Abstract
We report the results of a multicenter evaluation of a new assay for the detection of hepatitis C virus (HCV) RNA in human serum or plasma based on transcription-mediated amplification (HCV TMA). Analysis of combined data obtained from 15 independent sites, including 4 sites in the United States and 11 in Europe, by using preproduction kits showed a limit of detection of 9.8 IU/ml and an overall mean specificity of 97.9%. In addition, assay runs and samples were valid consistently (97.8% of assay runs and 98.0% of specimen results). Of the 15 sites that participated in the multicenter evaluation, 2 subsequently carried out additional performance studies with production kits in support of the assay's registration in France. Comparison of the findings from these two sites during the multicenter evaluation and during the registration studies showed an overall improvement in assay performance. A statistically significant (P < 0.001) improvement was achieved for both specificity and specimen validity in the registration studies, which were 99.4 and 98.1%, respectively. Combined data from the two sites showed a lower limit of detection of approximately 2.4 IU/ml and an improved assay validity of 100%, although the sample size was too small to show statistical significance at the 0.05 level. In summary, the performance characteristics of HCV TMA indicate that this assay is a reliable tool for the detection of HCV RNA in serum or plasma. Improvement in assay performance has been demonstrated with refinement of assay reagents, instrumentation, and operator experience.
Qualitative hepatitis C virus (HCV) RNA assays play a key role in both the diagnosis and monitoring of HCV infection. The enzyme immunoassays (EIAs) that detect antibodies to HCV cannot differentiate between active and resolved infection. Qualitative HCV RNA assays detect viral genomes and therefore can both confirm the presence of active infection and demonstrate its presence 4 to 6 weeks before antibody seroconversion takes place (1). Most qualitative HCV RNA tests are 10- to 100-fold more sensitive than are quantitative HCV RNA tests and up to 1,000-fold more sensitive than is the HCV core antigen assay, rendering them the method of choice both for confirming active infection and for assessing viral clearance in response to therapy (4, 7). Indeed, according to the recent National Institutes of Health consensus statement on management of hepatitis C (http://consensus.nih-gov/cons/116/116cdc_intro.htm), a qualitative HCV RNA assay with a lower limit of detection of 50 IU/ml or less should be used to confirm acute or chronic HCV infection in a patient with a positive EIA result as well as to indicate sustained virologic response, defined as the absence of detectable HCV RNA in serum at 24 weeks after the end of treatment.
A new assay for the detection of HCV RNA in human serum or plasma based on transcription-mediated amplification (HCV TMA) has recently been introduced. Developed and manufactured by Gen-Probe Inc. (San Diego, Calif.) and marketed by Bayer Corp. Diagnostics Division (Tarrytown, N.Y.), the VERSANT HCV RNA qualitative assay is available worldwide. An extensive preclinical evaluation of the assay conducted at both Bayer and Gen-Probe revealed a sensitivity of 5.3 IU/ml (1a), which is consistent with the French package insert of 50 copies/ml (9.6 IU/ml) detected at least 95% of the time (VERSANT HCV RNA qualitative assay product insert, Bayer Corp.). The preclinical evaluation also demonstrated the excellent specificity (99.6%) and high reproducibility of the assay, as well as its ability to detect different HCV genotypes, its robustness to different storage and handling conditions, and the lack of interference by endogenous substances as well as common pathogens and commonly used drugs (1a). Other studies have confirmed both the sensitivity and specificity of the assay and its ability to equally detect different HCV genotypes (5, 8).
Here we report the results of the first multicenter evaluation of HCV TMA. The purpose of the study was to assess the analytical performance of the assay in first-time-user laboratories and to assess and refine Bayer's ability to successfully transfer the product for consumer use. The study involved 15 independent sites, 4 in the United States and 11 in Europe. We analyzed data from all 15 sites to determine the sensitivity, specificity, and validity of assay runs and specimen results in the field. In addition, we evaluated the improvement in assay performance by comparing results of the multicenter evaluation to those of additional studies performed at two sites in support of the registration of the assay in France. We also assessed the ability of HCV TMA to detect different HCV genotypes and examined the concordance between the results of HCV TMA and the Roche AMPLICOR HCV 2.0 assay (Amplicor).
MATERIALS AND METHODS
Study sites.
