Abstract
The human immunodeficiency virus type 1 (HIV-1) particle contains two identical RNA strands, each corresponding to the entire genome. The 5′ untranslated region (UTR) of each RNA strand contains extensive secondary and tertiary structures that are instrumental in different steps of the viral replication cycle. We have characterized the 5′ UTRs of nine different HIV-1 isolates representing subtypes A through G and, by comparing their homodimerization and heterodimerization potentials, found that complementarity between the palindromic sequences in the dimerization initiation site (DIS) hairpins is necessary and sufficient for in vitro dimerization of two subtype RNAs. The 5′ UTR sequences were used to design donor and acceptor templates for a coupled in vitro dimerization-reverse transcription assay. We showed that template switching during reverse transcription is increased with a matching DIS palindrome and further stimulated proportional to the level of homology between the templates. The presence of the HIV-1 nucleocapsid protein NCp7 increased the template-switching efficiency for matching DIS palindromes twofold, whereas the recombination efficiency was increased sevenfold with a nonmatching palindrome. Since NCp7 did not effect the dimerization of nonmatching palindromes, we concluded that the protein most likely stimulates the strand transfer reaction. An analysis of the distribution of template-switching events revealed that it occurs throughout the 5′ UTR. Together, these results demonstrate that the template switching of HIV-1 reverse transcriptase occurs frequently in vitro and that this process is facilitated mainly by template proximity and the level of homology.
On the global scale of the AIDS epidemic, a considerable number of human immunodeficiency virus type 1 (HIV-1) viruses have evolved, all of which have been classified into a major group (M group) of at least 11 genetically distinct subtypes (A through K) or into more distantly related outlier groups (O and N groups) (42). However, this classification is complicated by the fact that 10% of HIV-1 strains are intersubtype recombinants (43), some of which have played a major role in the AIDS pandemic. Interestingly, only some pairs of subtypes have been observed to recombine (26) and factors affecting the frequency and specificity of recombination between subtypes remain important to define.
The 5′ untranslated region (UTR) of the HIV-1 genome is a multifunctional region that can fold into a conserved secondary structure. The functional RNA elements include the trans-activating region (TAR) that binds the transcription factor Tat, the splice donor (SD), and the polyadenylation (polyA) signal. RNA elements used later in the virus replication are the dimerization initiation site (DIS), the packaging signal (PSI), and the primer binding site (PBS). Secondary structure models for the 5′ UTR of the HIV-1 genome have been proposed based on phylogenetic comparison and enzymatic and chemical probing of the leader sequence (reviewed by Berkhout [2, 3]). It has recently been established that the HIV-1 leader can adopt two alternative conformations in vitro that regulate the dimerization potential (4, 16, 17).
In vitro studies have shown that the DIS hairpin displays in its loop a palindromic sequence that is believed to initiate the dimerization reaction by forming a kissing-loop complex (44). Two subtype-specific palindromes exist, GCGCGC and GUGCAC, denoted (CG) and (UA), respectively (38). Variant strains with the nonpalindromic sequence GUGCGC (UG) were observed to arise spontaneously in a subtype population carrying either the (CG) or (UA) palindrome (45). The nucleotides flanking the palindrome exhibit subtype-specific variation: AA(CG)A for subtype B; AA(UA)U for subtype C, in which an A-U base pair increases the palindrome length to 8 bp; and AG(UA)A for subtype A-MAL, where the guanine has been shown to coordinate Mg2+ ion binding (19, 40). From chemical modification studies and computer modeling, the purines have been proposed to be involved in noncanonical interactions stabilizing the kissing-loop complex (40). A nuclear magnetic resonance study of the DIS kissing-loop complex of subtype B-LAI has revealed a bent conformation (30), which is different from the straight conformation recently revealed by the crystal structure analysis of subtypes A-MAL and B-LAI (12).
It has been observed that the kissing-loop complex (loose dimer) of B-LAI is converted into an extended duplex (tight dimer) (23, 25, 33, 44), whereas this transition was not observed for A-MAL (39). The structure of an extended duplex has been investigated by nuclear magnetic resonance for short heat-annealed RNAs from subtype B-LAI virus (31) and by crystallography for short purified RNAs from subtype A-MAL virus (13), and both show an extended duplex with unpaired bulges. The transition from the loose to tight dimers for subtype B-LAI is further increased by the addition of Gag (14) or nucleocapsid protein NCp7 (32).
Interstrain recombination requires infection of the same cell by two different viral strains and packaging of heterologous viral RNA genomes into the same virion. At the reverse transcription step during the subsequent round of infection, template switching by reverse transcriptase (RT) results in a mosaic progeny virus. Template switching is required at the first and second strand-transfers of reverse transcription but can also repair breaks in the RNA templates, a process known as forced copy-choice recombination (7). Template switching was also observed in the absence of strand breaks, leading to the more general model of copy choice recombination (50) for which three different mechanisms have been proposed. (i) Pausing of RT due to either the sequence (49) or structure (46) has been proposed to induce template switching. An apparent paradox of this model was observed, since NCp7 decreases pausing yet enhances template switching (35). (ii) The interactive stem-loop mechanism suggests that the presence of a stem-loop in the RNA, being copied by RT, gives rise to the interaction between nascent cDNA and the acceptor stem-loop (20). (iii) The acceptor invasion mechanism argues that a stem-loop structure in the acceptor template initiates annealing to the nascent cDNA strand copied from the donor (34).
The DIS hairpin has been shown to influence reverse transcription of the HIV-1 genome since deletion of this region was found to severely impair second-strand transfer in vivo (36). However, two defective viruses with nonhomologous DIS palindromes can recombine to form replicating HIV-1 chimeras with the same efficiency as viruses with matching palindromes, implying that the DIS palindrome is dispensable for RNA dimerization and template switching in vivo (45). The influence of the DIS hairpin in template switching has also recently been investigated in vitro by a detailed mapping of switching events. This study yielded a recombination hot spot in the 5′ part of the DIS hairpin (1). This resembles the finding in a murine system that the DIS hairpin of murine leukemia virus is a robust hot spot for recombination, obtained from using an in vivo forced recombination system (27-29). In the present study, we investigated nine different strains of HIV-1, representing at least seven distinct subtypes, and found that template-switching efficiency correlates with both the ability to form the DIS-mediated kissing-loop complex and the level of homology between the templates. Although template switching is enhanced by a stable DIS kissing-loop complex or by the presence of NCp7, we did not find a template-switching hot spot within the DIS region itself.
MATERIALS AND METHODS
Subtype-specific RT-PCR on genomic RNA isolated from AIDS patients.
