Abstract
cAMP-dependent protein kinase has a central role in the control of mammalian sperm capacitation and motility. Previous protein biochemical studies indicated that the only cAMP-dependent protein kinase catalytic subunit (C) in ovine sperm is an unusual isoform, termed Cs, whose amino terminus differs from those of published C isoforms of other species. Isolation and sequencing of cDNA clones encoding ovine Cs and Cα1 (the predominant somatic isoform) now reveal that Cs is the product of an alternative transcript of the Cα gene. Cs cDNA clones from murine and human testes also were isolated and sequenced, indicating that Cs is of ancient origin and widespread in mammals. In the mouse, Cs transcripts were detected only in testis and not in any other tissue examined, including ciliated tissues and ovaries. Finally, immunohistochemistry of the testis shows that Cs first appears in pachytene spermatocytes. This is the first demonstration of a cell type–specific expression for any C isoform. The conservation of Cs throughout mammalian evolution suggests that the unique structure of Cs is important in the subunit's localization or function within the sperm.
INTRODUCTION
cAMP-dependent protein kinase (PKA) (for review, see Taylor et al., 1990) is a key enzyme in the control of mammalian sperm function (Garbers and Kopf, 1980). PKA-dependent protein phosphorylation is essential for rendering mammalian sperm capable of movement during epididymal maturation (Pariset et al., 1985; Jaiswal and Majumder, 1996; Yeung et al., 1999) and is critical for the maintenance of motility in mature sperm (Garbers et al., 1971; Lindemann, 1978; Tash and Means, 1982; Brokaw, 1987; San Agustin and Witman, 1994; Chaudhry et al., 1995). PKA also is important in the signaling events leading to capacitation and the acrosome reaction in sperm (Duncan and Fraser, 1993; Visconti et al., 1995, 1997, 1999a,b; Galantino-Homer et al., 1997; Aitken et al., 1998; Osheroff et al., 1999). Thus, an understanding of the proteins involved in sperm cAMP-dependent control pathways is a major goal of current research in reproductive biology (Cummings et al., 1994; Burton et al., 1999; Osheroff et al., 1999).
The PKA holoenzyme consists of two catalytic subunits (C) bound to two regulatory subunits (R) in a tetrameric complex (R2C2). There are three known genes encoding mammalian C. The Cα gene is expressed in most tissues (Showers and Maurer, 1986; Uhler et al., 1986a,b). The Cβ gene also is expressed in multiple tissues but generally at lower levels than Cα (Showers and Maurer, 1986; Uhler et al., 1986b). Cγ is a transcribed retroposon found only in primates and expressed only in testis (Beebe et al., 1990; Reinton et al., 1998).
We recently determined that the PKA catalytic subunit of ovine sperm (Cs) differs from that of bovine, murine, or human Cα1 (the predominant somatic isoform) in its amino terminus (San Agustin et al., 1998). A combination of tandem mass spectrometry and Edman degradation of Cs peptides indicated that the amino-terminal myristate and first 14 amino acids of the published Cα1 subunits are replaced by an amino-terminal acetate and 6 different amino acids in ovine Cs. However, short peptide sequences from more carboxyl-terminal portions of ovine Cs were identical to the published sequence of bovine Cα1. Although the complete sequence of neither the sperm nor the somatic form of ovine C was determined, the results indicated that ovine Cs is a novel isoform more closely related to Cα1 than to Cβ or Cγ.
The discovery that ovine sperm contain a novel isoform of C raised a number of important questions. First, how is the sperm isoform generated? Is it the product of a unique gene or of an alternative transcript derived from the same gene as Cα1? Second, how widely distributed is it phylogenetically? The unique isoform was not identified in previous biochemical, immunological, and molecular genetic analyses of sperm PKA or C RNAs and cDNAs from testis of rodents and primates (Beebe et al., 1990; Øyen et al., 1990; Reinton et al., 1998; Burton et al., 1999); was it simply overlooked, or did Cs evolve relatively recently in the sheep or its immediate ancestors? Third, in what tissues is Cs expressed? If it is expressed in a range of ciliated tissues, it may have been selected for assembly into ciliary and flagellar axonemes in general. If Cs is expressed in both male and female reproductive tissues, it may be specific to the germ line. If it is expressed only in testis, is it present in all testicular cells, only in the germ cells, or only in those germ cells producing protein for incorporation into the sperm? If the latter, Cs may have evolved for assembly or function in the unusual intracellular environment of the sperm.
We have now isolated cDNA clones encoding ovine testis Cs and Cα1 and determined their nucleotide sequences. In agreement with our previous amino acid sequence data, the cDNAs predict different amino-terminal sequences for Cs and Cα1. The differences extend from the subunits' amino termini to their presumptive exon 1/exon 2 boundaries. (Presumptive exon junctions for the ovine Cα1 and Cs cDNAs and the human Cs cDNA are based on the mouse Cα genomic sequence [Chrivia et al., 1988]). However, the nucleotide sequences of the Cs and Cα1 cDNAs downstream of these boundaries are identical. Moreover, the first exon for Cs (termed exon 1s) and the first exon for Cα1 (termed exon 1a) are spliced to the same 3′-untranslated region (UTR) in mature transcripts. These results provide conclusive evidence that Cs is the product of an alternative transcript of the Cα gene. We found that Cs also is present in murine and human testis, and we cloned and sequenced cDNAs encoding the Cs from these species. Thus, Cs is of ancient origin and widespread in mammals. We used reverse transcriptase (RT)-PCR to probe for Cs transcripts in a wide variety of murine tissues, including ciliated tissues and ovarian tissue, and found that Cs transcripts are present only in the testis. Finally, we generated an antibody specific for the amino terminus of murine Cs. Immunohistochemistry with the use of this antibody indicates that Cs is present only in spermatogenic cells and appears first in pachytene spermatocytes when many other proteins destined for assembly into the developing sperm are first synthesized. This is the first demonstration of a cell type–specific expression of any C isoform. Together, these findings indicate that Cs is a sperm-specific isoform of Cα that has been conserved throughout mammalian evolution. The unique structure of Cs may be important in the assembly, localization, or function of this key regulatory subunit in the sperm.
MATERIALS AND METHODS
PCR Primers
Oligonucleotide primers used in this work are listed in Table 1. Cαa, Cαb, CαcR, CαdR, and CαeR were derived from consensus sequences of bovine, murine, rat, and human Cα1 mRNAs (Uhler et al., 1986a; Chrivia et al., 1988; Maldonado and Hanks, 1988; Wiemann et al., 1991, 1992). oCα376 and oCα482R were derived from the composite ovine Cs and Cα1 cDNA sequences reported in this paper (Figure 1). The Cs-specific primers oCs(−11) and mCs(−188) were derived from the sequences of ovine Cs exon 1s and murine Cs exon 1s, respectively (Figure 1C; see also Figure 3A). mCα791R was from the murine Cα1 cDNA sequence (Uhler et al., 1986a; Chrivia et al., 1988), and hCα(−60) was from the human Cα1 cDNA sequence (Maldonado and Hanks, 1988). AP1 and nested AP1 (Marathon cDNA amplification kit, Clontech Laboratories, Palo Alto, CA) were adaptor-specific primers used in rapid amplification of cDNA ends (RACE) reactions.
Table 1.
Oligonucleotide primers used in amplifying Cα1 and Cs cDNAs
| Primer | Description | 5′ to 3′ nucleotide sequence |
|---|---|---|
| Cαa | Bovine Cα1 7–26 | AACGCCGCCGCCGCCAAGAA |
| Cαb | Bovine Cα1 328–347 | TCCTTCAAGGACAACTCAAA |
| CαcR | Complement of bovine Cα1 782–800 | TTCAAGTCAGAGCTGAAGT |
| CαdR | Complement of bovine Cα1 928–947 | ATGAAGGGAGCTTCCACCTT |
| CαeR | Complement of bovine Cα1 929–955 | ACTTTGGTATGAAGGGAGCTTCCACCT |
| oCα376 | Ovine Cα1 376–402 (equivalent to ovine Cs 352–378) | GGTGGGGAGATGTTCTCACACCTGCGA |
| oCα482R | Complement of ovine Cα1 456–482 (equivalent to ovine Cs 432–458) | AGCGAGTGCAGGTACTCTAAGGTCAGG |
| oCs(−11) | Ovine Cs −11 to 16 | AAGACTGAGTGATGGCTTCCAACCCCA |
| mCα791R | Complement of murine Cα1 771–791 | GAGCTGAAGTGGGATGGGAAC |
| mCs(−188) | Murine Cs −188 to −167 | GTTCTATCTGCCCCTACCCTGC |
| hCα(−60) | Human Cα1 −60 to −43 | GCCGCAGCCAGCACCCGC |
| AP1 | Adaptor primer | CCATCCTAATACGACTCACTATAGGGC |
| Nested AP1 | Adaptor primer | ACTCACTATAGGGCTCGAGCGGC |
Numbers represent nucleotide positions in Cα1 or Cs mRNA. Nucleotides upstream of a translation start site are numbered 3′ to 5′ beginning with −1; those downstream are numbered 5′ to 3′ beginning with +1.