A total of 15 independent sites participated in the study, including 4 sites in the United States and 11 in Europe. Each site had a dedicated operator and used preproduction kit lots of HCV TMA. Before testing was begun, dedicated operators at 14 sites received training both at facilities within Bayer Corp. and on-site. At one site, the operator received only on-site training. Each site conducted four assay runs (one run per day for 4 days) for assessment of analytical performance. In addition, 10 sites tested specimens with HCV TMA that had been tested previously with Amplicor (Roche Molecular Systems, Pleasanton, Calif.).
Two sites performed additional studies of sensitivity, specificity, validity, and HCV genotype detection in support of the application for approval of the assay in France. These registration studies were performed with production kit lots.
HCV TMA.
HCV TMA was performed as specified by instructions and with kits provided by Bayer Corp. To begin, 50 μl of target capture reagent (capture oligonucleotides and magnetic microparticles in HEPES buffer with detergent) was mixed with 1.0 ml of internal control reagent (RNA transcript in HEPES buffer with detergent). Then, 400 μl of target capture reagent with internal control was added to separate polypropylene tubes, each of which contained 500 μl of sample or calibrator, and the mixture was incubated at 60°C for 20 min and then at room temperature for 25 min. After the magnetic microparticles were washed twice with wash buffer (HEPES buffer with detergent), 75 μl of amplification reagent (primers, deoxynucleoside triphosphates [dNTPs], NTPs, and cofactors in Tris buffer) and 200 μl of 100% dimethyl silicone were added to each tube and the mixture was incubated at 60°C for 10 min and then at 41.5°C for 10 min. To produce RNA amplicons, 25 μl of enzyme reagent (Moloney murine leukemia virus reverse transcriptase and T7 RNA polymerase in HEPES-Tris buffer) was added to each tube and the mixture was incubated at 41.5°C for 1 h. For detection using the hybridization protection assay, 100 μl of HCV probe reagent (chemiluminescent oligonucleotide probe in succinate buffer with detergent) was added to each tube and the mixture was incubated at 60°C for 1 h; then 250 μl of selection reagent (borate buffer with surfactant) was added and the mixture was incubated at 60°C for 1 h. The tubes were allowed to cool at 19 to 27°C for at least 10 min, and then the relative light units (RLUs) for each sample and calibrator were read in either a LEADER high-count (HC) luminometer with exponential tail fit algorithm data reduction software version 1.30 (for multicenter evaluation by all 15 sites) or a LEADER HC+ luminometer with TMA data reduction software (for registration studies by two sites).
Validity of assay runs and sample results.
The validity of the assay runs and sample results was determined on the basis of criteria specified by Bayer Corp. Each assay run included three replicates each of a positive calibrator (inactivated HCV-positive plasma in defibrinated normal human plasma with gentamicin and sodium azide) and negative calibrator (defibrinated normal human plasma with gentamicin and sodium azide). For an assay run to be considered valid, two or more of the positive calibrators and two or more of the negative calibrators must be valid on the basis of the acceptance criteria specified for each luminometer.
For the LEADER HC luminometer used in the multicenter evaluation, positive calibrators must generate an analyte signal between 400,000 and 2,000,000 RLU, inclusive, and an internal-control signal less than or equal to 1,000,000 RLU to be valid. Negative calibrators must generate an analyte signal between 0 and 35,000 RLU, inclusive, and an internal-control signal between 75,000 and 300,000 RLU, inclusive, to be valid. A sample result was considered to be valid and reactive when the sample generated an analyte signal greater than or equal to the analyte cutoff and an internal-control signal less than or equal to 2,000,000 RLU.
For the LEADER HC+ luminometer used by two sites in the registration studies, positive calibrators must generate an analyte signal between 400,000 and 2,700,000 RLU, inclusive, and an internal-control signal less than or equal to 475,000 RLU to be valid. Negative calibrators must generate an analyte signal between 0 and 40,000 RLU, inclusive, and an internal-control signal between 75,000 and 300,000 RLU, inclusive, to be valid. A sample result was considered to be valid and reactive when the sample generated an analyte signal greater than or equal to the analyte cutoff and an internal-control signal less than or equal to 475,000 RLU.
For both luminometers, a sample result was considered to be valid and nonreactive when the sample generated an analyte signal less than the analyte cutoff and an internal-control signal greater than or equal to the internal-control cutoff.
Sensitivity.
