PEN MET PIIA LIN PEF | + (0.6 + 1.3) | − | − | − | − | − |
| 24aF | 1998 | Toulouse | PEN MET KAN NEO TOB PIIA LIN PEF | + (0.6 + 1.3) | − | − | − | − | − |
| 24aF | 1999 | Toulouse | PEN MET KAN NEO TOB CHL PIIA LIN PEF | + (0.6 + 1.3) | − | − | − | − | − |
| 24bF | 2000 | Toulouse | PEN MET KAN NEO TOB PIIA LIN PEF | + (0.6 + 1.3) | − | − | − | − | − |
| 24cF | 1998 | Toulouse | PEN MET KAN NEO TOB PIIA LIN PEF | + (0.6 + 1.3) | − | − | − | − | − |
| 24cF | 1999 | Paris-A | PEN MET SPT KAN NEO TOB MLSBc PRI PIIA PEF | + (0.6 + 1.3) | − | − | − | − | − |
| 24dF | 1999 | Paris-A | PEN MET KAN NEO TOB PIIA LIN PEF | + (0.6 + 1.3) | − | − | − | − | − |
| 24eF | 1998 | Toulouse | PEN MET KAN NEO TOB PIIA LIN PEF | + (0.6 + 1.3) | − | − | − | − | − |
| 24fF | 1999 | Toulouse | PEN MET KAN NEO TOB PIIA LIN PEF | + (0.6 + 1.3) | − | − | − | − | − |
| 24gF | 1999 | Toulouse | PEN MET KAN NEO TOB PIIA LIN PEF TECI | + (0.6 + 1.3) | − | − | − | − | − |
| 24hF | 1997 | Paris-A | PEN MET KAN NEO TOB GEN MLSBc PRI PIIA TMP PEF | + (0.6 + 1.3 + 4.2 + 8) | − | − | − | − | − |
| 25F | 2000 | Toulouse | PEN MET MLSBc PRI PIIA PEF | + (0.6 + 1.3 + 3) | − | − | − | − | − |
| 26F | 1993 | Paris-D | PEN MET KAN NEO TOB PRI PIB PIIA PEF RIF | − | − | + (9.5) | − | − | + (9.5) |
| 27 | 2000 | Toulouse | PEN MET SPT KAN NEO TOB MLSBc PRI PIIA PEF TECI | + (0.6 + 5) | − | − | − | − | − |
| 28G | 1997 | Paris-A | PEN PIIA LIN | + (0.6 + 1.3) | − | − | − | − | − |
| 29G | 1993 | Paris-A | PEN MET STR KAN NEO TOB GEN PIIA LIN RIF | + (0.6 + 1.3) | − | − | − | − | − |
| 30aH | 1999 | Toulouse | PEN PIIA LIN PEF | + (0.6 + 1.3) | − | − | − | − | − |
| 30bH | 1988 | Paris-A | PEN STR SPT KAN NEO TOB GEN PRI PIB PIIA LIN | + (0.6 + 1.3) | − | + (9.5) | − | − | + (9.5) |
| 31H | 1999 | Paris-E | PEN KAN TOB GEN TET MLSBc PRI PIIA FUC TECI | + (0.6 + 1.3) | − | − | − | − | − |
| 32 | 1999 | Paris-A | PEN TET ERY PRI PIB PIIA PEF | − | − | + (8.0) | − | − | + (8.0) |
| 33 | 1997 | Paris-A | PEN KAN TOB GEN PIIA LIN CLI TMP FUC | + (0.6 + 1.3) | − | − | − | − | − |
| 34 | 1999 | Paris-A | PEN MET STR KAN TOB GEN MLSBc PRI PIIA SUL PEF RIF FUC hVANI TEC | + (0.6 + 1.1) | − | − | + (7.5) | + (7.5) | − |
| 35 | 2001 | Villiers-St Denis | PEN MET STR KAN NEO TOB GEN MLSBc PRI PIIA SUL TMP PEF RIF FUC FOF VANI TEC | + (0.6 + 1.1 + 2.5) | − | − | + (9.5) | + (9.5) | − |
| 36a | 1996 | Paris-B | PEN MET STR KAN NEO PRI PIB PIIA | − | − | + (9.5) | − | − | + (9.5) |
| 36a | 1996 | Paris-B | PEN MET STR KAN NEO PRI PIB PIIA | − | − | + (9.5) | − | − | + (9.5) |
| 37 | 1995 | Paris-F | PEN STR KAN TOB GEN CHL MLSBc PRI PIIA SUL TMP PEF RIF FOF | + (0.6 + 1.1) | − | − | + (5.5) | + (5.5) | − |
| 38a | 2001 | Villiers-St Denis | PEN MET SPT KAN NEO TOB GEN MLSBc PRI PIIA PEF | + (0.6 + 1.1) | − | + (9.5) | + (5.0) | + (5.0) | + (9.5) |
| 38b | 2001 | Villiers-St Denis | PEN MET SPT KAN NEO TOB MLSBc PRI PIIA PEF | − | − | + (9.5) | − | − | + (9.5) |
Genotypes which include strains that are possibly related (less than seven band differences) are marked with the same superscript letter.