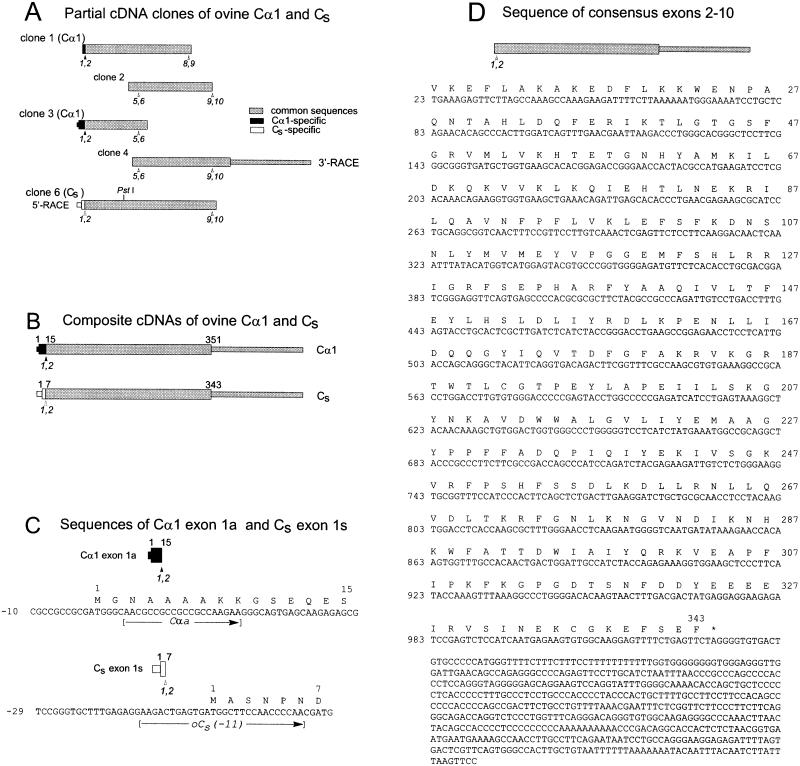
Figure 1.
Cloning and sequences of ovine Cα1 and Cs cDNAs. The initiating methionine is designated as amino acid residue 1, and the first base of the initiation codon ATG is designated as nucleotide 1. (A) Bars represent the five cDNA clones used to obtain the composite cDNAs and nucleotide sequences of ovine Cα1 and Cs. The ORFs are depicted as the wider portions of the bars. Sequences common to both Cα1 and Cs are gray, sequences specific for Cα1 are black, and sequences specific for Cs are white. For orientation, selected presumptive exon junctions based on the murine Cα genomic sequence (Chrivia et al., 1988) are marked below the bars (arrowheads). (B) Bars represent the composite cDNAs of ovine testis Cα1 and Cs. The numbers on top of the bars indicate the positions of amino acid residues encoded at the start (1) and ends (351 and 343) of the ORFs and the ends (15 and 7) of exon 1 of ovine Cα1 and Cs, respectively. Shading is as in A. (C) Partial nucleotide and predicted amino acid sequences of ovine Cα1 exon 1a and ovine Cs exon 1s. The positions of the forward primers Cαa and oCs(−11) also are shown. (D) The nucleotide sequence of exons 2–10, which are identical for ovine Cα1 and Cs cDNAs, and their predicted amino acid sequence. The amino acid and nucleotide positions indicated (right and left margins, respectively) are those for Cs. Sequence data for ovine Cα1 and ovine Cs have been deposited in GenBank/EMBL/DDBJ under accession numbers AF238979 and AF238980, respectively.
Figure 3.
Partial nucleotide and amino acid sequences of murine Cs and human Cs cDNA. (A and B) Murine Cs cDNA (clone 7) and human Cs cDNA (clone 8) were obtained by 5′-RACE with the use of murine and human testis cDNAs as template. The shading and numbering are as in Figure 1. The BglII and PstI sites that are present in clone 7 but not in clone 8 are indicated. The murine and human Cs sequences are available from GenBank/EMBL/DDBJ under the accession numbers AF239743 and AF239744, respectively. (C and D) The cDNA sequences of clones 7 (mCs) and 8 (hCs) are compared with the corresponding regions of the murine Cα1 (mCα1) (Uhler et al., 1986a) and human Cα1 (hCα1) (Maldonado and Hanks, 1988) sequences. Dashes indicate nonconsensus nucleotides.
Preparation of RNA and Synthesis of cDNA for RACE
Total RNA was prepared as described by Ausubel et al. (1989). The final preparation was suspended in 300 mM sodium acetate, 70% ethanol, and stored at −80°C. Murine oocyte total RNA was prepared from ∼30 oocytes kindly provided by Dr. Joyce Tay (University of Massachusetts Medical School). In more recent RNA preparations, tissues from mice were immersed immediately after excision in RNA Later (Ambion, Austin, TX), eliminating the need for immediate storage in liquid nitrogen. Ovine testis mRNA was prepared from 1.9 mg of total RNA (Clontech PT1353-1), yielding ∼100 μg of poly(A)+ RNA. About 350 μg of murine testis poly(A)+ RNA was obtained from 1 mg of murine testis total RNA. Marathon adaptor-ligated ovine and murine testis cDNAs for RACE were prepared as recommended (Clontech protocol PT 1115-1, with SuperScript II Rnase H− RT [Life Technologies, Grand Island, NY] used instead of avian myeloblastosis virus RT). Marathon-ready human testis cDNA was purchased from Clontech.
Cloning of Ovine Testis Cα1 cDNA (Clones 1, 2, 3, and 4)
PCR was carried out with the use of the Elongase enzyme mix (Life Technologies). Table 2 summarizes the amplification schemes used. In the RT reactions, first-strand cDNA was synthesized from ovine testis total RNA with the use of SuperScript II RT and oligo(dT)12–18 as primer (Life Technologies).
Table 2.
Generation of ovine Cα1 clones by RT-PCR and 3′-RACE
| Amplification | Primers | Thermocycler conditions | |
|---|---|---|---|
| Clone 1 | RT-PCR | Cαa, CαcR | Annealing at 50°C, extension at 68°C, 35 cycles, final 10-min extension at 68°C |
| Clone 2 | RT-PCR | Cαb, CαdR | |
| Clone 3 | RT-touchdown PCR (Don et al., 1991) | hCα(−60), oCα482R | 94°C, 72°C, 5 cycles; 94°C, 70°C, 5 cycles; 94°C, 68°C, 30 cycles |
| Clone 4 | Touchdown PCR (3′-RACE) | oCα376, AP1 | 94°C, 70°C, 5 cycles; 94°C, 68°C, 5 cycles; 94°C, 65°C, 30 cycles |
The PCR products were cloned by ligation to pBluescript II KS(−) phagemid (Stratagene, La Jolla, CA) followed by electroporation into Epicurean Coli XL1-Blue cells (Stratagene). Clones 1, 2, and 3 were identified by restriction mapping. Clone 4 was identified by hybridization to a 32P-labeled clone 2.
Cloning of Ovine, Murine, and Human Cs cDNAs (Clones 6, 7, and 8)
5′-RACE was performed on Marathon adaptor-ligated ovine, murine, and human testis cDNAs (Table 3). The 5′-RACE products were then subcloned as described above. Ovine Cs subclones (clone 6) were identified by hybridization to a 32P-labeled clone 1. Clone 7 was verified to be a murine C clone by high-stringency hybridization to 32P-labeled clone 1 and by its characteristic digestion patterns by specific restriction enzymes. Murine Cα cDNA is cut by BglII at position 218 of Cα1, whereas ovine Cα is not; both are cut by PstI at position 290. Clone 8 was verified to be a human Cs clone by high-stringency hybridization to 32P-labeled clone 1 and by its resistance to digestion by PstI.
Table 3.