Assay sensitivity was assessed at each of the 15 sites by using a six-member dilution panel tested in replicates of six per run, with one run performed each day over the course of 4 days to give a total of 2,160 results. The dilution panel members contained HCV RNA concentrations of 164, 82, 61, 41, 20, and 8 copies/ml. The dilution panel was created by diluting HCV genotype 1a stock material (ProMedDx, LLC, Norton, Mass.) into human HCV-seronegative EDTA plasma (Seracon II basepool; Intergen Co., Purchase, N.Y.). The HCV RNA level (in copies per milliliter) in the stock material was measured with the VERSANT HCV RNA 3.0 assay (bDNA) (Bayer Corp.), and the precise dilution of each panel member was determined by gravimetric means. Due to insufficient sample volume, dilution panel members were not retested in the event of an invalid result. Only valid results from valid runs were included in the sensitivity analysis. The limit of detection (LoD) was defined as the HCV RNA concentration at which reactive results were obtained for 95% of the tests. Logistic regression analysis (3) was used to determine the LoD from results combined from all of the sites after conversion of copies per milliliter to IU per milliliter using a conversion factor of 5.2 copies/IU. The 5.2-copies/IU conversion factor is used for converting VERSANT HCV RNA 3.0 assay (bDNA) values from copies per milliliter to IU per milliliter and was derived from testing the World Health Organization (WHO) International Standard for HCV RNA (NIBSC code 96/790) by using an RNA transcript reference standard in place of the kit standards.
Two sites performed additional sensitivity studies by testing dilutions of the WHO International Standard for HCV RNA (9, 10). At site 1, dilutions of the standard were calibrated at HCV RNA concentrations of 20, 10, 5, 2.5, and 1 IU/ml and each dilution was tested 10 times to yield a total of 50 results. At site 2, dilutions of the standard were calibrated at HCV RNA concentrations of 20, 10, 5, 2, and 1 IU/ml. The 20-, 5-, 2-, and 1-IU/ml dilutions were tested 10 times, and the 10-IU/ml dilution was tested 20 times, to yield a total of 60 results.
Specificity.
Assay specificity was evaluated at each site by testing seronegative plasma samples obtained from the Center for Blood Research (Sacramento, Calif.). A total of 1,600 unique seronegative plasma samples were tested. Fourteen sites tested 100 unique samples twice in different runs, and one site tested 200 unique samples once, thereby yielding 3,000 test results. Each site performed one run per day on four different days, with 50 samples included in each run. Samples that gave invalid or reactive results were retested at the discretion of the site. A total of 89 of 100 samples that initially gave invalid results and 57 of 62 samples that gave reactive results were retested. Only valid results from valid runs were included in the specificity analysis. Specificity was defined as the percentage of nonreactive test results among all valid results.
Additional specificity studies were performed by two sites. Site 1 tested 514 samples from blood donors, provided by the Hôpital Beaujon (Paris, France) transfusion center. Two of these samples were excluded from the analysis because they were found to be anti-HCV positive and HCV RNA positive by the COBAS Amplicor HCV monitor (Roche Molecular Systems). Site 2 tested 516 seronegative sera obtained from ProMedDx. Fourteen of these samples were excluded from the analysis because valid results were not obtained from them.
HCV genotype detection.
The ability of HCV TMA to detect HCV genotypes 1a, 1b, 2, 3, 4, 5, and 6 was assessed at two study sites. Serum samples were genotyped at site 1 by using the VERSANT HCV LiPA (Bayer Corp.) and at site 2 by sequencing the 5′-untranslated region using the TruGene hepatitis C assay (Visible Genetics, Inc., Toronto, Canada). The viral load of each sample was measured by the VERSANT HCV RNA 3.0 assay (bDNA), and samples were diluted to 50 copies/ml (site 1) or 10 IU/ml (site 2) into Basematrix (Boston Biomedica, Inc., West Bridgewater, Mass.). HCV genotypes were tested in replicates of 10 at each site.
Concordance studies.
Serum samples that had previously been tested for HCV RNA with Amplicor and stored at −20°C or below were tested by HCV TMA by 10 of the sites. Investigators were asked to select samples that had been previously tested by any quantitative and qualitative HCV RNA assay and, where possible, to select those whose results fell below the limit of detection of the quantitative assay. In addition, samples of interest such as those with EIA-negative and PCR-positive results, serial samples from liver transplant patients, and samples for which clinical information was available were selected. Each of the 10 sites tested serum samples in one run per day over the course of 2 days, with up to 92 serum samples included in each run. Samples with invalid or discrepant results were retested by one or both assays if sufficient volume was available. Only valid results from valid runs were included in the analysis.