Abbreviations: CHL, chloramphenicol; CLI, clindamycin; FUC, fucidic acid; FOF, fosfomycin; hVANI, heterogeneous vancomycin intermediate; KAN, kanamycin; LIN, lincomycin; MLSBc, macrolide-lincosamide-SGB constitutive resistance; MLSBi, macrolide-lincosamide-SGB inducible resistance; MET, methicillin; MIN, minocycline; NEO, neomycin; PEF, pefloxacin; PEN, penicillinase; PIB, pristinamycin IB (SGB); TEC, teicoplanin; TECI, TEC intermediate; TOB, tobramycin; TMP, trimethoprim; VANI, vancomycin intermediate.
The following genes were investigated by hybridization at high stringency with the intragenic probes: vatA, vatB, vatC, vatD, vatE, vgaA, vgaB, vgaAv, vgbA, and vgbB. The genes reported were those found in at least one strain. The presence or absence of the cited genes are indicated by + or −, respectively. “+” is followed by the size(s) of the hybridizing HindIII fragment(s) in kilobases.
Media.
Staphylococci were grown in brain heart infusion broth (Difco Laboratories, Detroit, Mich.). Susceptibility to antibiotics was tested on Mueller-Hinton agar (Bio-Rad, Hercules, Calif.).
Susceptibility to antimicrobial drugs.
All clinical isolates were stored at −80°C and were isolated on Mueller-Hinton agar containing 10 μg of pristinamycin IIA ml−1. Susceptibility to antibiotics was determined by a disk diffusion assay with commercially available antibiotic disks (Bio-Rad), according to the recommendations of the French Society of Microbiology and with disks prepared in our laboratory as described previously (1). The MICs of vancomycin and teicoplanin were determined by the E-test according to the manufacturer's recommendations (AES, Combourg, France). A population analysis on brain heart infusion broth containing 4 mg of vancomycin liter−1 was carried out as previously described to screen for heterogeneous vancomycin intermediate S. aureus (hetero-VISA) strains (11).
DNA isolation and analysis.
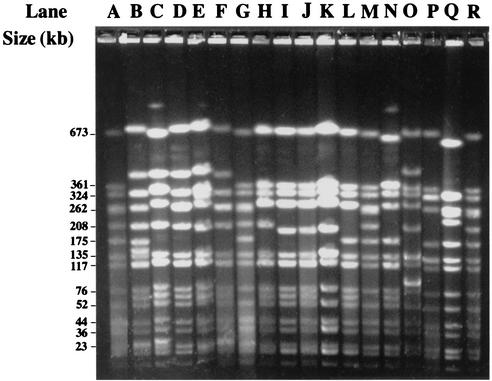
Total cellular DNA was isolated from staphylococcal strains and purified by using the QIAamps tissue kit from Qiagen (Hilden, Germany). S. aureus plasmid DNA was extracted and purified by using the QIAprep spin plasmid kit from Qiagen according to the manufacturer's instructions but with an additional step consisting of 1 h at 37°C in the presence of 10 μl of lysostaphin (1 mg ml−1). Restriction endonucleases were obtained from Amersham-Pharmacia Biotech, Inc. (Piscataway, N.J.) and used according to the manufacturer's instructions. Instagene matrix DNA preparations were used for PCR experiments according to the manufacturer's instructions (Bio-Rad). The DNA was digested with SmaI and subjected to pulsed-field gel electrophoresis as described previously (Fig. 1) (13). Concatameric bacteriophage lambda DNA molecules (48.5 kb; Bio-Rad) and the SmaI fragments of the cellular DNA from S. aureus NCTC 8325 were used as size standards. Macrorestriction fingerprints were compared visually and were scanned by using GelCompar II software (Applied Maths, Sint-Martens-Latem, Belgium). A similarity matrix was created by use of the band-based Dice similarity coefficient (tolerance of 1% and optimization of 2%). The unweighted pair-group method with average linkages was used to cluster the strains on the basis of the SmaI patterns. If the dendrograms revealed clusters that included strains with similarity of at least 80%, the patterns of the strains in the same cluster were compared visually on the same gel. The strains were clustered according to the following criteria proposed by Tenover et al. (31): (i) strains were grouped in the same major genotype if their patterns differed by no more than three bands (these strains were considered to be closely related and monoclonal); (ii) if patterns differed by between four and six bands, the strains were scored as being posssibly related but were, nevertheless, classified into distinct genotypes to discriminate them from the closely related strains; and (iii) if patterns differed by seven or more bands, strains were considered to be different. Major genotypes are designated by Arabic numerals. Strains with undistinguishable patterns were classified within the same subtype. Subtypes are designated by Arabic numerals with letter suffixes.