Primers were designed based on subtype TAR sequences and Gag open reading frame (ORF) sequences (9, 10, 18). Primer positions on the HIV-1 templates are given according to HXB-2 numbering, which is used throughout this paper. RNA purified from patient material was treated at 90°C for 1.5 min and snap-cooled on ice for 5 min. For cloning steps, reverse transcription was performed with OmniScript RT (Qiagen) according to the accompanying protocol with the BW-875 primer (5′-GGGTGGCTCCTTCTGATAATGCTGAAAACATG-3′). For the initial PCR amplification, the BW-875 primer was used together with the following forward primers specific for subtype TAR sequences (positions +3 to +33): subtype B/D, 5′-TCTCTCTGGTTAGACCAGATTTGAGCCTGGG-3′; subtype AC/C2, 5′-TCTCTCTAGGTAGACCAGATCTGAGCCTGGG; subtype C1, 5′-TCTCTCTAGGTAGACCAGATCTGAGCCCGGG-3′; subtype AE, 5′-TCTCTCTTGTTAGACCAGGTC-GAGCCCGGG-3′; subtype F1, 5′-TCTCTCTAGTTAGACCAGATTTGAGCCTGGG-3′; and subtype G/AG, 5′-TCTCTCTTGCTAGACCAGATCTGAGCCTGGG-3′ (subtype-specific differences are underlined). For the next round of PCR, a new set of TAR primers was used, containing an upstream EcoRI restriction site and a T7 promoter (5′-gcgaattctaatacgactcactata-3′), followed by the subtype-specific TAR sequence from positions +1 to +20 as indicated above (small letters indicate a non-HIV sequence). To obtain the subtype clones of the 5′ UTR, the BW-444 primer (5′-TCCCTGCTTGCCCATACT-3′) was used to prime reverse transcription, followed by BW-355-Acc65I (5′-gagggtaccACTGACGCTCTCGCACCCAT-3′). PCR products of all subtypes were digested with EcoRI and Acc65I and cloned into pUC18. To obtain 1-744 clones, BW-875 was used for reverse transcription, followed by BW-744-BamHI (5′-ctggatccCTATAGGGTAATTTTGGCTGACCT-3′). PCR products of subtypes A, B, C2, and D were digested with EcoRI and BamHI and cloned into pUC18. Sequencing was done on the ABI-3100 system (Applied Biosystems). The subtype letters used throughout this paper are as follows: AC (93UG66), B (HXB2), C1 (93ZM74), C2 (93ZM45), D (94ZR80), AE (97TH87), F1 (94BR77), G (93CB76), and AG (94LB12). The subtype lettering is updated relative to the designations used in previous papers (2, 9, 10, 18). The 93UG66 isolate is an AC recombinant with the leader derived from subtype C. Subtype AE is a circulating recombinant form (CRF-01-AE) of subtype A and a putative subtype E (15). Subtype AG is an AG recombinant termed CRF-02-AG (8).
Donor and acceptor constructs.
Acceptor constructs were generated by PCR amplification of the 1-355 subtype constructs with the primer set FW-EcoRI-aTAG-TAR (tcaggaattctagcgtcgta followed by a subtype-specific TAR sequence from positions +1 to +20 as indicated above) and BW-355-Acc65I, followed by digestion with EcoRI and Acc65I and cloning in pBluescript KS(+) (Stratagene). Donor constructs were generated by PCR amplification of the 1-744 subtype constructs with the FW-T7-dTAG-PBS primer (5′- gctaatacgactcactataggctgcgactcatgtctaGGCGCCCGAACAGGGAC-3′) and BW-444-BamHI primer (5′-cgggaTCCCTGCTTGCCCATACT-3′), followed by PCR amplification with the FW-EcoRI-T7 primer (5′-gcgaattctaatacgactcactata-3′) and BW-444-BamHI. The final PCR product was digested with EcoRI and BamHI and cloned in pUC18. The acceptor and donor constructs were verified by sequencing.
RNA transcription.
Runoff transcription on linearized plasmids was done with the T7-MEGAscript kit (Ambion) according to the accompanying protocol. Transcripts were internally labeled by adding [α-32P]UTP (10 μCi/μl; Hartmann Analytic) to the transcription mixture, purified on 4% denaturing polyacrylamide gels, and excised by UV shadowing or autoradiography. Overnight elution in TE buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA) was followed by phenol extraction and ethanol precipitation. Pellets were redissolved in water, quantified by UV absorbance measurements or scintillation counting, and diluted to 1 μM.
Dimerization assays.
Dimerization assays were performed at an RNA concentration of 100 nM. The RNA was denatured at 85°C for 5 min, snap-cooled on ice for 5 min, and incubated at 37°C for 30 min in 10 μl of dimerization buffer (50 mM Na-cacodylate [pH 7.5], 250 mM KCl, 5 mM MgCl2). The samples were placed on ice, 10 μl of loading buffer containing 5% glycerol and 5 mM MgCl2 was added, and the samples were analyzed on native gels containing either 5, 1, or 0.1 mM MgCl2 or 1 mM EDTA in addition to 100 mM Tris-HCl (pH 8.9) and 90 mM borate and run at 1 W at 4°C. All quantifications were done using Molecular Imager FX and Quantity One software (Bio-Rad). Heterodimerization assays were performed with transcripts corresponding to nucleotides 1 to 466 of subtype B and nucleotides 1 to 543 of subtype C2 which were generated by transcription on the 1-744 subtype constructs linearized with restriction enzymes TfiI and EarI, respectively. One hundred nanomoles of these unlabeled transcripts was mixed with 100 nmol of labeled leader RNA prior to denaturation and snap-cooling, followed by incubation in dimerization buffer at 37°C for 30 min at an RNA concentration of 200 nM. The samples were put on ice, and native loading buffer was added before loading on a native gel, keeping a MgCl2 concentration of 5 mM. Dimer species were visualized by autoradiography. Labeled transcripts corresponding to subtype AC nucleotides 183 to 381 and subtype B, C2, and D nucleotides 183 to 404 were generated by transcription on BstNI-linearized donor constructs. To increase the heterodimer species, these transcripts were mixed with unlabeled leader RNA at a molar ratio of 1:5. Denaturation, snap-cooling, dimerization, and electrophoresis were done as described above. Dimer species were quantified, and the percentage of heterodimer to homodimer was calculated by using the equation H = 2 × h/(d + 2 × h) × 100%, where the intensity of the dimer species d is given in counts · mm2 and the intensity of the heterodimer species h is multiplied by two since labeled and unlabeled RNAs are present at a 1:1 molar ratio.
Incubation with NCp7.
Donor and acceptor RNAs were mixed at a concentration of 200 nM and a molar ratio of 1:5. The mixtures were denatured at 85°C for 5 min and snap-cooled on ice for 5 min. RNA transcripts were incubated in 10 μl of NC buffer (20 mM Tris-HCl [pH 7.5], 5 mM dithiothreitol, 50 mM NaCl, 0.2 mM MgCl2) at 37°C for 15 min with 5 μM NCp7 (corresponding to a ∼100% coating of 100 nM RNA [51]); synthetic NCp7 (55 amino acids) from the LAV strain was provided by Jean-Luc Darlix (11). After incubation, the reaction was placed on ice for 2 min and then at 25°C for 2 min. Sodium dodecyl sulfate (0.2%) was added and left at room temperature for 5 min, followed by phenol extraction twice and 1× phenol-chloroform-1× chloroform. Ten microliters of 5% glycerol was added, and the gel was run at 5 W for 3 h at 4°C.
Coupled dimerization and template-switching assay.
Donor and acceptor RNAs were transcribed from donor constructs linearized with BamHI and acceptor constructs linearized with Acc65I. Primer BW-444 was 5′ end labeled with T4 polynucleotide kinase and [γ-32P]ATP (7,000 Ci/mmol; ICN Biomedicals). Primer was mixed with donor and acceptor transcripts at a molar ratio of 1:2:10. After denaturing and snap-cooling, dimerization buffer was added and incubation was allowed to proceed at 37°C for 30 min at an RNA concentration of 100 nM. Samples were placed on ice and subsequently used as substrates in reverse transcription reactions with HIV-1 RT from the LAV strain (provided by S. Stammes, Glaxo Wellcome, the Centralized Facility for AIDS reagents). Reverse transcription was allowed to proceed at 37°C for 30 min at an RNA concentration of 10 nM in a mixture containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 5 mM MgCl2, 10 mM dithiothreitol, 100 μM concentrations of each deoxynucleoside triphosphate, and 0.2 U of HIV-1 RT/μl. The reaction was terminated by the addition of 10 mM EDTA, precipitated, redissolved in Maxam-Gilbert loading solution (80% deionized formamide, 10 mM NaOH, 1 mM EDTA, 0.02% xylene cyanol, 0.02% bromphenol blue), and run at 70 W on a 4% denaturing polyacrylamide gel. Full-length donor and acceptor extension products were quantified, and the template-switching efficiency T was calculated as T = s/(f + s) × 100%, where s indicates the intensity of the switched product and f indicates the intensity of the full-length donor product. A subset of donor to acceptor combinations (donor D to acceptor B, donor C2 to acceptor AC, and donor B to acceptor AC) was chosen for investigation of the influence of dimerization on template-switching efficiency. Different dimerization conditions were used: dimerization at 37°C for 5 or 40 min was followed by 30-min reverse transcription at the same conditions described above. Finally, the RNA transcripts were incubated with NCp7 as described above, which was followed by 30 min of reverse transcription on 10 nM RNA with 0.5 μM NCp7 under the same conditions mentioned above.