RESULTS
Performance of HCV TMA at 15 sites.
The LoD, specificity, and validity of HCV TMA were assessed by combining the data from the 15 sites. These performance characteristics are summarized in Table 1.
TABLE 1.
Performance of HCV TMA at 15 sites
| Parameter | Value | No. of results |
|---|---|---|
| LoD | 9.8 IU/ml | 2,160a |
| Specificity | 97.9% | 2,985b |
| Run validity | 97.8% | 138c |
| Specimen validity | 98.0% | 11,975d |
Number of results obtained from testing of six replicates of a six-member dilution panel on four separate days at 15 sites.
Number of valid results obtained from testing of a total of 1,600 unique specimens.
Total number of assay runs performed by 15 sites in the multicenter evaluation.
Total number of specimen results obtained from 15 sites in the multicenter evaluation.
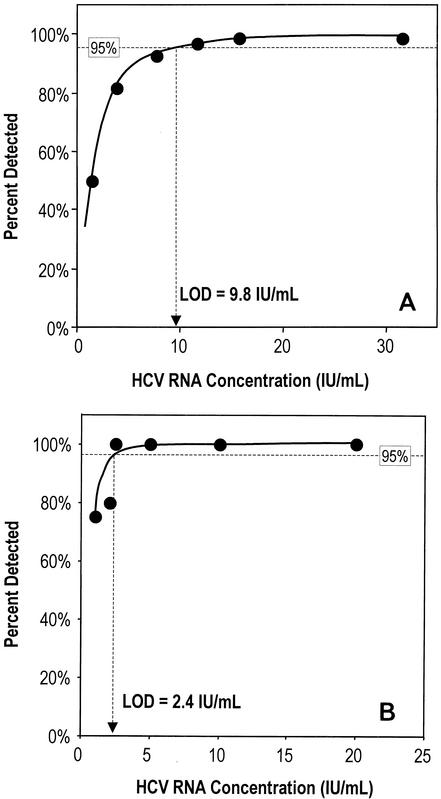
The LoD was derived by logistic regression analysis of 2,160 results obtained from testing of the six-member dilution panel in replicates of six in four separate runs at each of 15 sites. Results from these experiments are shown in Fig. 1 and indicate that the LoD was 9.8 IU/ml (95% confidence interval [CI], 8.5 to 11.5 IU/ml).
FIG. 1.
(A) Sensitivity of HCV TMA based on data from 15 laboratories participating in the multicenter evaluation. The LoD was calculated to be 9.8 HCV RNA IU/ml and is shown as the x coordinate where the calculated curve intercepts the 95% detection line. (B) Sensitivity of HCV TMA based on data from the registration studies performed in two laboratories. The LoD was calculated to be 2.4 HCV RNA IU/ml and is shown as the x coordinate where the calculated curve intercepts the 95% detection line.
Assay specificity was assessed by analyzing results from 1,600 seronegative samples. Of the 3,000 test results obtained on initial testing, 2,900 were valid and 100 were invalid. A total of 89 of the 100 initially invalid seronegative samples were retested, of which 85 were valid and 4 were invalid on retesting. Including the results of retesting, 2,922 of the total 2,985 valid test results obtained were nonreactive and 63 were reactive, thereby indicating an overall mean specificity of 97.9% (lower one-sided 95% CI, 97.3%). Considering only results from initial testing, 2,838 of the 2,900 valid results obtained were nonreactive and 62 were reactive, indicating a mean specificity of 97.9%.
To gain insight into assay performance at various sites, the validity of assay runs and specimen results was evaluated. A total of 138 assays runs were performed at the 15 sites participating in the multicenter evaluation. The number of runs performed at each site ranged from 7 to 13. At 12 sites 100% of the assay runs were valid, and at 14 sites more than 90% of the runs were valid. Overall, 135 of the 138 total assay runs performed were valid, thereby yielding an assay run validity of 97.8%. A total of 11,975 specimen results were obtained from the 15 sites participating in the multicenter evaluation. The number of specimen results obtained at each site ranged from 608 to 1,060. At 12 of the 15 sites, ≥98.4% of results from specimens were valid. Overall, 11,736 of the 11,975 specimen results were valid, yielding a specimen validity of 98.0%.