Generation of ovine, murine, and human Cs clones by 5′-RACE
| First
round (5′-RACE)
|
Second round
|
|||
|---|---|---|---|---|
| Primers | Thermocycler conditions | Primers | Thermocycler conditions | |
| Clone 6 (ovine) | AP1, CαeR | Annealing at 59°C, extension at 68°C, 40 cycles, final 10-min extension at 68°C | – | – |
| Clone 7 (murine) | AP1, mCα791R | Annealing at 56°C, extension at 68°C, 40 cycles, final 10-min extension at 68°C | Nested AP1, oCα482R | Annealing at 59°C, extension at 68°C, 35 cycles, final 10-min extension at 68°C |
| Clone 8 (human) | AP1, CαeR | Annealing at 59°C, extension at 68°C, 40 cycles, final 10-min extension at 68°C | ||
Sequencing of Clones 1 to 8
Sequencing of the cDNA clones was done at the Iowa State University DNA Sequencing Facility (Ames, IA). Analysis of sequences was carried out with the use of version 10.0-UNIX of the Wisconsin Package (Genetics Computer Group, Madison, WI). Nucleotides upstream of a translation start site are numbered 3′ to 5′ beginning with −1; those downstream are numbered 5′ to 3′ beginning with +1. Translation of Cs or Cα1 is presumed to begin with the methionine immediately upstream of the amino-terminal glycine or alanine, respectively (Uhler et al., 1986a; San Agustin et al., 1998) (Figure 1, B and C).
Detection of Cs and Cα1 mRNA in Murine and Human Tissues
RT-PCR was carried out on total RNA from murine and ovine testes. PCR was carried out on human testis cDNA (Marathon-Ready human testis cDNA, Clontech). Two sets of gene-specific primers were used: oCs(−11) and CαeR to detect the presence of Cs mRNA, and Cαa and CαeR to detect Cα1 mRNA. The thermocycler program was similar to that used for clone 1 except that the reaction was carried out for 35 cycles with annealing at 59°C.
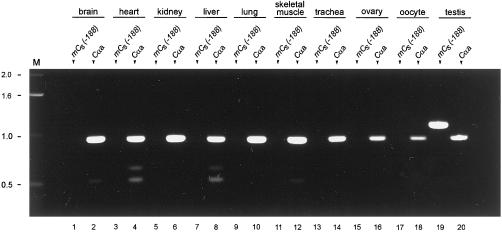
To determine the presence of Cs and Cα1 transcripts in various murine tissues, RT-PCR was performed on total RNA from murine brain, heart, kidney, liver, lung, ovary, oocytes, skeletal muscle, testis, and trachea with the use of two sets of primers: mCs(−188) and CαeR to detect Cs mRNA, and Cαa and CαeR to detect Cα1 mRNA. Thermocycler conditions were 30 cycles (35 cycles for oocytes) and annealing at 61°C.
Polyclonal Antibody against Murine Cs
The peptide Ac-ASSNDVK was synthesized and injected into rabbits (Research Genetics, Huntsville, AL). The first six residues of the peptide correspond to the predicted unique mCs amino terminus without the initiator methionine (see Figure 3A); the seventh residue, K, is shared by both murine Cs and Cα1. It was assumed that the amino-terminal alanyl residue of murine Cs is acetylated, as is the case with ovine Cs (San Agustin et al., 1998). The antibodies were affinity purified by a two-step procedure. The antisera first were applied to a column containing the synthetic acetylated peptide coupled to Sepharose 4B, and the bound antibodies were eluted by low pH. The released antibodies then were applied to a second column containing the unacetylated synthetic peptide coupled to Sepharose 4B, and the antibodies that did not bind were collected and retained. The concentration of the affinity-purified antibody was 0.83 mg/ml.
Preparation of Murine Testis and Brain Extracts
Testes (∼1.6 g) from six adult mice were excised, minced in 4-ml of cold testis homogenization buffer (10 mM potassium phosphate, pH 6.8, 1 mM EDTA, 0.1 mM DTT), and ground in a glass homogenizer. Brain tissue (∼1.3 g) from three mice was mixed with 1 ml of cold brain homogenization buffer (100 mM piperazine-N,N′-bis[2-ethanesulfonic acid], pH 6.8, 2 mM EGTA, 1 mM MgSO4, 2 mM DTT, 4 M glycerol) and ground in a glass homogenizer. The homogenates were centrifuged at 6500 × g for 15 min at 4°C. The supernatants were further clarified by centrifugation at 96,000 × g for 75 min at 4°C.
Isolation of mCs and mCα1 from Murine Testis
Because both murine Cs and Cα1 are expressed in testis (see RESULTS), both isoforms were present in the clarified testis extract. The two isoforms were copurified with the use of the protocol for the purification of ovine Cα1 from ram skeletal muscle as described previously (San Agustin et al., 1998). Fractions containing murine Cs and Cα1 eluted from the CM Fast Flow column (0.5 × 5 cm, Amersham Pharmacia Biotech, Piscataway, NJ) between 180 and 230 mM NaCl (see Figure 6). No other polypeptide was detected in the fractions containing these two proteins.
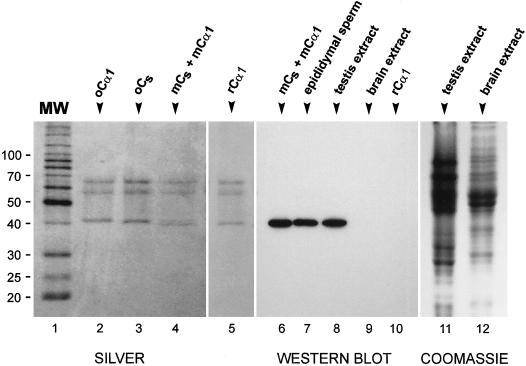
Figure 6.
Specificity of the anti-mCs antibody. (Left) Silver-stained SDS-polyacrylamide gels of purified ovine Cα1 (oCα1), purified ovine Cs (oCs), a mixture of murine Cs and Cα1 (mCs + mCα1) isolated from murine testis, and mouse recombinant Cα1 (rCα1). Molecular mass markers (MW) are in kilodaltons. As reported previously (San Agustin et al., 1998), ovine Cs migrates slightly faster than ovine Cα1. The partially purified murine Cα1 and Cs, which are resolved as two bands at ∼40 kDa, appear to migrate slightly faster than their ovine homologues. The bands in the 60- to 70-kDa range are human keratin contaminants (San Agustin et al., 1998). (Center) Western blot probed with an affinity-purified antibody generated against an acetylated peptide corresponding to the unique amino terminus of murine Cs. The antibody reacts with a single protein in the mixture of murine Cs and Cα1 (mCs + mCα1), in murine epididymal sperm (1 × 106 sperm), and in murine testis extract (50 μg of total protein) but does not react with any band in murine brain extract (30 μg of total protein) or with recombinant Cα1 (37 ng). (Right) SDS-polyacrylamide gel of murine testis extract (50 μg) and murine brain extract (30 μg) stained with Coomassie blue as loading control for lanes 8 and 9 of the Western blot.
Western Blotting
Protein samples were subjected to electrophoresis in a 10% polyacrylamide gel and blotted to polyvinylidene difluoride membrane (San Agustin et al., 1998). The blot was then treated with blocking solution (Tris-buffered saline [TBS] with 0.1% Tween-20, 1% cold fish scale gelatin [Sigma Chemical, St. Louis, MO], 5% nonfat dry milk) for 1 h at room temperature and incubated overnight at 4°C with the anti-murine Cs antibody diluted 1:4000 with the blocking solution. The blot was brought to room temperature, washed three times with blocking solution, and then incubated for 1 h with secondary antibody (HRP-conjugated goat anti-rabbit immunoglobulin G diluted 1:2000 with blocking solution). It was then washed twice with blocking solution and once with TBST (TBS with 0.1% Tween-20). Cross-reacting proteins were detected with the use of the ECL detection reagent (hydrogen peroxide/luminol; Amersham Life Science, Boston, MA). Exposure of the blot to film (AR X-Omat, Kodak, Rochester, NY) was usually between 10 and 50 s.
Immunohistochemistry
Mouse testes were excised from freshly killed adult mice and placed in 40 ml of chilled Bouin's fixative. Testes were punctured at several places with a needle (26 gauge) to allow quicker penetration of the fixative and agitated gently in an orbit shaker at 4°C. After 2 h of shaking, the testes were cut in half. Fixation was continued for an additional 24 h at 4°C. The fixed testes were washed five times with TBS, passed through a series of graded ethanol solutions followed by xylene, and then embedded in paraffin. Thin sections, typically 5 μm thick, were cut from the paraffin block, transferred to silanized coverslips, and dried overnight in an oven at 37°C. The testis sections were deparaffinized with xylene and then rehydrated by immersion in a graded series of aqueous isopropanol solutions.