Isolation and detection of template-switching events.
The cDNA corresponding to the switched product was purified by phenol-chloroform extraction, redissolved in water, and amplified with the primers FW-aTAG (5′-gggcgaattctagcgtcgtag-3′) and BW-444. The PCR products were cloned and sequenced by using the TOPO TA cloning kit (Invitrogen). The recombinant sequences were analyzed, and the template-switching events were identified by aligning with the parent donor and acceptor sequences with SeqEd software (Applied Biosystems).
Nucleotide sequence accession numbers.
The accession numbers created over the course of this study are as follows: for AC, AY118154; for C1, AY118157; for C2, AY118158; for D, AY118159; for AE, AY118155; for F1, AY118160; for G, AY118161; and for AG, AY118156. The alignment used in Fig. 1B is available at EMBL (ALIGN 000493).
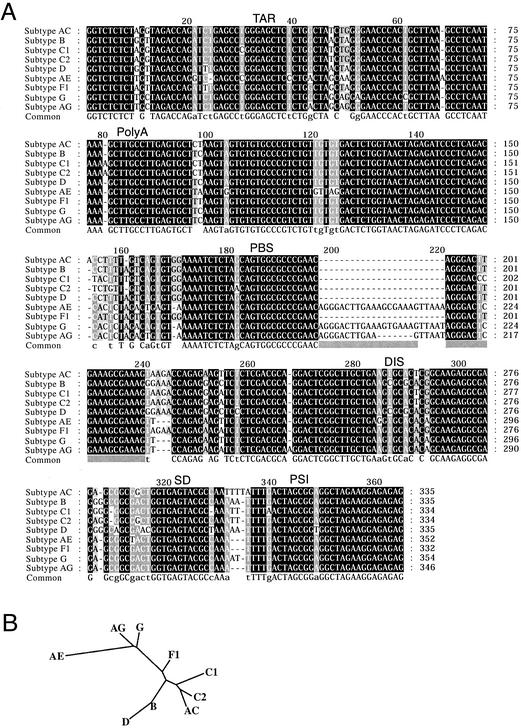
FIG. 1.
(A) Alignment of the cloned leader sequences of eight circulating HIV-1 isolates obtained from eight infected individuals. The sequence of subtype B-HXB2 is included as a reference sequence. The subtype assignment is shown to the left (see Material and Methods), and the positions of the functional RNA elements are denoted as follows: TAR; PolyA, hairpin containing polyA site used in the 3′ LTR; PBS; DIS; SD, major SD; PSI. Conservation of alignment positions is indicated by box shading: black; 100%; gray, 80%; light gray, 60%. (B) Unrooted phylogenetic tree based on the alignment of the subtype sequences shown in panel A. The branch lengths were calculated by using the PHYLIP program (J. Felsenstein, 1993. PHYLIP [Phylogeny Inference Package] version 3.5c. Distributed by the author. Department of Genetics, University of Washington, Seattle).
RESULTS
Cloning and characterization of 5′ leader sequences from nine isolates of HIV-1.
Leader sequences of nine different isolates of HIV-1 were isolated by RT-PCR from RNA purified from patient blood samples that were previously subtyped based on partial Gag and long terminal repeat (LTR) sequences (9, 10). Leader sequences (nucleotides 1 to 335) were cloned and sequenced for subtypes C1, AE, F1, G, and AG, and a larger fragment, including the 5′ part of the Gag ORF (nucleotides 1 to 744), was sequenced for subtypes AC, B, C2, and D. The leader sequences of all cloned subtypes are aligned in Fig. 1A. A phylogenetic comparison of the sequences with previously published LTR and Gag sequences confirms the subtype annotation (see Materials and Methods). The sequence similarity of the untranslated leaders was found to be as high as 85 to 95%, compared to only 45% in the Gag ORF, which illustrates the high conservation of the 5′ UTR. Based on the sequence alignment, a phylogenetic tree was constructed (Fig. 1B).
Subtypes AE and G contain a 24-nucleotide insert that is placed after position 196 within the PBS, as indicated in Fig. 1A (see Discussion). Subtype AG has the same characteristics, but with a 7-nucleotide deletion within the insert. In addition, these subtypes contain a 3-nucleotide deletion 20 nucleotides downstream of the PBS. A hypervariable region was also found in the single-stranded AU region situated between the SD and PSI hairpins.
The DIS palindrome can be divided into two groups, (CG) for subtypes B and D and (UA) for subtypes AC, C1, C2, AE, F1, G, and AG. Variants for subtypes AC, D, and F1 that exhibited differences in the DIS palindrome and flanking purines and in the hypervariable AU stretch (nucleotides 329 to 338) were isolated from the same patient. Notably, a variant of subtype F1, designated F1′, contained a nonpalindromic (UG) kissing-loop sequence.
Stabilization of homodimers by Mg2+ ions.
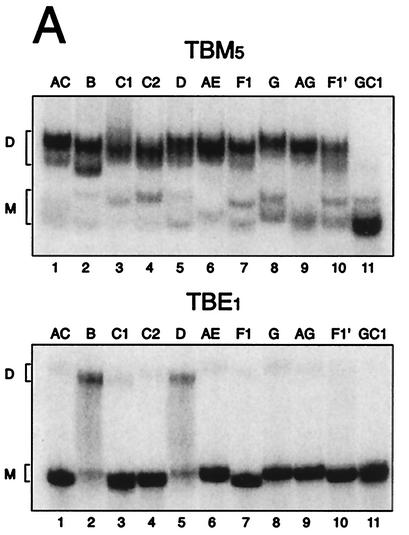
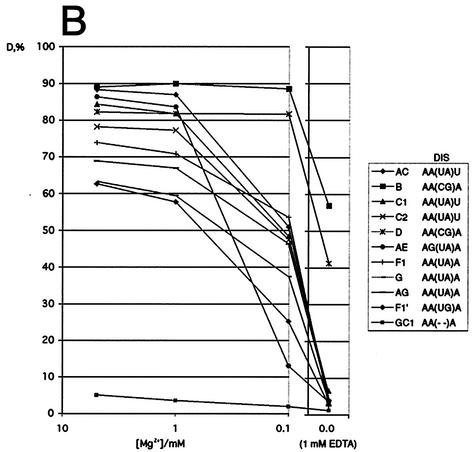
To test the ability of the subtype leader transcripts to dimerize, the RNA transcripts were incubated under conditions that promoted dimerization (250 mM KCl, 5 mM MgCl2) and analyzed on native gels containing different amounts of MgCl2. In the presence of 5 mM Mg2+ in the gel, all subtypes dimerized to a level of 63 to 90% (Fig. 2A, upper panel) and all subtypes formed one to two different dimer species. Subtype F1′, which contains an imperfect palindrome, has the lowest dimer-forming potential of the natural substrates. No dimer formation was observed in the GC1 control transcripts with a 4-nucleotide deletion in the DIS palindrome (5) (Fig. 2A). When decreasing the Mg2+ concentration in the native gel, the subtypes exhibited considerable differences in dimer stability (Fig. 2B). With the lowering of the concentration from 5 to 1 mM, which is comparable to the intracellular Mg2+ concentration (47), all subtypes displayed only a minor decrease in stability. At 0.1 mM, all subtypes with an (UA) palindrome exhibited a major drop in dimer stability, the subtype AE dimer responding most strongly (Fig. 2B). Removal of Mg2+ ions (1 mM EDTA) disrupted the dimers formed by subtypes with the (UA) palindromes but only reduced the amount of B and D dimers with the (CG) palindromes to about 50% (Fig. 2A, lower panel, and B). This implies that the dimerization of natural occurring subtype RNAs with (AU) palindromes requires Mg2+ ions. If not otherwise stated, all subsequent dimerization and reverse transcription assays were performed at 5 mM Mg2+.