Additional performance studies at two sites.
Following the multicenter evaluation, two of the sites performed additional studies of HCV TMA in support of the registration of the assay in France. A comparison of the performance characteristics observed by these two sites for the multicenter evaluation with the characteristics observed in the registration studies is shown in Table 2.
TABLE 2.
Improvement in performance of HCV TMA at two sites
| Parameter | Value (no. of results) in:
|
P | |
|---|---|---|---|
| Multicenter evaluation | Registration studies | ||
| LoDa | 9.5 IU/ml (288) | 2.4 IU/ml (110) | >0.05e |
| Specificityb | 95.5% (400) | 99.4% (1,014) | <0.001 |
| Run validityc | 95.5% (22) | 100% (16) | >0.05e |
| Specimen validityd | 96.1% (1,881) | 98.1% (1,340) | <0.001 |
LoD for multicenter evaluation at two sites based on 288 results obtained from testing of six replicates of a six-member dilution panel on four separate days at each site. LoD for registration studies based on 110 results obtained from 10 to 20 replicates of five-member dilution panels at two sites.
Specificity for multicenter evaluation at two sites based on 400 results obtained by testing 200 unique seronegative specimens twice at each site. Specificity for registration studies at two sites based on 1,014 valid results obtained from testing of 1,014 unique seronegative specimens.
Run validity based on 22 assay runs performed at two sites in multicenter evaluation and 16 assay runs in registration studies.
Specimen validity based on 1,881 specimen results obtained from two sites in multicenter evaluation and 1,340 specimens results from registration studies.
Sample is too small to show statistical significance at the 0.05 level.
Assay sensitivity was determined in the registration studies by testing dilutions of the WHO International Standard for HCV RNA. Results at both sites showed detection of HCV RNA in 100% of the replicate samples that contained 20, 10, and 5 IU/ml. Site 1 also detected 100% of replicate samples with HCV RNA at 2.5 IU/ml and 60% of samples with HCV RNA at 1 IU/ml. Site 2 detected HCV RNA in 80% of replicate samples at 2.0 IU/ml and in 90% of samples at 1 IU/ml. Due to the small number of samples tested, the difference between site 1 and site 2 in detection of samples with HCV RNA at 1.0 IU/ml was not statistically significant (P = 0.244). Logistic regression analysis of these results indicated a LoD of 2.4 IU/ml (Fig. 1B). By comparison, testing of the six-member dilution panel during the multicenter evaluation by the same two sites indicated a LoD of 9.5 IU/ml, although the sample was too small to show statistical significance at the 0.05 level (Table 2).
The registration studies performed at the two sites also showed a significant improvement in assay specificity and in the validity of specimen results (Table 2). Valid results from testing of 1,014 unique seronegative specimens in the registration studies yielded a specificity of 99.4%, whereas 400 results from testing of 200 unique seronegative specimens in the multicenter evaluation yielded a significantly lower (P < 0.001) specificity of 95.5%. Similarly, 98.1% of the specimen results in the registration studies were valid, compared with 96.1% of specimen results that were valid when tested by these two sites in the multicenter evaluation. The difference in specimen validity was found to be statistically significant (P < 0.001). Run validity also was higher in the registration studies than in the multicenter evaluation (100% and 95.5%, respectively), although the sample was too small to show statistical significance at the 0.05 level.
HCV genotype detection.
To assess the sensitivity of HCV TMA for different HCV genotypes, 10 replicates of each of genotypes 1a, 1b, 2, 3, 4, 5, and 6 were tested at two sites. Two clinical isolates of each genotype, one at each site, were tested at either 50 copies/ml or 10 IU/ml. Site 1 tested replicates at a dilution that contained 50 copies/ml (equivalent to 9.6 IU/ml), and site 2 tested replicates at a dilution of 10 IU/ml. At both sites, 100% of replicates of each HCV genotype were detected.
Concordance between HCV TMA and Amplicor.