Antigens were retrieved by boiling the coverslips for 20 min in 10 mM citrate, pH 6 (Polak and Van Noorden, 1997). The coverslips were rinsed in water and then transferred to individualized humidors, i.e., a Petri dish with moistened filter paper and Parafilm on top to hold the coverslip (Sanders and Salisbury, 1995). The testis sections were incubated with 250 μl of blocker solution (TBS containing 5% BSA, 20% normal swine serum) for 1 h at room temperature. The blocker solution was removed by blotting and replaced with 250 μl of anti-murine Cs antibody diluted 1:1000 to 1:2000 with one-fifth blocker solution (TBS, 1% BSA, 4% normal swine serum). The sections were incubated with the antibody overnight at 4°C, returned to room temperature, washed with TBST, and treated for 40 min with 250 μl of biotinylated swine anti-rabbit immunoglobulin G (DAKO, Carpinteria, CA) diluted 1:200 with TBS, 1% BSA, 10% normal mouse serum. After washing with TBST, the sections were incubated for 40 min with 250 μl of alkaline phosphatase–conjugated streptavidin (DAKO) diluted 1:300 with TBS, 0.5% BSA. The sections were washed with TBST and then exposed to the BCIP/NBT/INT (5-bromo-4-chloro-3-indoxyl phosphate/nitroblue tetrazolium chloride/iodonitrotetrazolium violet) substrate system (DAKO). Color was allowed to develop for 30 min, after which the coverslips were rinsed with water. The sections were then counterstained with Harris' hematoxylin (5 min) and finally mounted on glass slides with an aqueous-based mountant (Glycergel, DAKO).
RESULTS
Cloning of Ovine Testis Cα1 and Cs cDNAs
To determine the relationship of ovine Cs to ovine Cα1, we cloned and sequenced the complete ORFs of their cDNAs. Figure 1A illustrates the overlapping cDNA clones, arranged to scale and position, that were used to assemble the composite cDNAs (Figure 1B) of ovine testis Cα1 and Cs.
Clone 1, corresponding to a portion of the Cα1 mRNA extending from exon 1 to exon 9, was obtained with the use of the Cα1-specific primer Cαa and the reverse primer CαcR. Sequencing confirmed that this clone encoded amino acids specific to the amino terminus of Cα1 (Figure 1C). Clone 2, obtained with the use of consensus primers based on published mammalian Cα1 sequences, was 100% identical with clone 1 in the region of overlap.
The remaining sequence of the 5′ end of the ORF of ovine Cα1 mRNA was obtained from clone 3, which was generated with the use of hCα(−60) as the forward primer and oCα482R as the reverse primer. A forward primer based on the 5′-UTR of human Cα1 mRNA was used because the 5′-UTR of bovine Cα1 mRNA is not known, and we reasoned that the 5′-UTR of human Cα1 was likely to be similar to that of ovine Cα1. Clone 3 encoded amino acids specific to the amino terminus of Cα1 and was identical to clones 1 and 2 in the regions of overlap.
Clone 4, containing the 3′ end of the ORF and the 3′-UTR of ovine Cα mRNA, was obtained as a 3′-RACE product of ovine testis cDNA. Clone 4 was 100% identical to clones 1, 2, and 3 in their regions of overlap.
The 5′ end of the ORF and the 5′-UTR of ovine Cs were obtained by 5′-RACE with the use of CaeR as gene-specific primer and ovine testes cDNA as template. A single band of product was observed in agarose gels, and a number of subclones of this PCR band were isolated for nucleotide sequencing. Although the CaeR primer could have amplified both Cs and Cα1 cDNAs, all subclones contained sequences coding for the unique amino terminus of Cs contiguous to sequences identical to exons 2–10 of the Cα1 clones. The finding that the cDNA sequences of exons 2–10 of Cα1 and Cs are identical at the nucleotide level provided strong evidence that Cs is the product of an alternatively spliced mRNA in which a unique Cs exon (hereafter referred to as exon 1s) is spliced to exon 2 of the Cα gene.
Further proof that exon 1 (hereafter referred to as exon 1a) of the Cα1 mRNA and exon 1s of the Cs mRNA are spliced to the same downstream sequence was obtained by carrying out RT-PCR of ovine testis mRNA with the use of the forward primers Cαa and oCs(−11), based on sequences located in exons 1a and 1s, respectively, with the reverse primer oCα1402R, which is complementary to sequence located in the 3′ noncoding region of exon 10. Both primer pairs yielded products of the expected size (our unpublished results), confirming that the 3′-UTR of exon 10 is common to both Cs and Cα1 mRNAs.
Nucleotide and Predicted Amino Acid Sequences of Ovine Cα1 and Cs cDNAs
Figure 1C shows the partial sequences of Cα1 exon 1a and Cs exon 1s obtained from the ovine cDNA clones. The ORF of Cα1 exon 1a codes for 15 amino acids, whereas that of Cs exon 1s codes for 7 different amino acids. The amino acid residues encoded by ovine Cα1 exon 1a (minus the initiator methionine) are identical to those reported for bovine (Shoji et al., 1983; Wiemann et al., 1992), murine (Uhler et al., 1986a; Chrivia et al., 1988), rat (Wiemann et al., 1991), hamster (Howard et al., 1991), and human (Maldonado and Hanks, 1988) Cα1, whereas the amino acid sequence predicted from Cs exon 1s (minus the initiator methionine) exactly matches the amino-terminal sequence for ovine Cs obtained through protein biochemistry (San Agustin et al., 1998). The nucleotide sequence of exons 2–10, which are identical for both the Cα1 and Cs cDNAs, is presented in Figure 1D; the predicted amino acid sequence is 100% identical (78 of 78 residues) with the partial amino acid sequence of this portion of ovine Cs obtained from Edman analysis of its cyanogen bromide and tryptic fragments (San Agustin et al., 1998).
The ovine Cs cDNA predicts a protein of 343 amino acids (including the initiating methionine) with a mass of 39,858 Da, whereas the ovine Cα1 cDNA predicts a protein of 351 amino acids with a mass of 40,589 Da. Because the amino terminus of Cα1 is myristylated and that of Cs is acetylated (San Agustin et al., 1998), the mass of the modified Cα1 is predicted to be 899 Da greater than the mass of modified Cs, in excellent agreement with the difference of 890 Da determined empirically by mass spectrometry (San Agustin et al., 1998).
Similar Cs mRNAs Are Present in Murine and Human Testis
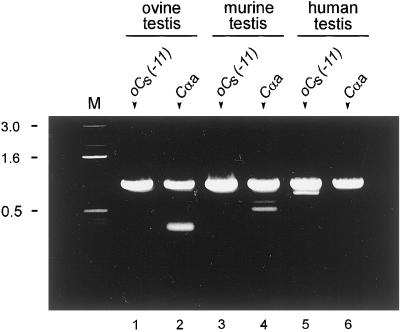
To determine if Cs mRNAs are present in the testes of other mammalian species, we carried out PCR with the use of ovine, murine, and human testicular cDNA as template and forward primers (Figure 1C) specific for either Cs [oCs(−11)] or Cα1 [Cαa]. In all cases, the reverse primer was CαeR. In all three species, the Cs-specific primer yielded PCR product of the expected size (Figure 2, lanes 1, 3 and 5). Therefore, Cs is widespread in mammals. Cα1 transcripts also were found in the testes of all three species (Figure 2, lanes 2, 4, and 6), confirming that both C isoforms occur in the testis. The Cα1 and Cs PCR products had very similar sizes (slightly less than 1 kilobase), which agrees with the calculated sizes of 949 bases for the Cα1 PCR product and 942 bases for the Cs PCR product.
Figure 2.
Detection of Cα1 and Cs mRNA in various species. RT-PCR of total RNA from ovine and murine testes and PCR of cDNA from human testis. The forward primers used to generate the PCR products are shown at the top of the lanes: oCs(−11) to amplify Cs and Cαa to amplify Cα1. The reverse primer in all cases was CαeR. The PCR products were subjected to electrophoresis in an 0.8% agarose gel and stained with ethidium bromide. Cα1 and Cs products were obtained from all three species. Controls in which the RT was omitted yielded no bands. Lane M, DNA molecular mass markers (in kilobases).
Nucleotide Sequences of cDNAs Encoding the Amino Termini of Murine and Human Cs
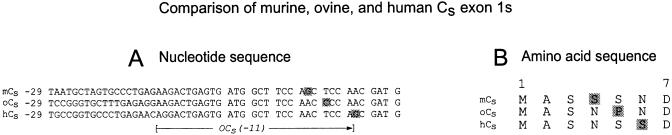
To confirm that murine and human testes have Cs, and to determine the degree of similarity between the amino termini of these proteins and that of ovine Cs, cDNAs of murine and human Cs were amplified from testis cDNA by 5′-RACE with the use of identical sets of primers (nested AP1 and oCα482R). Only one PCR band was observed in each case. These products were cloned and sequenced. As with the ovine 5′-RACE cDNA, all of the clones encoded Cs. Figure 3, A and B, show the partial nucleotide and predicted amino acid sequences of murine Cs cDNA (clone 7) and human Cs cDNA (clone 8), respectively. Figure 3, C and D, show the alignment of murine Cα1 with murine Cs and human Cα1 with human Cs. As in the sheep, exon 1s of the murine Cs cDNA and exon 1s of the human Cs cDNA showed very little identity with their Cα1 counterparts, whereas Cs nucleotides downstream of the exon 1/exon 2 junction were 100% identical to the published sequences for the Cα1 cDNAs. However, exon 1s of murine Cs and exon 1s of human Cs were very similar to the ovine Cs exon 1s (Figure 4A). The coding region of exon 1s of each of the three cDNAs differs from the others at only 2 of 22 positions. Each of these substitutions would result in the incorporation of a different amino acid residue into the Cs molecule (Figure 4B). The first three amino acid residues are predicted to be identical for all three species, but the next three residues are S or N at positions 4 and 6 and P or S at position 5.