FIG. 2.
Effect of Mg2+ ions on the stability of subtype dimers. (A) Dimerization of all subtype leader transcripts analyzed on native polyacrylamide gels containing 5 mM MgCl2 (TBM5) or 1 mM EDTA (TBE1). The positions of the monomer and dimer species are indicated to the left of the gels by the letters M and D, respectively. (B) Quantification of dimerization efficiency (D = [number of dimers]/[number of dimers + number of monomers]) for all subtypes by a similar gel retardation assay of subtype homodimers as shown in panel A in the presence of decreasing amounts of MgCl2 (5, 1, and 0.1 mM) and in the presence of EDTA.
DIS-dependent heterodimerization.
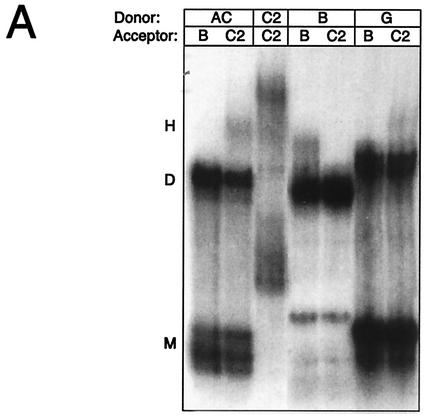
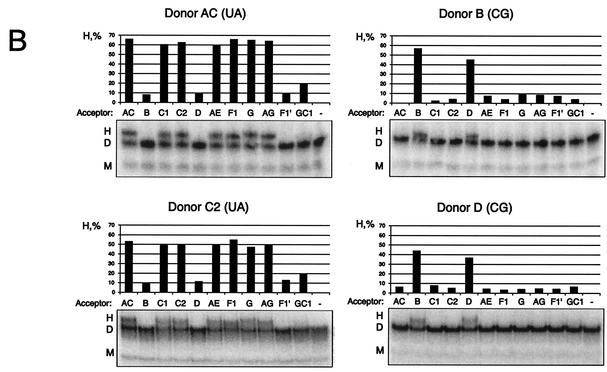
To investigate the heterodimerization between different subtypes, dimerization was performed by using RNA transcripts of different lengths. Radiolabeled transcripts of subtypes AC, B, and G, including the entire 5′ UTR, were incubated at a 1:1 molar ratio with the unlabeled leader RNA of subtype B or C2 spanning nucleotides 1 to 460 and 1 to 540, respectively, and the dimerization efficiency was assayed on native polyacrylamide gels containing 5 mM Mg2+ (Fig. 3A). Heterodimerization was observed for AC/C2 and more weakly for G/C2 combinations that contained matching DIS palindromes, whereas no dimerization was observed between HIV-1 RNAs with divergent palindromes (AC/B, B/C2, and G/B).
FIG. 3.
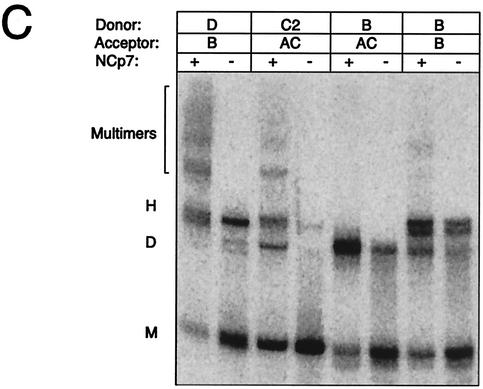
Heterodimerization between subtypes is dependent exclusively on the DIS palindrome. (A) Heterodimerization between labeled transcripts of subtypes AC, B, or G (Donor; nucleotides 183 to 404) and longer unlabeled leader transcripts of subtypes B (Acceptor; nucleotides 1 to 466) or C2 (Acceptor; nucleotides 1 to 543) at a molar ratio of 1:1. (B) Heterodimerization between labeled transcripts of subtype AC (nucleotides 183 to 381) or subtypes B, C2, and D (Donor; nucleotides 183 to 404) with a 20-nucleotide 5′-tag sequence and longer unlabeled transcripts of subtypes AC, B, C1, C2, D, AE, F1, G, AG, F1′, and GC1 (Acceptor; nucleotides 1 to 355) at a molar ratio of 1:5. The percentage of heterodimers compared to that of homodimers was calculated as follows: H = 2 × h/(d + 2 × h) × 100%, where d and h are the intensities of the dimer and heterodimer, respectively. The DIS palindrome classes corresponding to donors AC, B, C2, and D are indicated in brackets. (C) Heterodimerization in the presence and absence of NCp7 by using a selected set of the same RNAs as shown in panel B. NCp7 was removed by phenol extraction prior to loading. The multimers that were formed only when using RNAs with matching DIS palindromes and in the presence of NCp7 are marked. The positions of the monomer, dimer, and heterodimer species are indicated with M, D, and H, respectively.
Heterodimerization was also studied by using shorter RNAs corresponding to the substrates used for the template-switching assay described below. An RNA, spanning half the leader sequence and the Gag ORF sequence from subtypes AC, C2, B, and D (nucleotides 183 to 400), was mixed with a fivefold molar excess of unlabeled leader transcripts spanning nucleotides 1 to 360 from all other subtypes and analyzed by native gel electrophoresis (Fig. 3B). The result was essentially the same as that for the longer transcripts, showing that heterodimerization was observed only between RNAs containing a matching palindrome. The variant F1′, with the (UG) palindrome, did not show significant heterodimer formation, even though potential base pairings are possible with both the (GC)- and (AU)-type palindromes through G-U base pairing. This implies that a perfect symmetry of the initial palindrome kissing is important to make a stable kissing-loop complex.
The effect of NCp7 on heterodimerization was investigated by using the short-RNA substrates at a 1:5 molar ratio as shown in Fig. 3B. Incubation with NCp7 stimulated heterodimerization between transcripts with matching palindromes (D/B, C2/AC, and B/B), but substrates with nonmatching palindromes (B/AC) remained unable to form heterodimers (Fig. 3C). Note that these experiments were performed under a low Mg2+ concentration (0.2 mM), which explains the low yield of dimers in the absence of NCp7 compared to that shown in Fig. 3B. Interestingly, several multimeric species were observed when RNAs with matching palindromes were incubated in the presence of NCp7. This suggests that elements in the region from nucleotides 1 to 183 (present only in the unlabeled acceptor substrate) are able to dimerize in an NCp7-dependent manner, forming multimeric RNA complexes.
Characterization of intersubtype template switching.