Serum samples that previously had been tested for HCV RNA with Amplicor were selected by the site investigators and then tested at 10 of the sites. The concordance between results of HCV TMA and Amplicor is shown in Table 3. Of the 1,012 samples tested, HCV RNA was detected in 514 (50.8%) of the specimens and was not detected in 447 (44.2%) of the specimens by both assays. Thus, the overall concordance was 94.9%. Of the 51 samples that were discordant, HCV RNA was detected in 46 (4.5%) by HCV TMA but not by Amplicor and HCV RNA was detected in 5 (0.5%) by Amplicor but not by HCV TMA.
TABLE 3.
Concordance between HCV TMA and Amplicor
| Sample type | No. of samples | Concordance (%) | Amplicor result | HCV TMA result
|
|
|---|---|---|---|---|---|
| No. + | No. − | ||||
| All samples | 1,012 | 94.9 | + | 514 | 5 |
| − | 46 | 447 | |||
| EIA indeterminate, Amplicor negative | 6 | 66.7 | − | 2 | 4 |
| EIA positive, Amplicor negative | 71 | 91.5 | − | 6 | 65 |
| EIA positive, Amplicor positive | 32 | 100 | + | 32 | 0 |
| − | 0 | 0 | |||
Of the 46 discordant samples that were positive for HCV RNA by HCV TMA and negative by Amplicor, patient information and results of retesting by both assays was available for 4. As shown in Table 4, all four patients had a clinical history that was consistent with HCV infection. Three of the patients were EIA positive and on retesting were positive for HCV RNA by HCV TMA and negative by Amplicor. The fourth patient was an EIA-negative individual who had received an accidental needlestick containing blood from an HCV-positive patient. Samples from this patient were consistently negative for HCV RNA by Amplicor. On initial testing by HCV TMA, this patient tested positive for HCV RNA with a relatively low signal-to-cutoff ratio of 1.47. On retesting, a negative result was obtained with a signal-to-cutoff ratio of 0.26. Unfortunately, no additional clinical information was available for this patient, and due to limited sample volume, no further testing was performed.
TABLE 4.
Resolution of discordant samples
| Sample no. | HCV TMA result
|
Amplicor result
|
Patient history | ||
|---|---|---|---|---|---|
| Initial test | Retest | Initial test | Retest | ||
| 1 | + | + | − | − | EIA positive, liver transplant recipient |
| 2 | + | + | − | − | EIA positive, receiving interferon treatment for HCV infection; positive for HCV RNA by Amplicor at month 3 of treatment |
| 3 | + | + | − | − | EIA positive, dialysis patient; positive for HCV RNA by PCR for 3 consecutive years |
| 4 | + | − | − | − | Accidental needlestick from HCV-positive patient; baseline sample was EIA negative |
The concordance between HCV TMA and Amplicor was analyzed further in a subset of samples for which EIA results were available (Table 3). Six samples were scored as EIA indeterminate/Amplicor negative, of which two were positive for HCV RNA by HCV TMA. Seventy-one specimens were scored as EIA positive/Amplicor negative, of which six had HCV RNA detectable by HCV TMA (concordance, 91.5%). HCV RNA was detected by HCV TMA in all 32 specimens that were scored as EIA positive/Amplicor positive (100% concordance).
DISCUSSION
Careful evaluation by outside laboratories following the manufacturer's assessment is key to the routine use of a commercial assay in clinical laboratories (7). This study is the first multicenter evaluation of HCV TMA, and the results presented here represent those of first-time operators using preproduction kits. Combined data from the 15 independent laboratories in the United States and Europe that participated in the study showed an overall mean sensitivity of 9.8 IU/ml (95% CI, 8.5 to 11.5 IU/ml), which is consistent with the French package insert claim of 50 copies/ml (9.6 IU/ml) detected at least 95% of the time. Analysis of 2,985 valid test results from 1,600 seronegative specimens by the 15 sites yielded an overall mean specificity of 97.9% (lower one-sided 95% CI, 97.3%). In addition, consistently valid assay runs and sample results were obtained, with 97.8% of assay runs and 98.0% of specimen results from all sites considered valid.