FIG. 1.
Pulsed-field gel electrophoresis of SmaI-digested total DNA from SGAr S. aureus clinical isolates. Lanes A, G, M, and R, NCTC 8325 used as standard; lane B, genotype 17a strain; lane C, genotype 15a strain; lane D, genotype 17c strain; lane E, genotype 16b strain; lane F, genotype 17b strain; lane H, genotype 24h strain (Paris-A, 1997); lane I, genotype 24a strain; lane J, genotype 24a strain (Toulouse, 1998); lane K, genotype 24c strain (Paris-A, 1999); lane L, genotype 30a strain; lane N, genotype 30b strain; lane O, BM3318; lane P, BM3093; lane Q, BM10692.
Labeling of DNA probes, blotting, and hybridization.
Hybridization experiments were performed at 65°C as described previously (19).
PCR.
DNA was amplified by PCR by using the Ready-To-Go kit (Amersham) according to the manufacturer's instructions in a Crocodile III apparatus (Appligène, Illkirch, France). The primers used to detect antibiotic resistance genes were those described previously: vatA (8), vatB (4), vgaB (2), and vgbA (7). A DNA fragment from within vgaAv (19) was amplified with primer A (5′-CTCCGTGTTGAAGATGTTTCG-3′; nucleotides [nt] 5881 to 5901; accession no. AF186237) and primer B (5′-GGATTCAAACGCCTCTATAGCC-3′; nt 6339 to 6318; accession no. AF186237). The pIP1680 DNA fragment extending from pAMβ1 repE-like gene to vatA was amplified with the primers RepE (5′-ATTGCTGAAGGTACTGAAGG-3′; nt 4343 to 4362; accession no. AF007787) and VatA2 (5′-CAATGACCATGGACCTGATC-3′; nt 269 to 288; accession no. L07778). PCR experiments were carried out at high stringency (initial cycle of 5 min at 95°C and 2 min at 55°C, followed by 35 cycles of 1 min at 72°C, 30 s at 95°C, and 1 min at 55°C, with a final extension step of 5 min at 72°C).
RESULTS
Analysis of antibiotypes.
As shown in Table 1, 48 of the 62 SGAr isolates tested (77.4%) were also resistant to the mixtures of A and B compounds, including pristinamycin. Of these 48 isolates, 46 were resistant to B compounds. These 46 SGBr isolates were distributed into three MLSB phenotypes: MLSBc (31 isolates), SGB (11 isolates), and MLSBiSGB (4 isolates). Of the 16 SGAr isolates susceptible to B compounds, 14 were also resistant to lincomycin, and 2 of these 14 isolates were also resistant to clindamycin.
A total of 46 of the 62 SGAr S. aureus isolates studied were MRSA (74.2%) (Table 1). These MRSA isolates could be divide into gentamicin-sensitive (GENs) and GENr phenotypes (30 and 16 strains, respectively), whereas 9 of the 16 methicillin-susceptible S. aureus (MSSA) isolates were GENs. Eighteen of the MRSA isolates (39.1%) and none of the MSSA isolates were spectinomycin resistant. Pefloxacin-resistant and rifampin-resistant strains were more prevalent among MRSA than among MSSA. Indeed, 40 of the 46 (87%) MRSA isolates were pefloxacin-resistant compared to just 7 of the 16 MSSA isolates (43.8%), and 9 of the MRSA isolates (19.6%) were rifampin resistant compared to one of the MSSA isolates (6.3%).