Figure 4.
Comparison of exon 1s–encoded regions of ovine, murine, and human Cs. Partial cDNA nucleotide (A) and predicted amino acid (B) sequences of the murine (mCs), ovine (oCs), and human (hCs) versions of Cs exon 1s are aligned. The nonconsensus bases of the ORFs are highlighted, as are the amino acid residues that will result from these substitutions. The position of the primer oCs(−11) also is shown.
Cs mRNA Is Found Exclusively in the Testis
To investigate the tissue distribution of Cs, we carried out RT-PCR with the use of murine total RNA from various tissues as template. mCs- and Cα1-specific forward primers were chosen to yield different-sized PCR products with CαeR as the reverse primer. Cα1 mRNA was detected in all tissues assayed (Figure 5), whereas Cs mRNA was detected only in testis (Figure 5, lane 19). It is important to note that Cs mRNA was not detected in ciliated tissues such as brain, lung, and trachea, indicating that Cs is not a component of cilia. Moreover, Cs mRNA was not detected in ovarian tissue or oocytes, indicating that Cs is not expressed in the female germ line. These results strongly suggest that Cs is expressed only in the testis, where the translated protein becomes integrated into the sperm tail.
Figure 5.
Detection of Cα1 and Cs mRNA in murine tissues. RT-PCR of total RNA from various murine tissues. The forward primers used to generate the PCR products are shown at the top of the lanes: mCs(−188) to amplify Cs and Cαa to amplify Cα1. The reverse primer in all cases was CαeR. These primer sets are predicted to yield PCR products of 949 bases for murine Cα1 and 1119 bases for murine Cs. The PCR products were subjected to electrophoresis in an 0.8% agarose gel and stained with ethidium bromide. Transcripts encoding the Cα1 isoform are present in all the tissues analyzed, whereas Cs transcripts are detected only in the testis. Lane M, DNA molecular mass markers (in kilobases).
Cs Is Expressed Only in Germ Cells and First Appears in Mid Pachytene Spermatocytes
To determine the pattern of expression of Cs in the testis, a rabbit anti-peptide antibody was made against the unique amino-terminal sequence of murine Cs. The specificity of the antibody was demonstrated in Western blots. Fractions of purified C from murine testes contain two proteins that migrate with mobilities very similar to those of pure ovine Cs and ovine Cα1 in SDS-polyacrylamide gels (Figure 6, lanes 1–4). These proteins are presumed to represent Cs and Cα1, both of which are expressed in the testis (Figures 2 and 5). When Western blots of this mixture were probed with the antibody, a single protein of ∼40 kDa was detected (Figure 6, lane 6). The antibody reacted strongly with a single band of the same size in murine epididymal sperm, which are presumed to contain Cs but not Cα1 (San Agustin et al., 1998), and in murine testis extract, but it did not recognize any protein in murine brain extract, which contains Cα1 and Cβ but not Cs. The antibody also did not recognize purified ovine Cα1, which has the same amino-terminal sequence as murine Cα1 (our unpublished results), nor murine recombinant Cα1 (kindly provided by Dr. S. Taylor, University of California, San Diego) (Figure 6, lanes 5 and 10). Therefore, the antibody is highly specific for Cs and does not appear to recognize any other protein in the testis.
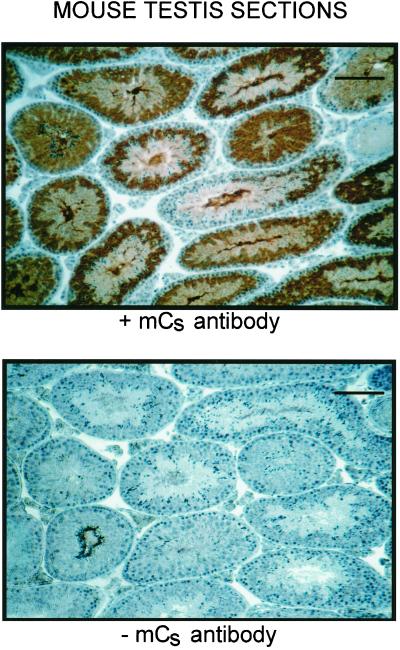
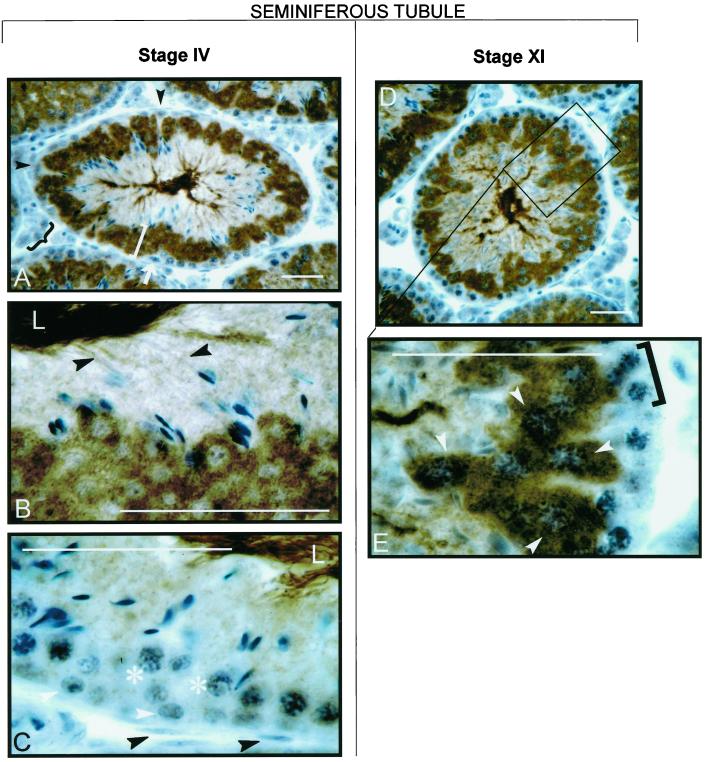
In sections of murine testes (Figures 7 and 8), the antibody stained only germ cells and did not react with Sertoli cells, Leydig cells, or any other non-germ cells. It also did not stain spermatogonia, zygotene spermatocytes, or early pachytene spermatocytes. The antibody stained mid pachytene spermatocytes of stage VI tubules very weakly, stained mid pachytene spermatocytes of stage VIII tubules slightly more strongly (our unpublished results), and stained late pachytene spermatocytes of stage XI tubules very strongly (Figure 8). Therefore, Cs appears to be synthesized first in mid pachytene and is highly expressed by late pachytene. The antibody also stained round spermatids, elongating spermatids, and mature sperm present in the lumen of the seminiferous tubules (Figure 8). Cs was present in the cytosol of round spermatids and appeared to move from the cytosol into the developing flagella as the spermatids matured. Controls in which the primary antibody was omitted did not exhibit any staining.
Figure 7.
Immunohistochemical staining of murine testis sections with the use of the anti-mouse Cs antibody. Cells stained brown are positive for Cs. Only germ cells at later stages of spermatogenesis stain with the antibody (top); because the cell associations seen in cross-sections of the seminiferous tubules vary depending on their stage in the spermatogenic cycle, the different tubules display different staining patterns. No staining is detected in the absence of the primary antibody (bottom). Bars, 100 μm.
Figure 8.
Higher magnification of testis sections stained with anti-mouse Cs antibody. Bars, 20 μm. Tubules shown correspond to stages IV and XI of the seminiferous epithelium cycle according to the system of Leblond and Clermont (Leblond et al., 1963; Clermont and Bustos-Obregon, 1968). In the stage IV tubule, staining is absent from interstitial cells (A, black brace), Sertoli cells (A, black arrowheads), peritubular cells (C, black arrowheads), spermatogonia (C, white arrowheads), and early pachytene spermatocytes (C, asterisks). A spermatogonium undergoing mitosis is also shown (A, white arrow). Round spermatids have intensely stained cytosol (A, white bracket). In the previous generation of elongated spermatids that have moved farther toward the lumen (L), the cytoplasm now stains less intensely but the developing flagella (B, black arrowheads) are darkly stained. Darkly stained tails of mature sperm are visible in the lumens (L) of the stage IV tubules (B and C). In the stage XI tubule, staining of the cytosol of the spermatids occupying the inner portion of the tubule diminishes as they elongate (D). Staining is absent from zygotene spermatocytes (E, black bracket) but is prominent in the cytoplasm of late pachytene spermatocytes (E, white arrowheads).