Of the 15 sites that participated in the multicenter evaluation, 2 subsequently carried out additional performance studies; results from these studies are more representative of findings of more experienced operators using production kits. Comparison of the findings from these two sites during the multicenter evaluation and during the registration studies showed an overall improvement in assay performance. In the multicenter evaluation, the combined data from the two sites showed a LoD of 9.5 IU/ml. By comparison, a LoD of approximately 2.4 IU/ml was obtained in the registration studies, although the sample was too small to show statistical significance at the 0.05 level. A statistically significant (P < 0.001) improvement was achieved for both specificity and specimen validity. Whereas the combined data from the two sites showed a 95.5% specificity in the multicenter evaluation, a specificity of 99.4% was obtained in the registration studies. Similarly, significant improvement was observed in specimen result validity, which increased from 96.1% in the multicenter evaluation to 98.1% in the registration studies. The validity of the assay runs also improved from 95.5 to 100%, but the sample was too small to show statistical significance at the 0.05 level.
Several factors might account for the overall improvement in assay performance at the two sites, including differences in assay reagents, instrumentation, and operator experience. The multicenter evaluation was conducted with preproduction kits, whereas the registration studies were performed with production kits that included reagents refined for optimal assay performance. Both in-house studies and studies by independent laboratories using production kits have shown assay sensitivity values (i.e., LoDs) that exceed those stated in the French package insert. A preclinical evaluation conducted at Bayer and Gen-Probe showed an assay sensitivity of 5.3 IU/ml (1a). Although this study combined results from both production (n = 3) and preproduction (n = 4) kits, the investigators found a trend toward performance improvement when production kits were used later in the study (Bayer Corp., internal communication). Independent studies by Krajden et al. (5) and Ross et al. (8) using only production kits reported sensitivity values of 6.0 and 5 IU/ml, respectively. Studies by Ross et al. (8) and Gorrin et al. (1a) noted assay specificities of >98% and ≥99.5%, respectively, slightly better than those observed in this multicenter evaluation. In addition to assay reagents, improvements in the luminometers used may have contributed to the improvement in assay performance. The LEADER HC+ luminometer used by the two sites in the registration studies not only allows better tracking of specimens through the use of bar codes but also has an improved pump system that is less prone to clogging.
Other factors that may have contributed to improvement in assay performance include operator experience and attention to critical details in the assay protocol. During the course of training operators for the multicenter evaluation, two steps in the assay protocol were refined to improve assay performance. One was the addition of enzyme to reagents in the reaction tube, and the other was thorough vortexing of reagents. Indeed, sites where operators were trained after identification of the proper execution of these two key steps achieved better assay performance than did sites where operators were trained before potential errors in these two key steps were known.
Another aspect of assay performance evaluated by the two sites in the registration studies was the ability of HCV TMA to detect all HCV genotypes equally. In panels constructed with HCV RNA levels at or near the LoD of the assay (50 copies/ml or 9.6 IU/ml at site 1 and 10 IU/ml at site 2), both sites obtained 100% detection of replicates of HCV genotypes 1a, 1b, 2, 3, 4, 5, and 6. These results are likely to reflect results obtained with additional clinical isolates, since the target region for HCV TMA is the highly conserved 5′ untranslated region. Indeed, these findings are supported by those of Ross et al. (8) and Gorrin et al. (1a) of equivalent high-level detection of all HCV genotypes.
Finally, an overall concordance of 94.9% between the results of HCV TMA and those of Amplicor also was found by using data from the 10 sites that participated in the concordance study. This result is similar to the overall concordance of 97.3% reported by Ross et al. (8) between the results of HCV TMA and COBAS Amplicor HCV 2.0. Among EIA-positive/PCR-negative samples, the 91.5% concordance reported here is lower than the 96.2% concordance reported by Krajden et al. (5). Among EIA-positive/PCR-positive samples, the 100% concordance reported here is identical to that reported by Krajden et al. (5). Interestingly, as observed in the previous studies, the greater discordance observed in the present study appears to be due mostly to the identification of specimens that were positive for HCV RNA by HCV TMA but negative by Amplicor. Unfortunately, results of retesting by both assays and limited patient history were available for only four discrepant samples. Retesting confirmed the initial results for three of these samples, and in all four cases the patient history indicated either confirmed or possible HCV infection. The fourth sample was positive for HCV RNA by HCV TMA on initial testing but negative on retesting. The signal-to-cutoff ratio for this sample was relatively low in both the initial test and the retest, indicating either that the initial test result was a false-positive result or that the amount of HCV RNA in the sample was close to the detection limit of the assay. In such cases, clarification of the patient's clinical history might aid in interpreting the test results (2). Of note, testing of samples for the concordance analysis by HCV TMA was done retrospectively on samples previously tested by other assays, and details of sample handling and freeze-thaw cycles are unknown. Taken together, the discordant specimens that were positive for HCV RNA by HCV TMA but negative by Amplicor may be explained by the greater sensitivity of HCV TMA.