Of the 62 SGAr strains, 5 had reduced susceptibility to teicoplanin and vancomycin; three of these strains were hetero-VISA, and two were glycopeptide-intermediate S. aureus. These strains were isolated in three different hospitals between 1991 and 2001. All of these strains were MRSA and were either resistant or susceptible to GEN and related aminoglycosides. We also detected six teicoplanin-intermediate strains, four of which were MRSA.
Analysis of the SmaI patterns.
As shown in Table 1, the 62 SGAr isolates were distributed into 38 distinct genotypes based on the comparison of SmaI patterns, and each genotype included between 1 and 11 isolates. Although grouped into distinct genotypes, the strains in the following genotypes may have been related because their SmaI patterns differed by six bands or less: genotypes 1 and 2; 16 and 17; 18 and 19; 20 and 21; 22 and 23; 24, 25, and 26; 28 and 29; and 30 and 31. It is noteworthy that major clonal diversity (20 genotypes) was observed among the 30 isolates from one hospital (Paris-A). Some of the isolates belonging to the same genotype were isolated in distinct hospitals located in different cities (12a and 12b, 17a and 17b, 24a to 24h, and 30a and 30b) and/or had different antibiotypes (14a and 14b; 15a to 15c; 24a, 24c, 24g, and 24h; 30a and 30b). Some strains grouped in the same genotype were also disseminated over fairly large time scales (15a in 1991 and 15b in 1999; 30b in 1988 and 30a in 1999).
Distribution of the streptogramin resistance genes tested.
In addition to the eight SGAr genes tested, we looked for the presence of vgbA and vgbB, which are frequently localized on SGAr plasmids. We detected vgaAv in 37 of the 62 SGAr strains analyzed (59.7%). These strains were isolated between 1981 and 2001. vgaAv was found alone in 21 strains, in association with vgaB-vatB in 14 strains, in association with vatA-vgbA in 1 strain, and in association with vgaB-vatB and vatA-vgbA in 1 strain. In this latter strain, we confirmed the presence of each of these resistance genes by using PCR to amplify the cellular DNA of a single colony. Of the 25 isolates that did not carry vgaAv, 24 contained vatA-vgbA and 1 contained vgaA alone. Thus, we detected at least one SGAr determinant in each of the 62 SGAr clinical isolates investigated. None of the isolates carried vatC, vatD, vatE, or vgbB.
We measured the sizes of the HindIII fragments that hybridized with the intragenic streptogramin resistance probes. Clinical isolates with the same hybridization pattern did not necessarily belong to the same genotype. In the 21 strains carrying only vgaAv, this gene was carried by HindIII fragments of 0.6 + 1.3 kb (18 isolates), 0.6 + 1.3 + 3 kb (1 isolate), 0.6 + 1.3 + 4.2 + 8 kb (1 isolate), and 0.6 + 5 kb (1 isolate). Two of these profiles were identical to those of S. aureus strains BM3327 (0.6 + 1.3 kb) and BM3252 (0.6 + 1.3 + 3 kb) (19), which carry one and two chromosomal copies of Tn5406, respectively (20). In the 14 strains carrying vgaAv and vatB-vgaB, we found three HindIII patterns with vgaAv: 0.6 + 1.1 kb (10 strains), 0.6 + 1.1 + 1.3 kb (3 strains), and 0.6 + 1.1 + 2.5 (1 strain). Two of these profiles were not distinguishable from those of S. aureus strains BM3318 (0.6 + 1.1 kb) and BM3385 (0.6 + 1.1 + 1.3 kb) (19), which carry either one plasmid copy or one chromosomal copy and one plasmid copy of Tn5406, respectively (20). In each of the 14 strains containing vgaAv-vatB-vgaB, a single HindIII fragment hybridized with both vatB and vgaB. The size of this fragment varied (4.5, 5, 5.5, 7.5, and 9.5 kb). In S. aureus BM3385, in which vatB and vgaB were shown to be cotranscribed (2), the two genes were located on a 7-kb HindIII fragment (results not shown).
In each of the 26 strains containing vatA and vgbA, associated or not with other genes (vgaAv in one isolate and vgaAv-vatB-vgaB in one isolate), vatA and vgbA were found on a single HindIII fragment of 8 or 9.5 kb, whereas in the seven native vgaA-vatA-vgbA plasmids tested previously (3), the cotranscribed genes, vatA-vgbA, were on a 4.8-kb HindIII fragment.
Plasmid content of the SGAr strains carrying vatA-vgbA.