DISCUSSION
Cs Is the Product of an Alternative Transcript of the Cα Gene
Cs originally was characterized by protein biochemistry as an ovine sperm PKA catalytic subunit differing from ovine somatic Cα1 in its electrophoretic mobility, mass, and amino-terminal sequence up to the presumptive exon 1/exon 2 junction (San Agustin et al., 1998). The current study provides definitive molecular genetic evidence that ovine Cs is the product of an alternative transcript of the Cα gene. First, the nucleotide sequences of Cs and Cα1 cDNAs downstream of the exon 1/exon 2 junction are absolutely identical. If the proteins were the products of different genes, at least some substitutions would have occurred at the nucleotide level since the divergence of the two genes at least 65 million years ago (see below). Second, exon 1s of Cs and exon 1a of Cα1 are both spliced to the same 3′-UTR.
Examination of the mouse genome sequence (GenBank accession number M18241) indicates that the mouse exon 1s sequence (see below) is not contiguous with the 5′ sequence of exon 2 of Cα. Therefore, the Cs mRNA must result from alternative splicing of a Cα transcript. Production of the Cs transcript also may depend on an alternative initiation site within the Cα gene.
Cs is the third Cα isoform to be reported. Thomis et al. (1992) described a partial human cDNA that was identical with human Cα1 cDNA sequence at its 5′ end but that contained sequences derived from introns flanking both sides of exon 8. This cDNA predicts a Cα isoform, termed Cα2, that would be substantially truncated at its carboxyl-terminal end. The Cα2 cDNA appeared to be expressed in at least two human cell lines.
Similar Cs Isoforms Are Widespread in Mammals
PCR with the use of a primer based on the nucleotide sequence of exon 1s of ovine Cs indicated that Cs is expressed in the testes of mouse and human as well as sheep. The nucleotide sequences of partial cDNAs encoding the murine and human Cs isoforms revealed that Cs exon 1s is very similar in all three species, each differing from the other at only two positions. In the mouse and human, as in the sheep, the sequences indicate that the 15 amino acids encoded by Cα1 exon 1a are replaced by 7 different amino acids in Cs. In all three species, an alanine replaces the glycine that follows the first methionine in Cα1. In Cα1, this methionine is cleaved off posttranslationally, and the newly exposed amino-terminal glycine is myristylated (Shoji et al., 1983). Because the glycine is replaced with alanine in murine and human Cs, they probably are not myristylated but rather are acetylated, as is ovine Cs (San Agustin et al., 1998).
The presence of Cs in primates, rodents, and ungulates indicates that this isoform arose early in evolution, at least before the divergence of these mammalian orders more than 65 million years ago (Young, 1962).
The Murine Cx Pseudogene Likely Arose from a Cs mRNA
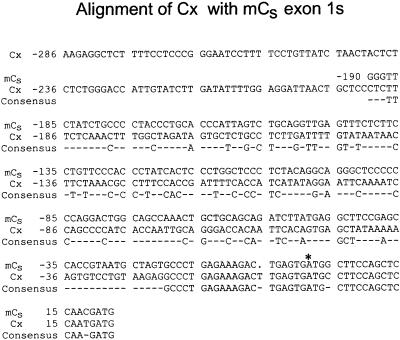
A PKA catalytic subunit–related sequence, Cx, is present in the murine genome (Cummings et al., 1994). This sequence was reported to be most closely related to that of the Cα gene, but it lacks introns and, relative to Cα, contains frame-shift mutations, premature termination codons, and missense mutations. It is not transcribed. Therefore, it appears to be a pseudogene of the retroposon class (Weiner et al., 1986). Cx is closely related to Cα downstream of the Cα exon 1/exon 2 junction but does not resemble the Cα sequence upstream of this site, leading to speculation that the mRNA intermediate that gave rise to Cx may have been incompletely spliced (Cummings et al., 1994). However, a comparison of the murine Cs exon 1s nucleotide sequence with the Cx 5′ sequence reveals near identity from Cs nucleotide −20 to the Cs exon 1/exon 2 junction (Figure 9). Therefore, Cx probably arose by reverse transcription of a Cs mRNA followed by nonhomologous recombination of the cDNA into the genome of a male germ cell.
Figure 9.
Comparison of the 5′ sequence of murine Cx pseudogene with that of exon 1s of murine Cs. The Cx nucleotide sequence is nearly identical to that extending from Cs nucleotide −20 downstream to the end of Cs exon 1s. An asterisk indicates the translation start site of Cs. Dashes indicate nonconsensus nucleotides.
Tissue and Cell Distribution of Cs
Using a RT-PCR assay and primers specific for Cs or Cα1, we detected Cs transcripts in murine testis but not in murine brain, heart, kidney, liver, lung, ovary, oocytes, trachea, or skeletal muscle. In contrast, Cα1 transcripts were present in all tissues tested. Therefore, Cs appears to be expressed only in the testis.
It is significant that Cs is not expressed in highly ciliated tissues such as the lung, trachea, and brain. PKA is important in the control of somatic cilia (for review, see Witman, 1990), and it was possible that Cs is an isoform specific for cilia and flagella in general. However, the current results indicate that this is not the case. Similarly, the absence of Cs expression in ovaries and oocytes rules out the possibility that Cs is expressed in all germ cells. Rather, it appears to be present only in the male. In oocytes, PKA is believed to play a major role in the maintenance of meiotic arrest (Schultz, 1988; Rose-Hellekant and Bavister, 1996). This important function probably is performed by Cα1, which our results indicate is present in ovaries and oocytes.
Immunohistochemistry of murine testis sections with the use of an anti-peptide antibody against the unique amino terminus of murine Cs indicated that Cs is present only in germ cells. Synthesis of Cs appears to be initiated during mid pachytene. Therefore, transcription of Cs must be directed, at least initially, by the diploid nucleus. This finding is consistent with previous studies showing that synthesis of SDS-soluble sperm proteins is highest during meiosis (O'Brien and Bellvé, 1980) and that transcription and translation during spermatogenesis both peak in mid pachytene (Monesi, 1965). Subsequently, Cs is localized to the developing flagellum of the elongating spermatids. It should be noted that this is the first demonstration of a cell type–specific expression of any C isoform.
The fact that Cs does not appear to be present in spermatogonia and prepachytene spermatocytes suggests that any cAMP-dependent functions in these cells are mediated by Cα1 or some other isoform of C. It will be of interest to determine if Cα1 is present together with Cs in meiotic and postmeiotic cells or if Cs mediates all cAMP-dependent functions (Amat et al., 1990; Delmas et al., 1993) during spermiogenesis. It was reported that Cα mRNA is present in pachytene spermatocytes (Øyen et al., 1990; Landmark et al., 1993), but the probes used would not have distinguished between Cα1 and Cs mRNAs, so this should be reexamined. In any case, Cs was the only C isoform detected in Western blots of ovine ejaculated, epididymal, and rete testis sperm (San Agustin et al., 1998), and it was the only isoform isolated from ovine sperm flagella (San Agustin et al., 1998), despite the fact that Cα1 would have copurified with Cs had it been present in the flagella. Therefore, if Cs and Cα1 occur together in spermatids, Cs must be specifically targeted to the developing sperm structures.
Function of Unique Cs Structure
The fact that Cs is present in a wide range of mammals raises the possibility that its unique structure has an important role in the assembly or function of the subunit. Cs is not released from demembranated ovine sperm in the presence of cAMP (San Agustin and Witman, 1994; San Agustin et al., 1998), indicating that it is attached to structures within the sperm even when activated. The unique structure of Cs may be responsible for this behavior. In Cα1, the exon 1a–encoded residues form the first two turns of a long α-helix that extends across the surface of the catalytic core of the enzyme. This helix is anchored to the hydrophobic core by the amino-terminal myristate (Zheng et al., 1993). In the absence of this myristate, the Cα1 exon 1a residues are unstructured (Knighton et al., 1991). In contrast to the situation in Cα1, the residues encoded by exon 1s of Cs form a shorter domain, are not predicted to form an α-helix (Chou and Fasman, 1978), and lack a terminal myristate to serve as an anchor (San Agustin et al., 1998). Such a short, probably unstructured amino-terminal domain is likely to leave the catalytic subunit's hydrophobic core exposed, possibly allowing Cs to bind to hydrophobic sites within the sperm. Alternatively, a flexible amino-terminal tail might itself bind to a structure within the sperm and tether Cs to that structure. In either case, the attachment of Cs to the sperm tail by cAMP–insensitive bonds would explain the inability of cAMP to release Cs from demembranated sperm.