In summary, the performance characteristics of HCV TMA indicate that this assay is a reliable tool for the detection of HCV RNA in serum or plasma. In multiple laboratories using preproduction kits and performing the assay for the first time, the assay sensitivity was consistent with the claim in the French package insert of 50 copies/ml (9.6 IU/ml) detected at least 95% of the time. Moreover, the improvement in assay performance observed in laboratories with experienced workers using production kits illustrates the possibility that assay sensitivity may even exceed that stated in the French package insert. HCV TMA is easy to use and offers a number of practical advantages. All assay steps, including sample processing, amplification, and detection, are performed in a single tube, and up to 92 samples can be processed in a single run within approximately 5 h. As a highly sensitive, specific, and reliable assay for detection of HCV RNA, HCV TMA may be suitable for routine use in the clinical laboratory.
Acknowledgments
We thank all the individuals at Bayer Corp. that provided support during the study, including Sherrie Ransom, Aylin Asatourians, Kathy Benjamin, Catherine Lemagne, Jonathan Miller, Ed Salanga, Colin Hill, Astrid Klein, Mariella Zainetti, Laura Cattozi, and Marie-Pierre Branger. We also thank Kristi Whitfield (Posterdocs, Oakland, Calif.) for graphics and Linda Wuestehube (SciScript, Lafayette, Calif.) for writing and editorial assistance. We also thank the 15 participating study sites.
REFERENCES
- 1.Carithers, R. L., Jr., A. Marquardt, and D. R. Gretch. 2000. Diagnostic testing for hepatitis C. Semin. Liver Dis. 20:159-171. [DOI] [PubMed] [Google Scholar]
- 1a.Gorrin, G., M. Friesenhahn, P. Lin, M. Sanders, R. Pollner, B. Eguchi, J. Pham, G. Roma, J. Spidle, S. Nicol, C. Wong, S. Bhade, and L. Comanor. 2003. Performance evaluation of the VERSANT HCV RNA qualitative assay by using transcription-mediated amplification. J. Clin. Microbiol. 41:310-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Irving, W. L. 2002. The role of the virology laboratory in the management of hepatitis C virus infection. J. Clin. Virol. 25:3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kleinbaum, D. G. 1992. Logistic regression: a self-learning text, p. 101-119. Springer-Verlag, New York, N.Y.
- 4.Krajden, M. 2000. Hepatitis C virus diagnosis and testing. Can. J. Public Health 91(Suppl. 1):S34-S39. [PubMed] [Google Scholar]
- 5.Krajden, M., R. Ziermann, A. Khan, A. Mak, K. Leung, D. Hendricks, and L. Comanor. 2002. Qualitative detection of hepatitis C virus RNA: comparison of analytical sensitivity, clinical performance, and workflow of the COBAS AMPLICOR HCV test version 2.0 and the HCV RNA transcription-mediated amplification (TMA) qualitative assay. J. Clin. Microbiol. 40:2903-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pawlotsky, J. M. 2000. Hepatitis C viral markers and quasispecies, p. 25-52. In T. J. Liang and J. H. Hoofnagle (ed.), Hepatitis C: biomedical research reports. Academic Press, Inc., San Diego, Calif.
- 7.Pawlotsky, J. M. 2002. Molecular diagnosis of viral hepatitis. Gastroenterology 122:1554-1568. [DOI] [PubMed] [Google Scholar]
- 8.Ross, R. S., S. O. Viazov, S. Hoffmann, and M. Roggendorf. 2001. Performance characteristics of a transcription-mediated nucleic acid amplification assay for qualitative detection of hepatitis C virus RNA. J. Clin. Lab. Anal. 15:308-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saldanha, J. 2001. Validation and standardisation of nucleic acid amplification technology (NAT) assays for the detection of viral contamination of blood and blood products. J. Clin. Virol. 20:7-13. [DOI] [PubMed] [Google Scholar]
- 10.Saldanha, J., N. Lelie, A. Heath, and the WHO Collaborative Study Group. 1999. Establishment of the first international standard for nucleic acid amplification technology (NAT) assays for HCV RNA. Vox Sang. 76:149-158. [DOI] [PubMed] [Google Scholar]