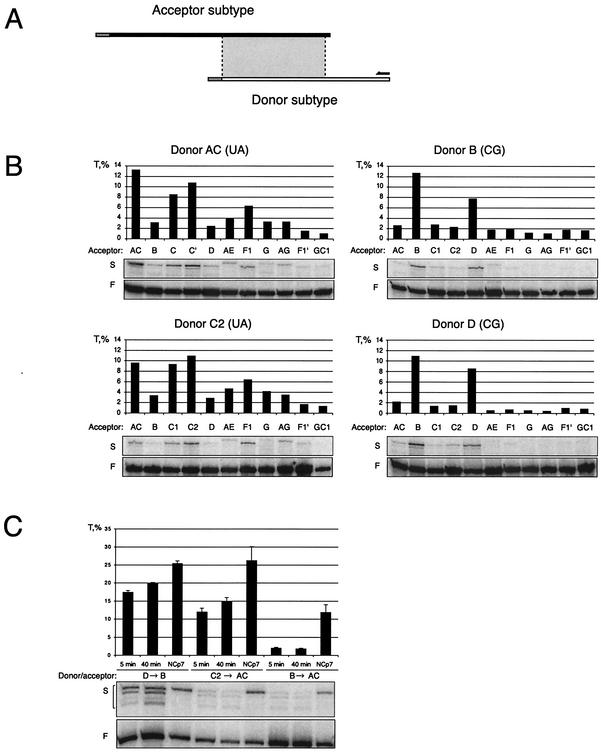
A combined dimerization-template-switching assay was developed with donor and acceptor templates sharing a region of homology of approximately 170 nucleotides. Reverse transcription was initiated from a primer that anneals only to the donor. To ensure that only internal template switching was assayed, we introduced a unique 5′ tag on the donor template, which prevented template switching when RT reached the end of the donor template (Fig. 4A). Donor substrates of subtypes AC, B, C2, and D were tested against acceptors of all subtypes, including DIS variant F1′ and the DIS mutant GC. A molar ratio of 1:5 of the donor to acceptor was used to increase the amount of heterodimer template in the reaction. If donors and acceptors were derived from the same subtype, template-switching efficiency ranged from 8.5 to 13.2%, whereas template switching between subtype heterodimers with matching DIS palindromes ranged from 3.3 to 10.8% for (UA) and 7.7 to 10.9% for (CG) (Fig. 4B). For nonmatching palindromes and variant F1′, (UG) template-switching efficiencies were always below 2%. Next, we investigated the effect of varying the preincubation time for RNA dimer formation and the effect of recombinant NCp7 protein on template switching between selected sets of templates (Fig. 4C). Increasing the dimerization incubation time from 5 to 40 min raised the D-to-B template-switching efficiency only slightly from 18 to 20%. The C2-to-AC switching was increased from 13 to 15%, and no effect was observed for B-to-AC switching, which remained low at 2% (Fig. 4C). Adding NCp7 to the reaction increased the template-switching efficiency approximately 1.5- to 2-fold for templates with matching palindromes (D to B and C2 to AC [Fig. 4C]) but sevenfold for unmatched templates (B to AC). Thus, the stimulatory effect of NCp7 on template switching is most dramatic for subtype templates with a nonmatching DIS palindrome.
FIG. 4.
Template-switching efficiency. (A) Experimental setup showing the donor and acceptor transcripts drawn to scale with unique tags at their 5′ end, the region of homology (shaded box), and the RT primer. (B) Template-switching efficiency T for all combinations of AC, B, C2, and D (Donor; nucleotides 183 to 444) and AC, B, C1, C2, D, AE, F1, G, AG, F1′, and GC1 (Acceptor; nucleotides 1 to 355) at a molar ratio of 1:5. Only elongated products are shown from the primer extension gels. F indicates the full-length donor product; S indicates primers that have switched to the acceptor template. The template-switching efficiency (T) was calculated as s/(f + s) × 100%, where s is the intensity of the template-switching product in the primer extension reaction and f is the intensity of the band corresponding to the full-length product of the donor. (C) Template-switching efficiencies for selected combinations of the donor to the acceptor under different conditions. The donor and acceptor templates were incubated under dimerization conditions for 5 and 40 min as indicated or treated with NCp7 in NC buffer for 15 min prior to reverse transcription (NCp7).
Mapping of recombination sites.
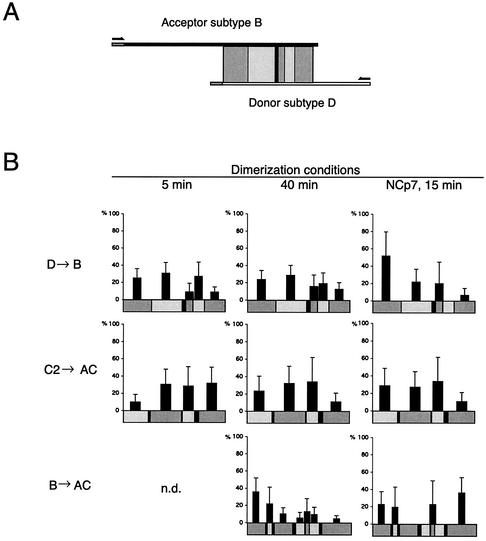
Based on the sequence differences between the subtypes, the region of homology can be divided into regions of identity (Fig. 5A). When RT switches templates during cDNA synthesis, it creates a progeny cDNA sequence that harbors sequence identity to both templates. cDNA strands produced from template switching were selectively amplified with PCR primers specific for the donor and acceptor templates, and 16 to 41 clones were sequenced for each condition. By sequence comparison, the positions of template switching were mapped to defined regions. The number of template-switching events within each region of identity is shown for D to B, C2 to AC, and B to AC in Table 1. The number was normalized by dividing it by the number of nucleotides in each region, and this value is illustrated in Fig. 5B (regions shorter than 5 nucleotides are listed in Table 1 but not included in Fig. 5B due to statistical deviation). Only cDNAs produced by a single template-switching event were included in the analysis. The D-to-B and C2-to-AC combinations with matching palindromes exhibited no particular hot spot for template switching, and the addition of NCp7 did not alter the distribution significantly (Fig. 5B). For switching between B-to-AC templates with nonmatching palindromes, we mapped the distribution of template switches after 40 min of dimerization. Under these conditions, the template switching was significantly more frequent toward the 5′ end of the template. Notably, two clones switched templates at 1 nucleotide located in the nonmatching DIS palindromes (Table 1). The addition of NCp7 resulted in a more even distribution of switching between the 5′ and 3′ regions of the template, whereas the region including the 5′ part of the DIS hairpin showed no template-switching events at all (Fig. 5B).
FIG. 5.
Distribution of template-switching events for different subtype combinations and conditions. (A) Illustration of the experimental setup with indication of regions of identity (shaded boxes) and phylogenetic markers (black lines) between subtypes B and D. The transcripts, primers, regions of identity, and markers are drawn to scale. (B) Distribution of template-switching events. Prior to RT transcription, the RNA substrates were dimerized for either 5 or 40 min or incubated for 15 min in the presence of NCp7. Analysis of recombinant clones was performed, and the identified template-switching events are summarized in Table 1. The y axis indicates the frequency of template switching normalized to the number of nucleotides within each region of identity. Standard error bars are estimated as Δpi = pi[(ni)−1/2 + (N)−1/2], where N is the total number of recombinants cloned and sequenced in each independent experiment and ni is the number of recombinants occurring in the interval i. All regions of identity less than 5 nucleotides long are omitted from the figure but are shown in Table 1. Three independent experiments were performed for the donor D-to-acceptor B combination (5 and 40 min) with similar distribution. All three experiments were included in the data analysis. Only single template-switching events were included. n.d., not determined.
TABLE 1.