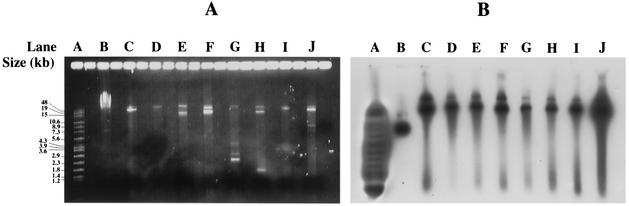
Plasmids were extracted from the 26 strains carrying vatA-vgbA and probed with vgbA (pIP1654) and IS257tnp (pIP1644). The extrachromosomal bands (Fig. 2A) that hybridized with both probes had the same apparent sizes and were located just below the chromosomal DNA fragments of the uncleaved total cellular DNA (Fig. 2B). The hybridization patterns with vgbA of these plasmids cleaved by HindIII (results not shown) were identical to those observed with the cellular DNA cleaved by the same enzyme (8 and 9.5 kb). In 5 isolates collected in two hospitals in 1998 and 1999, the cohybridizing band was 8 kb, whereas in 21 isolates collected in seven hospitals between 1988 and 2001 the cohybridizing band was 9.5 kb. The plasmids generating 8-kb HindIII fragments hybridizing with vgbA seem to have emerged later.
FIG. 2.
Agarose gel electrophoresis patterns of plasmid DNA from SGAr S. aureus clinical isolates carrying vatA-vgbA (lanes C to J). (A) Plasmid migration patterns; (B) hybridization patterns with the vgbA probe (pIP1654). Lanes A, the Raoul marker (Appligene) used as a standard; lanes B, total DNA from BM3093; lanes C, plasmid DNA from genotype 22a strain (Paris-A, 1998); lanes D, plasmid DNA from genotype 36a strain; lanes E, plasmid DNA from genotype 8a strain; lanes F, plasmid DNA from genotype 32 strain; lanes G, plasmid DNA from genotype 3 strain; lanes H, plasmid DNA from genotype 19 strain; lanes I, plasmid DNA from genotype 20 strain; lanes J, plasmid DNA from genotype 16a strain.
PCR experiments were carried out with primers pairing with pAMβ1 repE-like gene and vatA from pIP680 harbored by S. aureus strain BM3093. The cellular DNA of BM3093 and of five isolates of the present study carrying vatA-vgbA plasmids (16b, 17a, 30b, 32, and 22a isolated in 1998; Table 1) were used as substrates. Fragments of the same size (2.6 kb [data not shown]) were amplified from the DNA of BM3093 and the two isolates (16b and 30b) carrying plasmids generating 9.5-kb HindIII fragments hybridizing with vgbA. In contrast, none of the three other isolates that carried plasmids generating 8-kb HindIII fragments hybridizing with vgbA generated amplicons. Thus, the plasmids generating 9.5-kb HindIII fragments hybridizing with vgbA seem to be related to pIP680, whereas such a conclusion cannot be drawn for the other plasmid type.
DISCUSSION
The analysis of 62 SGAr S. aureus clinical isolates revealed numerous clones: 38 different genotypes were observed, each including closely related isolates whose SmaI patterns differed by no more than three fragments. Twenty-nine unrelated genotypes included possibly related isolates whose SmaI patterns differed by no more than six fragments. This multiplicity of unrelated clones is not surprising since these isolates were not necessarily collected during outbreaks, unlike those analyzed previously (15). Indeed, the clinical isolates included in the present study were mainly sent to us because of their uncommon antibiotic resistance patterns. Nevertheless, we observed the spread of monoclonal isolates both within the same hospital (Paris-A, Paris-B, Toulouse, or Villiers-St Denis) or in distinct hospitals, occasionally located in distinct cities (Villiers-St Denis, Paris-G, and Orsay; Paris-A and Toulouse; Paris-A and Grenoble), or at time intervals of up to 11 years, such as the genotype 30 strains collected in 1988 and 1999 (Table 1).