Such anchoring of activated Cs in the sperm could be advantageous. First, the phosphorylation of its substrates could be accomplished more efficiently. By maintaining the activated catalytic subunit in close proximity to its target substrates, rapid phosphorylation of these proteins upon activation of Cs would be ensured. Conversely, if cAMP levels decreased, Cs would be able to rapidly rebind to R, which itself would be anchored in the same general vicinity by A-kinase–anchoring proteins. Second, by limiting the distance that activated Cs can travel, promiscuous phosphorylation of other flagellar proteins and its potentially deleterious effects would be avoided. This type of spatial arrangement has been observed in other signal transduction complexes, in which the components of the signaling pathway are assembled on scaffold proteins for more effective physical interaction between enzyme and substrate and for enhanced specificity (Faux and Scott, 1996; Whitmarsh et al., 1998).
Recently, it was found that the majority of Cα was mislocalized in sperm of a knockout mouse lacking RIIα, the predominant PKA regulatory subunit in sperm (Burton et al., 1999). If the Cα isoform monitored in that study was indeed Cs, this result suggests that the unique structure of Cs is insufficient to properly localize the subunit in the absence of RIIα. However, it is quite possible that correct localization of Cs requires interactions with both R and another protein that interacts with Cs via an exposed hydrophobic site.
ACKNOWLEDGMENTS
We are grateful to Dr. Susan Taylor for her generous gift of murine recombinant Cα1. This study was supported by National Institutes of Health grant HD23858 and by grants from the George F. Booth Fund at the Greater Worcester Community Foundation and the Campbell and Hall Charity Fund.
Abbreviations used:
- C
catalytic subunit of PKA
- PKA
cAMP-dependent protein kinase
- R
regulatory subunit of PKA
- RACE
rapid amplification of cDNA ends
- RT
reverse transcriptase
- TBS
Tris-buffered saline, pH 7.5
- UTR
untranslated region
REFERENCES
- Aitken RJ, Harkiss D, Knox W, Paterson M, Irvine DS. A novel signal transduction cascade in capacitating human spermatozoa characterized by a redox-regulated, cAMP-mediated induction of tyrosine phosphorylation. J Cell Sci. 1998;111:645–656. doi: 10.1242/jcs.111.5.645. [DOI] [PubMed] [Google Scholar]
- Amat JA, Fields KL, Schubart UK. Stage-specific expression of phosphoprotein p19 during spermatogenesis in the rat. Mol Reprod Dev. 1990;26:383–390. doi: 10.1002/mrd.1080260414. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Protocols in Molecular Biology. Vol. 1. New York: John Wiley & Sons; 1989. [Google Scholar]
- Beebe SJ, Øyen O, Sandberg M, Frøysa A, Hansson V, Jahnsen T. Molecular cloning of a tissue-specific protein kinase (Cγ) from human testis: representing a third isoform for the catalytic subunit of cAMP-dependent protein kinase. Mol Endocrinol. 1990;4:465–475. doi: 10.1210/mend-4-3-465. [DOI] [PubMed] [Google Scholar]
- Brokaw CJ. Regulation of sperm flagellar motility by calcium and cAMP-dependent phosphorylation. J Cell Biochem. 1987;35:175–184. doi: 10.1002/jcb.240350302. [DOI] [PubMed] [Google Scholar]
- Burton KA, Treash-Osio B, Muller CH, Dunphy EL, McKnight GS. Deletion of type IIα regulatory subunit delocalizes protein kinase A in mouse sperm without affecting motility or fertilization. J Biol Chem. 1999;274:24131–24136. doi: 10.1074/jbc.274.34.24131. [DOI] [PubMed] [Google Scholar]
- Chaudhry PS, Creagh S, Yu N, Brokaw CJ. Multiple protein kinase activities required for activation of sperm flagellar motility. Cell Motil Cytoskeleton. 1995;32:65–79. doi: 10.1002/cm.970320108. [DOI] [PubMed] [Google Scholar]
- Chou PY, Fasman GD. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Chrivia JC, Uhler MD, McKnight GS. Characterization of genomic clones coding for the Cα and Cβ subunits of mouse cAMP-dependent protein kinase. J Biol Chem. 1988;263:5739–5744. [PubMed] [Google Scholar]
- Clermont Y, Bustos-Obregon E. Re-examination of spermatogonial renewal in the rat by means of seminiferous tubules mounted “in toto.”. Am J Anat. 1968;122:237–247. doi: 10.1002/aja.1001220205. [DOI] [PubMed] [Google Scholar]
- Cummings DE, Edelhoff S, Disteche CM, McKnight GS. Cloning of a mouse protein kinase A catalytic subunit pseudogene and chromosomal mapping of C subunit isoforms. Mamm Genome. 1994;5:701–706. doi: 10.1007/BF00426076. [DOI] [PubMed] [Google Scholar]
- Delmas V, van der Hoorn F, Mellstrom B, Jegou B, Sassone-Corsi P. Induction of CREM activator proteins in spermatids: down-stream targets and implications for haploid germ cell differentiation. Mol Endocrinol. 1993;11:1502–1514. doi: 10.1210/mend.7.11.8114765. [DOI] [PubMed] [Google Scholar]
- Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AE, Fraser LR. Cyclic AMP-dependent phosphorylation of epididymal mouse sperm proteins during capacitation in vitro: identification of an M(r) 95,000 phosphotyrosine-containing protein. J Reprod Fertil. 1993;97:287–299. doi: 10.1530/jrf.0.0970287. [DOI] [PubMed] [Google Scholar]
- Faux MC, Scott JD. Molecular glue: kinase anchoring and scaffold proteins. Cell. 1996;85:9–12. doi: 10.1016/s0092-8674(00)81075-2. [DOI] [PubMed] [Google Scholar]
- Galantino-Homer HL, Visconti PE, Kopf GS. Regulation of protein tyrosine phosphorylation during bovine sperm capacitation by a cyclic adenosine 3′5′-monophosphate-dependent pathway. Biol Reprod. 1997;56:707–719. doi: 10.1095/biolreprod56.3.707. [DOI] [PubMed] [Google Scholar]
- Garbers DL, Kopf GS. The regulation of spermatozoa by calcium cyclic nucleotides. Adv Cyclic Nucleotide Res. 1980;13:251–306. [PubMed] [Google Scholar]
- Garbers DL, Lust WD, First NL, Lardy HA. Effects of phosphodiesterase inhibitors and cyclic nucleotides on sperm respiration and motility. Biochemistry. 1971;10:1825–1831. [Google Scholar]
- Howard P, Day KH, Kim KE, Richardson J, Thomas J, Abraham I, Fleischman RD, Gottesman MM, Maurer RA. Decreased catalytic subunit mRNA levels and altered catalytic subunit mRNA structure in a cAMP-resistant Chinese hamster ovary cell line. J Biol Chem. 1991;266:10189–10195. [PubMed] [Google Scholar]
- Jaiswal BS, Majumder GC. Cyclic AMP phosphodiesterase: a regulator of forward motility initiation during epididymal sperm maturation. Biochem Cell Biol. 1996;74:669–674. doi: 10.1139/o96-072. [DOI] [PubMed] [Google Scholar]
- Knighton DR, Zheng J, Ten Eyck LF, Ashford VA, Xuong N, Taylor SS, Sowadski JM. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:407–420. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- Landmark BF, Øyen O, Skålhegg BS, Fauske B, Jahnsen T, Hansson V. Cellular location and age-dependent changes of the regulatory subunits of cAMP-dependent protein kinase in rat testis. J Reprod Fertil. 1993;99:323–334. doi: 10.1530/jrf.0.0990323. [DOI] [PubMed] [Google Scholar]
- Leblond CP, Steinberger E, Roosen-Runge EC. Spermatogenesis. In: Hartman CG, editor. Mechanisms Concerned with Conception. Oxford, UK: Pergamon Press;; 1963. pp. 1–72. [Google Scholar]
- Lindemann CB. A cAMP-induced increase in the motility of demembranated bull sperm models. Cell. 1978;13:9–18. doi: 10.1016/0092-8674(78)90133-2. [DOI] [PubMed] [Google Scholar]
- Maldonado F, Hanks SK. A cDNA clone encoding human cAMP-dependent protein kinase catalytic subunit Cα. Nucleic Acids Res. 1988;16:8189–8190. doi: 10.1093/nar/16.16.8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monesi V. Synthetic activities during spermatogenesis in the mouse RNA and protein. Exp Cell Res. 1965;39:197–224. doi: 10.1016/0014-4827(65)90023-6. [DOI] [PubMed] [Google Scholar]