Template-switching events
| Recombination partners and regions of identity (nucleotide position) | Size (nucleotides)a | No. of recombination events observed
|
||
|---|---|---|---|---|
| Dimerization
|
15-min NCp7 incubationd | |||
| 5 minb | 40 minc | |||
| Donor D to acceptor B | ||||
| 183-229 | 47 | 12 | 13 | 11 |
| 231-281 | 51 | 16 | 17 | 5 |
| 283-286 | 4 | 2 | 1 | 0 |
| 289-299 | 11 | 1 | 2 | 1 |
| 301-318 | 18 | 5 | 4 | 0 |
| 320-353 | 34 | 3 | 5 | 1 |
| Donor C2 to acceptor AC | ||||
| 183-223 | 41 | 2 | 4 | 5 |
| 228-279 | 52 | 8 | 7 | 6 |
| 281-301 | 21 | 3 | 3 | 3 |
| 310-353 | 44 | 7 | 2 | 2 |
| Donor B to acceptor AC | ||||
| 183-212 | 30 | ND | 14 | 5 |
| 217-223 | 7 | ND | 2 | 1 |
| 228-257 | 30 | ND | 4 | 0 |
| 262 | 1 | ND | 2 | 0 |
| 264-277 | 14 | ND | 1 | 0 |
| 280-285 | 6 | ND | 1 | 1 |
| 287-302 | 16 | ND | 2 | 0 |
| 307-355 | 49 | ND | 3 | 13 |
The sizes of the template regions were as follows (in nucleotides): 165 for donor D to acceptor B, 158 for donor C2 to acceptor AC, and 153 for donor B to acceptor AC.
The total number of recombination events for template regions under 5-min dimerization were 37 for donor D to acceptor B and 20 for donor C2 to acceptor AC. ND, not determined.
The total number of recombination events for template regions under 40-min dimerization were as follows: 41 for donor D to acceptor B, 16 for donor C2 to acceptor AC, and 27 for donor B to acceptor AC.
The total number of recombination events for template regions under 15-min incubation with NCp7 were as follows: 18 for donor D to acceptor B, 16 for donor C2 to acceptor AC, and 20 for donor B to acceptor AC.
DISCUSSION
The leader sequences of nine different isolates of HIV-1 were isolated by RT-PCR from AIDS patient blood samples. Based on an alignment, we were able to categorize the sequences according to subtypes and construct an evolutionary tree. A comprehensive comparison of their homo- and heterodimerization capabilities was performed, and an in vitro reverse transcription assay was applied to characterize the efficiency and position of template switching during RT transcription.
Constructing the alignment.
A main difficulty in constructing the subtype leader alignment is positioning the duplication insert in the PBS region. The alignment found in the Los Alamos HIV sequence database has two possible placements, both of which have been suggested in the literature (9, 21). We propose a new solution where the insert is placed within the PBS sequence (Fig. 1A). In this alignment, subtypes AE, G, and AG contain a 17- to 24-nucleotide insert in the PBS sequence as well as a 3-nucleotide deletion 20 nucleotides downstream of the PBS. Among the CRF-01-AE isolates, intermediates exist with the 3-nucleotide deletion but not the 24-nucleotide insertion (21). The 3-nucleotide deletion has arisen in a region recognized by a transcription factor, interferon responsive factor (48), and the insertion may restore this binding site. Furthermore, the upstream placement seems most appropriate if this insert is a result of an HIV-1-RT-mediated jump to a homologous sequence. The hypervariable region between the SD and PSI hairpins can probably be explained by RT slippage on homopolymeric stretches.
The differences in the sequences obtained in this report and in a previous publication (9) by using the same patient material might be the result of a viral quasispecies present in the infected patient and not caused by PCR errors. In support of this suggestion, the variations were all clustered in the DIS and in the hypervariable region between the SD and PSI. Moreover, we isolated two different species from the same material, F (UA) and F1′ (UG), both of which were found to be functional in the in vitro dimerization assay.
The subtype clones were compared with other known leader sequences in a phylogenetic analysis (data not shown). This showed that all subtypes were correctly assigned except for AC that was only A-like in the U3 sequence but subtype C-like in the leader and Gag ORF. Noticeably, the F1 sequence appears to be the first F-subtype U5 sequence to have been submitted to the public HIV sequence database.
Subtype RNA homo- and heterodimerization.
Homodimerization is highly efficient for all subtype leader transcripts in the presence of 1 to 5 mM Mg2+. However, only subtypes B and D with (CG) palindromes form dimers at lower Mg levels (0.1 mM or no Mg2+). Homodimerization of subtype AE exhibited a particular strong Mg2+ dependency, which can probably be attributed to the nucleotides flanking the DIS palindrome. Subtype AE has the unique composition, AG(UA)A, where the guanine has been shown to coordinate an Mg2+ ion (19, 40). The nonpalindromic (UG) subtype F1′ dimerizes with an efficiency of 63%, which is in agreement with previous studies on the Mg2+ requirements for the composition of the DIS palindrome (6, 37, 39). In conclusion, we found that the Mg2+ dependence of homodimerization among individual subtype DIS palindromes increases in the following order: AA(CG)A < AA(UA)A < AA(UA)U < AG(UA)A < AA(UG)A. The transition from kissing-loop complex to extended duplex (tight dimer) is evidenced for subtype B and, to a lesser extent, for subtype D by a faster migrating dimer species on Mg2+ gels and a more stable dimer on EDTA gels. This is in agreement with a previous study that observed two different dimer species for subtype B (25).
Heterodimerization experiments of 44 combinations of subtype RNA revealed that perfect complementarity between the DIS palindromic sequences is both necessary and sufficient for dimerization. Interestingly, no heterodimer species were observed when the subtype with the (UG) palindrome was mixed with a subtype with either the (CG) or (UA) palindromes, although these combinations should be able to form the kissing-loop complex with a single G-U base pair. This suggests that the composition or symmetry of the interaction is important for stable complex formation. The nonpalindromic (UG) can be viewed as an evolutionary intermediate between the (CG) and (UA) palindromes (24), and it has been shown to arise in subtype populations with either the (CG) or (UA) palindromes (45). Indeed, we found two subtype F1 variants with a single nucleotide substitution that changes the (UA) palindrome to the nonpalindrome (UG) type.
Template switching.
This study indicates that, in the absence of NCp7, the formation of the kissing-loop-induced RNA dimer is a prerequisite for efficient template switching, and frequencies up to 15% can be measured under such conditions. The importance of a dimeric template for template switching is further evidenced by the observation that increased dimerization time improves the template-switching efficiency of donors and acceptors with matching, but not nonmatching, DIS palindromes. However, by analyzing different combinations of subtype donors and acceptors, we observed significant variation in the template-switching efficiency among matching subtype pairs with comparable heterodimerization yields (compare Fig. 3B and 4B). This suggests that other factors influence the mechanism of template switching. One explanation is suggested by inspection of the phylogenetic tree in Fig. 1B. With one exception, namely that the template switching between two D templates is less efficient than that between D and B, the decrease in the template-switching efficiency between subtypes with matching palindromes seems to correlate with evolutionary distances and thus sequence homology. These observations suggest that template switching is driven primarily by RNA dimerization that brings the two templates in close proximity, but also by sequence homology, which may increase the annealing efficiency of the nascent cDNA tail onto the acceptor template. The same effect of sequence homology between donor and acceptor templates on recombination has recently been reported for murine leukemia virus in a forced copy choice in vivo assay (41).
The presence of NCp7 in the dimerization-reverse transcription assay further enhances the template-switching efficiency, in particular, for nonmatching palindromes. We set out to test whether this effect was on the dimerization or reverse transcription step. Interestingly, we found that NCp7 was unable to stimulate the dimerization of templates with unmatched palindromes but still resulted in a dramatic increase in template switching. A likely interpretation of this result is that NCp7 may increase the annealing efficiency of the emerging cDNA tail with the acceptor template and that this connection induces template switching as suggested previously by Lapadat-Tapolsky et al. (22). For matching templates, we measured only a twofold effect on switching efficiency by NCp7, which may have been due to similar effects. It was established by St. Louis et al. that in vivo recombination between HIV-1 templates with unmatched DIS palindromes occurs with the same efficiency as that for templates with matching sequences (45). Our result suggests that the presence of NCp7 in the capsid is able to promote efficient template switching independent of DIS dimerization but raises the question of how these templates are dimerized and packaged together in the same virion. It is possible that dimerization signals, which are not present in our constructs, may cooperate with the DIS region in vivo. Noticeably, the St. Louis study was performed with isolated DIS mutations within the same laboratory B-strain clone that has been selected for rapid in vitro replication in T cells, whereas this study was based on dimerization and template switching among divergent primary HIV-1 isolates replicating in peripheral blood mononuclear cells.