All of the staphylococcal isolates tested contained at least one SGAr gene. This finding is consistent with our previous findings concerning another collection of 52 SGAr independent staphylococci belonging to five species (1, 19). Since some of the clinical isolates in both collections carried several SGAr genes, we cannot rule out the possibility that staphylococci contain unknown genes, whereas among E. faecium strains new genes have to be characterized. In the study by Werner et al. (34), 7 of the 148 E. faecium isolates resistant to the mixtures did not contain any of the SGAr genes described to date and in the study by Soltani et al. (29), 8 of the 28 E. faecium mixture-resistant isolates did not contain any of these genes. Despite the multiplicity of unrelated clones detected in the present study, only six distinct SGr patterns were found. This may be due to the dissemination of structurally related plasmids in independent clones, as reported previously (3, 9). The vatA and vgbA combination was not detected in our previous study (1), in which vgaA was always associated with vatA-vgbA. A comparative analysis of seven native vatA-vgbA-vgaA plasmids (15 to 45 kb) revealed the presence of a common 12.1-kb fragment carrying the three streptogramin resistance genes (3). This probably resulted from integration of a pAMβ1-like plasmid harboring vatA-vgbA in which the replication gene is inactivated by IS257 insertion and a functional vgaA plasmid. A deletion between IS257 flanking vgaA may have resulted in the formation of vatA-vgbA plasmids (Fig. 2A). The fact that these plasmids have the same size and are found in 42% of the isolates studied here is striking.
Zarrouk et al. (36) reported that the staphylococcal strains resistant to A compounds but susceptible to B compounds remain susceptible to mixtures in vitro and in vivo. According to our previous (19) and present results, this is true when the strains carry vgaAv alone but not when they carry three SGAr genes. Indeed, the 14 SGBs isolates harboring vgaAv alone and originating from two hospitals (Paris-A and Toulouse) had the lincosamide and streptogramin A resistance phenotype. Like BM3385, the two lincosamide and streptogramin A-resistant isolates (genotypes 2 and 14a) (Table 1) that were susceptible to SGB and carried vgaAv, vatB, and vgaB were resistant to the mixtures (Table 1). Four of the seven HindIII hybridization patterns detected with vgaAv were identical to those of previously analyzed S. aureus clinical isolates in which vgaAv was shown to be carried by Tn5406 (19, 20). These results reflect a multiplicity of Tn5406 insertion sites in the collection of isolates tested.
We have found 45 antibiotypes among the SGAr clinical isolates tested. Some of them, such as genotypes 3 and 28 (Table 1), were suceptible to almost all other antibiotics, whereas genotype 15a, 15c, and 35 isolates were multiply resistant to various drugs, including glycopeptides (Table 1). The SmaI pattern of the genotype 15a isolate was identical to that of the major clone with reduced glycopeptide susceptibility found in European cities (17, data not shown). Coresistance to quinupristin-dalfopristin and glycopeptides (33), as in 5 of the 62 clinical isolates tested (8%), is of great concern since it may lead to therapeutic failure.
Since 1975, monoclonal staphylococcal isolates resistant to mixtures of the A and B compounds have occasionally been detected in the same hospitals or in hospitals located in geographically distinct French cities (15). However, in most French hospitals the prevalence of such isolates does not exceed 5% (14). The moderate use of natural mixtures in intensive care units, in which MRSA clinical isolates are easily spread, and the instability of SGAr plasmids, which are frequently lost or rearranged in the absence of selective pressure (1, 3), may explain why the numerous distinct isolates found in French hospitals have not disseminated often. The possible dissemination of such clones with the increased selective pressure of streptogramins after the recent licensing of quinupristin-dalfopristin has to be prevented by improving hygiene and patient isolation measures.
Acknowledgments
We thank the microbiologists who sent us clinical isolates.
For part of this work, J. Haroche received grants from the Fondation Bayer Santé and from CANAM (Caisse Nationale d'Assurance Maladie et Maternité des Travailleurs Non Salariés des Professions Non Agricoles).