- O'Brien DA, Bellvé AR. Protein constituents of the mouse spermatozoon. II. Temporal synthesis during spermatogenesis. Dev Biol. 1980;75:405–418. doi: 10.1016/0012-1606(80)90172-4. [DOI] [PubMed] [Google Scholar]
- Osheroff JE, Visconti PE, Valenzuela JP, Travis AJ, Alvarez J, Kopf GS. Regulation of human sperm capacitation by a cholesterol efflux-stimulated signal transduction pathway leading to protein kinase A-mediated up-regulation of protein tyrosine phosphorylation. Mol Hum Reprod. 1999;5:1017–1026. doi: 10.1093/molehr/5.11.1017. [DOI] [PubMed] [Google Scholar]
- Øyen O, Myklebust F, Scott JD, Cadd GG, McKnight GS, Hansson V, Jahnsen T. Subunits of cyclic adenosine 3′,5′-monophosphate-dependent protein kinase show differential and distinct expression patterns during germ cell differentiation: alternative polyadenylation in germ cells gives rise to unique smaller-sized mRNA species. Biol Reprod. 1990;43:46–54. doi: 10.1095/biolreprod43.1.46. [DOI] [PubMed] [Google Scholar]
- Pariset CC, Feinberg JM, Dacheaux JL, Weinman SJ. Changes in calmodulin level and cAMP-dependent protein kinase activity during epididymal maturation of ram spermatozoa. J Reprod Fertil. 1985;74:105–112. doi: 10.1530/jrf.0.0740105. [DOI] [PubMed] [Google Scholar]
- Polak JM, Van Noorden S. Introduction to Immunocytochemistry. 2nd ed. Oxford, UK: Bios Scientific Publishers; 1997. [Google Scholar]
- Reinton N, Haugen TB, Ørstavik S, Skålhegg BS, Hansson V, Jahnsen T, Taskén K. The gene encoding the Cγ catalytic subunit of cAMP-dependent protein kinase is a transcribed retroposon. Genomics. 1998;49:290–297. doi: 10.1006/geno.1998.5240. [DOI] [PubMed] [Google Scholar]
- Rose-Hellekant TA, Bavister BD. Roles of protein kinase A and C in spontaneous maturation and in forskolin or 3-isobutyl-1-methylxanthine maintained meiotic arrest of bovine oocytes. Mol Reprod Dev. 1996;44:250–255. doi: 10.1002/(SICI)1098-2795(199606)44:2<241::AID-MRD14>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- San Agustin JT, Leszyk JD, Nuwaysir LM, Witman GB. The catalytic subunit of the cAMP-dependent protein kinase of ovine sperm flagella has a unique amino-terminal sequence. J Biol Chem. 1998;273:24874–24883. doi: 10.1074/jbc.273.38.24874. [DOI] [PubMed] [Google Scholar]
- San Agustin JT, Witman GB. Role of cAMP in the reactivation of demembranated ram spermatozoa. Cell Motil Cytoskeleton. 1994;27:206–218. doi: 10.1002/cm.970270303. [DOI] [PubMed] [Google Scholar]
- Sanders MA, Salisbury JL. Immunofluorescence microscopy of cilia and flagella. Methods Cell Biol. 1995;47:163–169. doi: 10.1016/s0091-679x(08)60805-5. [DOI] [PubMed] [Google Scholar]
- Schultz RM. Regulatory functions of protein phosphorylation in meiotic maturation of mouse oocytes in vitro. Prog Clin Biol Res. 1988;267:137–151. [PubMed] [Google Scholar]
- Shoji S, Ericsson LH, Walsh KA, Fischer EH, Titani K. Amino acid sequence of the catalytic subunit of bovine type II adenosine cyclic 3′,5′-phosphate dependent protein kinase. Biochemistry. 1983;22:3702–3709. doi: 10.1021/bi00284a025. [DOI] [PubMed] [Google Scholar]
- Showers MO, Maurer RA. A cloned bovine cDNA encodes an alternate form of the catalytic subunit of cAMP-dependent protein kinase. J Biol Chem. 1986;261:16288–16291. [PubMed] [Google Scholar]
- Tash JS, Means AR. Regulation of protein phosphorylation and motility of sperm by cyclic adenosine monophosphate and calcium. Biol Reprod. 1982;26:745–763. doi: 10.1095/biolreprod26.4.745. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Buechler JA, Yonemoto W. cAMP-dependent protein kinase: framework for a diverse family of regulatory enzymes. Annu Rev Biochem. 1990;59:971–1005. doi: 10.1146/annurev.bi.59.070190.004543. [DOI] [PubMed] [Google Scholar]
- Thomis DC, Floyd-Smith G, Samuel CE. Mechanism of interferon action: cDNA structure and regulation of a novel splice-site variant of the catalytic subunit of human protein kinase A from interferon-treated human cells. J Biol Chem. 1992;267:10723–10728. [PubMed] [Google Scholar]
- Uhler MD, Carmichael DF, Lee DC, Chrivia JC, Krebs EG, McKnight GS. Isolation of cDNA clones coding for the catalytic subunit of mouse cAMP-dependent protein kinase. Proc Natl Acad Sci USA. 1986a;83:1300–1304. doi: 10.1073/pnas.83.5.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhler MD, Chrivia JC, McKnight GS. Evidence for a second isoform of the catalytic subunit of cAMP-dependent protein kinase. J Biol Chem. 1986b;261:15360–15363. [PubMed] [Google Scholar]
- Visconti PE, Johnson LR, Oyaski M, Fornes M, Moss SB, Gerton GL, Kopf GS. Regulation, localization, and anchoring of protein kinase A subunits during mouse sperm capacitation. Dev Biol. 1997;192:351–363. doi: 10.1006/dbio.1997.8768. [DOI] [PubMed] [Google Scholar]
- Visconti PE, Moore GD, Bailey JL, Leclerc P, Connors SA, Pan D, Olds-Clarke P, Kopf GS. Capacitation of mouse spermatozoa. II. Protein tyrosine phosphorylation and capacitation are regulated by a cAMP-dependent pathway. Development. 1995;121:1139–1150. doi: 10.1242/dev.121.4.1139. [DOI] [PubMed] [Google Scholar]
- Visconti PE, Ning X, Fornes MW, Alvarez JG, Stein P, Connors SA, Kopf GS. Cholesterol efflux-mediated signal transduction in mammalian sperm: cholesterol release signals an increase in protein tyrosine phosphorylation during mouse sperm capacitation. Dev Biol. 1999a;214:429–443. doi: 10.1006/dbio.1999.9428. [DOI] [PubMed] [Google Scholar]
- Visconti PE, Stewart-Savage J, Blasco A, Battaglia L, Miranda P, Kopf GS, Tezon JG. Roles of bicarbonate, cAMP, and protein tyrosine phosphorylation on capacitation and the spontaneous acrosome reaction of hamster sperm. Biol Reprod. 1999b;61:76–84. doi: 10.1095/biolreprod61.1.76. [DOI] [PubMed] [Google Scholar]
- Weiner AM, Deininger PL, Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- Whitmarsh AJ, Cavanagh J, Tournier C, Yasuda J, Davis RJ. A mammalian scaffold complex that selectively mediates MAP kinase activation. Science. 1998;281:1671–1674. doi: 10.1126/science.281.5383.1671. [DOI] [PubMed] [Google Scholar]
- Wiemann S, Kinzel V, Pyerin W. Cloning of the Cα catalytic subunit of the bovine cAMP-dependent protein kinase. Biochim Biophys Acta. 1992;1171:93–96. doi: 10.1016/0167-4781(92)90144-o. [DOI] [PubMed] [Google Scholar]
- Wiemann S, Voss H, Kinzel V, Pyerin W. Rat Cα catalytic subunit of the cAMP-dependent protein kinase: cDNA sequence and evidence that it is the only isoform expressed in myoblasts. Biochim Biophys Acta. 1991;1089:254–256. doi: 10.1016/0167-4781(91)90018-h. [DOI] [PubMed] [Google Scholar]
- Witman GB. Introduction to cilia and flagella. In: Bloodgood RA, editor. Ciliary and Flagellar Membranes. New York: Plenum Publishing;; 1990. pp. 1–29. [Google Scholar]
- Yeung CH, Weinbauer GF, Cooper TG. Responses of monkey epididymal sperm of different maturational status to second messengers mediating protein tyrosine phosphorylation, acrosome reaction, and motility. Mol Reprod Dev. 1999;54:194–202. doi: 10.1002/(SICI)1098-2795(199910)54:2<194::AID-MRD12>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Young JZ. The Life of Vertebrates. New York: Oxford University Press; 1962. [Google Scholar]
- Zheng J, Knighton DR, Xuong N, Taylor SS, Sowadski JM, Ten Eyck LF. Crystal structures of the myristylated catalytic subunit of cAMP-dependent protein kinase reveal open and closed conformations. Protein Sci. 1993;2:1559–1573. doi: 10.1002/pro.5560021003. [DOI] [PMC free article] [PubMed] [Google Scholar]