From sequencing a large number of cDNA clones that derived from individual template-switching events, we observed a similar template-switching frequency throughout the leader sequence for both (CG)- and (UA)-matched transcripts. This observation contrasts the recent work by Balakrishnan et al., who found a hot spot in the 5′ part of the DIS hairpin by using a similar in vitro template-switching assay (1). However, the experimental design differed, which may explain the discrepancy: they used two B strains with a different pattern of genetic markers, and the experimental conditions during dimerization and reverse transcription were significantly different. A preference for template-switching events has also been demonstrated in the 5′ part of the TAR hairpin (20), but this region is outside the region that was investigated in this report.
In conclusion, template switching occurs throughout the subtype leader sequences and appears to be stimulated by the level of RNA template dimerization and sequence homology. Even in the absence of matching DIS palindromes, NCp7 can mediate the transfer of RT enzyme between templates.
Acknowledgments
We thank M. P. de Baar for providing the patient material of HIV-1 subtypes; S. Stammers, Glaxo Wellcome (the Centralized Facility for AIDS Reagents), for HIV-1 RT; Jean-Luc Darlix for the generous donation of synthetic NCp7; Ray Brown for critical reading of the manuscript; and Rita Rosendahl Hansen for excellent technical assistance.
The work was supported in part by grants from the Danish National Research Foundation, The Carlsberg Foundation, The Danish AIDS Fund, The Dutch AIDS Fund, and the Karen Elise Jensen Foundation. R.E.J. was sponsored by a short-term EMBO fellowship, and C.K.D. was supported by the University of Aarhus.
REFERENCES
- 1.Balakrishnan, M., P. J. Fay, and R. A. Bambara. 2001. The kissing hairpin sequence promotes recombination within the HIV-I 5′ leader region. J. Biol. Chem. 276:36482-36492. [DOI] [PubMed] [Google Scholar]
- 2.Berkhout, B. 2000. Multiple biological roles associated with the repeat (R) region of the HIV-1 RNA genome. Adv. Pharmacol. 48:29-73. [DOI] [PubMed] [Google Scholar]
- 3.Berkhout, B. 1996. Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acid Res. Mol. Biol. 54:1-34. [DOI] [PubMed] [Google Scholar]
- 4.Berkhout, B., and J. L. van Wamel. 2000. The leader of the HIV-1 RNA genome forms a compactly folded tertiary structure. RNA 6:282-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkhout, B., and J. L. van Wamel. 1996. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J. Virol. 70:6723-6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clever, J. L., M. L. Wong, and T. G. Parslow. 1996. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J. Virol. 70:5902-5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coffin, J. M. 1979. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J. Gen. Virol. 42:1-26. [DOI] [PubMed] [Google Scholar]
- 8.Cornelissen, M., R. van Den Burg, F. Zorgdrager, and J. Goudsmit. 2000. Spread of distinct human immunodeficiency virus type 1 AG recombinant lineages in Africa. J. Gen. Virol. 81:515-523. [DOI] [PubMed] [Google Scholar]
- 9.De Baar, M. P., A. De Ronde, B. Berkhout, M. Cornelissen, K. H. Van Der Horn, A. M. Van Der Schoot, F. De Wolf, V. V. Lukashov, and J. Goudsmit. 2000. Subtype-specific sequence variation of the HIV type 1 long terminal repeat and primer-binding site. AIDS Res. Hum. Retrovir. 16:499-504. [DOI] [PubMed] [Google Scholar]
- 10.De Baar, M. P., A. M. van der Schoot, J. Goudsmit, F. Jacobs, R. Ehren, K. H. van der Horn, P. Oudshoorn, F. de Wolf, and A. de Ronde. 1999. Design and evaluation of a human immunodeficiency virus type 1 RNA assay using nucleic acid sequence-based amplification technology able to quantify both group M and O viruses by using the long terminal repeat as target. J. Clin. Microbiol. 37:1813-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Rocquigny, H., D. Ficheux, C. Gabus, M. C. Fournie-Zaluski, J. L. Darlix, and B. P. Roques. 1991. First large scale chemical synthesis of the 72 amino acid HIV-1 nucleocapsid protein NCp7 in an active form. Biochem. Biophys. Res. Commun. 180:1010-1018. [DOI] [PubMed] [Google Scholar]
- 12.Ennifar, E., P. Walter, B. Ehresmann, C. Ehresmann, and P. Dumas. 2001. Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nat. Struct. Biol. 12:12.. [DOI] [PubMed] [Google Scholar]
- 13.Ennifar, E., M. Yusupov, P. Walter, R. Marquet, B. Ehresmann, C. Ehresmann, and P. Dumas. 1999. The crystal structure of the dimerization initiation site of genomic HIV-1 RNA reveals an extended duplex with two adenine bulges. Structure Fold Des. 7:1439-1449. [DOI] [PubMed] [Google Scholar]
- 14.Feng, Y. X., S. Campbell, D. Harvin, B. Ehresmann, C. Ehresmann, and A. Rein. 1999. The human immunodeficiency virus type 1 Gag polyprotein has nucleic acid chaperone activity: possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J. Virol. 73:4251-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao, F., D. L. Robertson, S. G. Morrison, H. Hui, S. Craig, J. Decker, P. N. Fultz, M. Girard, G. M. Shaw, B. H. Hahn, and P. M. Sharp. 1996. The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J. Virol. 70:7013-7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huthoff, H., and B. Berkhout. 2002. Multiple secondary structure rearrangements during HIV-1 RNA dimerization. Biochemistry 41:10439-10445. [DOI] [PubMed]
- 17.Huthoff, H., and B. Berkhout. 2001. Two alternating structures of the HIV-1 leader RNA. RNA 7:143-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeeninga, R. E., M. Hoogenkamp, M. Armand-Ugon, M. de Baar, K. Verhoef, and B. Berkhout. 2000. Functional differences between the long terminal repeat transcriptional promoters of human immunodeficiency virus type 1 subtypes A through G. J. Virol. 74:3740-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jossinet, F., J. C. Paillart, E. Westhof, T. Hermann, E. Skripkin, J. S. Lodmell, C. Ehresmann, B. Ehresmann, and R. Marquet. 1999. Dimerization of HIV-1 genomic RNA of subtypes A and B: RNA loop structure and magnesium binding. RNA 5:1222-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, J. K., C. Palaniappan, W. Wu, P. J. Fay, and R. A. Bambara. 1997. Evidence for a unique mechanism of strand transfer from the transactivation response region of HIV-1. J. Biol. Chem. 272:16769-16777. [DOI] [PubMed] [Google Scholar]
- 21.Kurosu, T., T. Mukai, W. Auwanit, P. I. Ayuthaya, S. Saeng-Aroon, and K. Ikuta. 2001. Variable sequences in the long terminal repeat and Its downstream region of some of HIV type 1 CRF01_AE recently distributing among Thai carriers. AIDS Res. Hum. Retrovir. 17:863-866. [DOI] [PubMed] [Google Scholar]
- 22.Lapadat-Tapolsky, M., C. Gabus, M. Rau, and J. L. Darlix. 1997. Possible roles of HIV-1 nucleocapsid protein in the specificity of proviral DNA synthesis and in its variability. J. Mol. Biol. 268:250-260. [DOI] [PubMed] [Google Scholar]