REFERENCES
- 1.Allignet, J., S. Aubert, A. Morvan, and N. El Solh. 1996. Distribution of genes encoding resistance to streptogramin A and related compounds among staphylococci resistant to these antibiotics. Antimicrob. Agents Chemother. 40:2523-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allignet, J., and N. El Solh. 1997. Characterization of a new staphylococcal gene, vgaB, encoding a putative ABC transporter conferring resistance to streptogramin A and related compounds. Gene 202:133-138. [DOI] [PubMed] [Google Scholar]
- 3.Allignet, J., and N. El Solh. 1999. Comparative analysis of staphylococcal plasmids carrying three streptogramin-resistance genes: vat-vgb-vga. Plasmid 42:134-138. [DOI] [PubMed] [Google Scholar]
- 4.Allignet, J., and N. El Solh. 1995. Diversity among the gram-positive acetyltransferases inactivating streptogramin A and structurally related compounds and characterization of a new staphylococcal determinant, vatB. Antimicrob. Agents Chemother. 39:2027-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allignet, J., N. Liassine, and N. El Solh. 1998. Characterization of a staphylococcal plasmid related to pUB110 and carrying two novel genes, vatC and vgbB, encoding resistance to streptogramins A and B and similar antibiotics. Antimicrob. Agents Chemother. 42:1794-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allignet, J., V. Loncle, and N. El Solh. 1992. Sequence of a staphylococcal plasmid gene, vga, encoding a putative ATP-binding protein involved in resistance to virginiamycin A-like antibiotics. Gene 117:45-51. [DOI] [PubMed] [Google Scholar]
- 7.Allignet, J., V. Loncle, P. Mazodier, and N. El Solh. 1988. Nucleotide sequence of a staphylococcal plasmid gene, vgb, encoding a hydrolase inactivating the B components of virginiamycin-like antibiotics. Plasmid 20:271-275. [DOI] [PubMed] [Google Scholar]
- 8.Allignet, J., V. Loncle, C. Simenel, M. Delepierre, and N. El Solh. 1993. Sequence of a staphylococcal gene, vat, encoding an acetyltransferase inactivating the A-type compounds of virginiamycin-like antibiotics. Gene 130:91-98. [DOI] [PubMed] [Google Scholar]
- 9.Aubert, S., K. G. Dyke, and N. E. Solh. 1998. Analysis of two Staphylococcus epidermidis plasmids coding for resistance to streptogramin A. Plasmid 40:238-242. [DOI] [PubMed] [Google Scholar]
- 10.Barriere, J. C., N. Berthaud, D. Beyer, S. Dutka-Malen, J. M. Paris, and J. F. Desnottes. 1998. Recent developments in streptogramin research. Curr. Pharm. Des. 4:155-180. [PubMed] [Google Scholar]
- 11.Chesneau, O., A. Morvan, and N. E. Solh. 2000. Retrospective screening for heterogeneous vancomycin resistance in diverse Staphylococcus aureus clones disseminated in French hospitals. J. Antimicrob. Chemother. 45:887-890. [DOI] [PubMed] [Google Scholar]
- 12.Derbise, A., S. Aubert, and N. El Solh. 1997. Mapping the regions carrying the three contiguous antibiotic resistance genes aadE, sat4, and aphA-3 in the genomes of staphylococci. Antimicrob. Agents Chemother. 41:1024-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derbise, A., K. G. Dyke, and N. El Solh. 1996. Characterization of a Staphylococcus aureus transposon, Tn5405, located within Tn5404 and carrying the aminoglycoside resistance genes, aphA-3 and aadE. Plasmid 35:174-188. [DOI] [PubMed] [Google Scholar]
- 14.El Solh, N., and J. Allignet. 1998. Staphylococcal resistance to streptogramins and related antibiotics. Drug Resist. Updates 1:169-175. [DOI] [PubMed] [Google Scholar]
- 15.El Solh, N., M. Casetta, S. Aubert, A. Morvan, and J. Allignet. 1995. Typing of methicillin-resistant Staphylococcus aureus (MRSA) based on genome analysis, p. 45-64. In C. Brun-Buisson, M. W. Casewell, N. El Solh, and B. Régnier (ed.), Methicillin-resistant staphylococci. Flammarion, Paris, France.
- 16.Geisel, R., F. J. Schmitz, A. C. Fluit, and H. Labischinski. 2001. Emergence, mechanism, and clinical implications of reduced glycopeptide susceptibility in Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 20:685-697. [DOI] [PubMed] [Google Scholar]
- 17.Guerin, F., A. Buu-Hoi, J. L. Mainardi, G. Kac, N. Colardelle, S. Vaupre, L. Gutmann, I. Podglajen, K. P. Koteva, D. W. Hughes, and G. D. Wright. 2000. Outbreak of methicillin-resistant Staphylococcus aureus with reduced susceptibility to glycopeptides in a Parisian hospital. J. Clin. Microbiol. 38:2985-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haroche, J., J. Allignet, S. Aubert, A. E. Van Den Bogaard, and N. El Solh. 2000. satG, conferring resistance to streptogramin A, is widely distributed in Enterococcus faecium strains but not in staphylococci. Antimicrob. Agents Chemother. 44:190-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haroche, J., J. Allignet, C. Buchrieser, and N. El Solh. 2000. Characterization of a variant of vga(A) conferring resistance to streptogramin A and related compounds. Antimicrob. Agents Chemother. 44:2271-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haroche, J., J. Allignet, and N. El Solh. 2002. Tn5406, a new staphylococcal transposon conferring resistance to streptogramin A and related compounds, including dalfopristin. Antimicrob. Agents Chemother. 46:2337-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiramatsu, K. 1998. Vancomycin resistance in staphylococci. Drug Resist. Updates 1:135-150. [DOI] [PubMed] [Google Scholar]
- 22.Lepelletier, D., and Richet, H. 2001. Surveillance et contrôle des infections a Staphylococcus aureus resistants à la meticilline dans les hôpitaux français. Bull. Epidemiol. Hebd. 2001:25-27. [Google Scholar]
- 23.Malbruny, B., A. Canu, B. Bozdogan, B. Fantin, V. Zarrouk, S. Dutka-Malen, C. Feger, and R. Leclercq. 2002. Resistance to quinupristin-dalfopristin due to mutation of L22 ribosomal protein in Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2200-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukhtar, T. A., K. P. Koteva, D. W. Hughes, and G. D. Wright. 2001. Vgb from Staphylococcus aureus inactivates streptogramin B antibiotics by an elimination mechanism not hydrolysis. Biochemistry 40:8877-8886. [DOI] [PubMed] [Google Scholar]
- 25.Rende-Fournier, R., R. Leclercq, M. Galimand, J. Duval, and P. Courvalin. 1993. Identification of the satA gene encoding a streptogramin A acetyltransferase in Enterococcus faecium BM4145. Antimicrob. Agents Chemother. 37:2119-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ross, J. I., A. M. Farrell, E. A. Eady, J. H. Cove, and W. J. Cunliffe. 1989. Characterisation and molecular cloning of the novel macrolide-streptogramin B resistance determinant from Staphylococcus epidermidis. J. Antimicrob. Chemother. 24:851-862. [DOI] [PubMed] [Google Scholar]
- 27.Singh, K. V., K. Malathum, and B. E. Murray. 2001. Disruption of an Enterococcus faecium species-specific gene, a homologue of acquired macrolide resistance genes of staphylococci, is associated with an increase in macrolide susceptibility. Antimicrob. Agents Chemother. 45:263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh, K. V., G. M. Weinstock, and B. E. Murray. 2002. An Enterococcus faecalis ABC homologue (Lsa) is required for the resistance of this species to clindamycin and quinupristin-dalfopristin. Antimicrob. Agents Chemother. 46:1845-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soltani, M., D. Beighton, J. Philpott-Howard, and N. Woodford. 2000. Mechanisms of resistance to quinupristin-dalfopristin among isolates of Enterococcus faecium from animals, raw meat, and hospital patients in Western Europe. Antimicrob. Agents Chemother. 44:433-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srinivasan, A., J. D. Dick, and T. M. Perl. 2002. Vancomycin resistance in staphylococci. Clin. Microbiol. Rev. 15:430-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tenover, F. C., M. V. Lancaster, B. C. Hill, C. D. Steward, S. A. Stocker, G. A. Hancock, C. M. O'Hara, S. K. McAllister, N. C. Clark, and K. Hiramatsu. 1998. Characterization of staphylococci with reduced susceptibilities to vancomycin and other glycopeptides. J. Clin. Microbiol. 36:1020-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weisblum, B. 2000. Resistance to the macrolide-lincosamide-streptogramin antibiotics, p. 694-710. In V. A. Fischetti et al. (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 33.Werner, G., C. Cuny, F. J. Schmitz, and W. Witte. 2001. Methicillin-resistant, quinupristin-dalfopristin-resistant Staphylococcus aureus with reduced sensitivity to glycopeptides. J. Clin. Microbiol. 39:3586-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Werner, G., I. Klare, H. Heier, K. H. Hinz, G. Bohme, M. Wendt, and W. Witte. 2000. Quinupristin/dalfopristin-resistant enterococci of the satA (vatD) and satG (vatE) genotypes from different ecological origins in Germany. Microb. Drug Resist. 6:37-47. [DOI] [PubMed] [Google Scholar]
- 35.Werner, G., and W. Witte. 1999. Characterization of a new enterococcal gene, satG, encoding a putative acetyltransferase conferring resistance to streptogramin A compounds. Antimicrob. Agents Chemother. 43:1813-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zarrouk, V., B. Bozdogan, R. Leclercq, L. Garry, C. Carbon, and B. Fantin. 2000. Influence of resistance to streptogramin A type antibiotics on the activity of quinupristin-dalfopristin in vitro and in experimental endocarditis due to Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1168-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]