- 23.Laughrea, M., and L. Jette. 1996. HIV-1 genome dimerization: formation kinetics and thermal stability of dimeric HIV-1Lai RNAs are not improved by the 1-232 and 296-790 regions flanking the kissing-loop domain. Biochemistry 35:9366-9374. [DOI] [PubMed] [Google Scholar]
- 24.Laughrea, M., and L. Jette. 1997. HIV-1 genome dimerization: kissing-loop hairpin dictates whether nucleotides downstream of the 5′ splice junction contribute to loose and tight dimerization of human immunodeficiency virus RNA. Biochemistry 36:9501-9508. [DOI] [PubMed] [Google Scholar]
- 25.Laughrea, M., and L. Jette. 1996. Kissing-loop model of HIV-1 genome dimerization: HIV-1 RNAs can assume alternative dimeric forms, and all sequences upstream or downstream of hairpin 248-271 are dispensable for dimer formation. Biochemistry 35:1589-1598. [DOI] [PubMed] [Google Scholar]
- 26.McCutchan, F. E., M. O. Salminen, J. K. Carr, and D. S. Burke. 1996. HIV-1 genetic diversity. AIDS 10(Suppl. 3):S13-S20. [PubMed] [Google Scholar]
- 27.Mikkelsen, J. G., A. H. Lund, M. Duch, and F. S. Pedersen. 2000. Mutations of the kissing-loop dimerization sequence influence the site specificity of murine leukemia virus recombination in vivo. J. Virol. 74:600-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mikkelsen, J. G., A. H. Lund, M. Duch, and F. S. Pedersen. 1998. Recombination in the 5′ leader of murine leukemia virus is accurate and influenced by sequence identity with a strong bias toward the kissing-loop dimerization region. J. Virol. 72:6967-6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mikkelsen, J. G., A. H. Lund, K. D. Kristensen, M. Duch, M. S. Sorensen, P. Jorgensen, and F. S. Pedersen. 1996. A preferred region for recombinational patch repair in the 5′ untranslated region of primer binding site-impaired murine leukemia virus vectors. J. Virol. 70:1439-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mujeeb, A., J. L. Clever, T. M. Billeci, T. L. James, and T. G. Parslow. 1998. Structure of the dimer initiation complex of HIV-1 genomic RNA. Nat. Struct. Biol. 5:432-436. [DOI] [PubMed] [Google Scholar]
- 31.Mujeeb, A., T. G. Parslow, A. Zarrinpar, C. Das, and T. L. James. 1999. NMR structure of the mature dimer initiation complex of HIV-1 genomic RNA. FEBS Lett. 458:387-392. [DOI] [PubMed] [Google Scholar]
- 32.Muriaux, D., H. De Rocquigny, B. P. Roques, and J. Paoletti. 1996. NCp7 activates HIV-1 LAI RNA dimerization by converting a transient loop-loop complex into a stable dimer. J. Biol. Chem. 271:33686-33692. [DOI] [PubMed] [Google Scholar]
- 33.Muriaux, D., P. Fosse, and J. Paoletti. 1996. A kissing complex together with a stable dimer is involved in the HIV-1 LAI RNA dimerization process in vitro. Biochemistry 35:5075-5082. [DOI] [PubMed] [Google Scholar]
- 34.Negroni, M., and H. Buc. 2000. Copy-choice recombination by reverse transcriptases: reshuffling of genetic markers mediated by RNA chaperones. Proc. Natl. Acad. Sci. USA 97:6385-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Negroni, M., and H. Buc. 1999. Recombination during reverse transcription: an evaluation of the role of the nucleocapsid protein. J. Mol. Biol. 286:15-31. [DOI] [PubMed] [Google Scholar]
- 36.Paillart, J. C., L. Berthoux, M. Ottmann, J. L. Darlix, R. Marquet, B. Ehresmann, and C. Ehresmann. 1996. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J. Virol. 70:8348-8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paillart, J. C., R. Marquet, E. Skripkin, B. Ehresmann, and C. Ehresmann. 1994. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J. Biol. Chem. 269:27486-27493. [PubMed] [Google Scholar]
- 38.Paillart, J. C., R. Marquet, E. Skripkin, C. Ehresmann, and B. Ehresmann. 1996. Dimerization of retroviral genomic RNAs: structural and functional implications. Biochimie 78:639-653. [DOI] [PubMed] [Google Scholar]
- 39.Paillart, J. C., E. Skripkin, B. Ehresmann, C. Ehresmann, and R. Marquet. 1996. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc. Natl. Acad. Sci. USA 93:5572-5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paillart, J. C., E. Westhof, C. Ehresmann, B. Ehresmann, and R. Marquet. 1997. Non-canonical interactions in a kissing loop complex: the dimerization initiation site of HIV-1 genomic RNA. J. Mol. Biol. 270:36-49. [DOI] [PubMed] [Google Scholar]
- 41.Pfeiffer, J. K., and A. Telesnitsky. 2001. Effects of limiting homology at the site of intermolecular recombinogenic template switching during Moloney murine leukemia virus replication. J. Virol. 75:11263-11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robertson, D. L., J. P. Anderson, J. A. Bradac, J. K. Carr, B. Foley, R. K. Funkhouser, F. Gao, B. H. Hahn, M. L. Kalish, C. Kuiken, G. H. Learn, T. Leitner, F. McCutchan, S. Osmanov, M. Peeters, D. Pieniazek, M. Salminen, P. M. Sharp, S. Wolinsky, and B. Korber. 2000. HIV-1 nomenclature proposal. Science 288:55-56. [DOI] [PubMed] [Google Scholar]
- 43.Robertson, D. L., P. M. Sharp, F. E. McCutchan, and B. H. Hahn. 1995. Recombination in HIV-1. Nature 374:124-126. [DOI] [PubMed] [Google Scholar]
- 44.Skripkin, E., J. C. Paillart, R. Marquet, B. Ehresmann, and C. Ehresmann. 1994. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc. Natl. Acad. Sci. USA 91:4945-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.St. Louis, D. C., D. Gotte, E. Sanders-Buell, D. W. Ritchey, M. O. Salminen, J. K. Carr, and F. E. McCutchan. 1998. Infectious molecular clones with the nonhomologous dimer initiation sequences found in different subtypes of human immunodeficiency virus type 1 can recombine and initiate a spreading infection in vitro. J. Virol. 72:3991-3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suo, Z., and K. A. Johnson. 1997. RNA secondary structure switching during DNA synthesis catalyzed by HIV- 1 reverse transcriptase. Biochemistry 36:14778-14785. [DOI] [PubMed] [Google Scholar]
- 47.Traut, T. W. 1994. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 140:1-22. [DOI] [PubMed] [Google Scholar]
- 48.van Lint, C., C. A. Amella, S. Emiliani, M. John, T. Jie, and E. Verdin. 1997. Transcription factor binding sites downstream of the human immunodeficiency virus type 1 transcription start site are important for virus infectivity. J. Virol. 71:6113-6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu, W., B. M. Blumberg, P. J. Fay, and R. A. Bambara. 1995. Strand transfer mediated by human immunodeficiency virus reverse transcriptase in vitro is promoted by pausing and results in misincorporation. J. Biol. Chem. 270:325-332. [DOI] [PubMed] [Google Scholar]
- 50.Xu, H., and J. D. Boeke. 1987. High-frequency deletion between homologous sequences during retrotransposition of Ty elements in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 84:8553-8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.You, J. C., and C. S. McHenry. 1994. Human immunodeficiency virus nucleocapsid protein accelerates strand transfer of the terminally redundant sequences involved in reverse transcription. J. Biol. Chem. 269:31491-31495. [PubMed] [Google Scholar]