Abstract
The synthesis of plus and minus RNA strands of several RNA viruses requires as a first step the interaction of some viral regulatory sequences with cellular and viral proteins. The dengue 4 virus genome, a single-stranded, positive-polarity RNA, is flanked by two untranslated regions (UTR) located in the 5′ and 3′ ends. The 3′UTR in the minus-strand RNA [3′UTR (−)] has been thought to function as a promoter for the synthesis of plus-strand RNA. To study the initial interaction between this 3′UTR and cellular and viral proteins, mobility shift assays were performed, and four ribonucleoprotein complexes (I through IV) were formed when uninfected and infected U937 cells (human monocyte cell line) interacted with the 3′UTR (−) of dengue 4 virus. Cross-linking assays with RNAs containing the complete 3′UTR (−) (nucleotides [nt] 101 to 1) or a partial sequence from nt 101 to 45 and nt 44 to 1 resulted in specific binding of some cellular proteins. Supermobility shift and immunoprecipitation assays demonstrated that the La protein forms part of these complexes. To determine the region in the 3′ UTR that interacted with the La protein, two deletion mutants were generated. The mutant (del-96), with a deletion of nt 96 to 101, was unable to interact with the La protein, suggesting that La interacted with the 5′ portion of the 3′UTR (−). Complex I, which was the main ribonucleoprotein complex formed with the 3′UTR (−) and which had the fastest electrophoretic migration, contained proteins such as calreticulin and protein disulfide isomerase, which constitute important components of the endoplasmic reticulum.
Dengue virus (DEN), a mosquito-borne member of the Flaviviridae family, contains a single-stranded, positive-polarity RNA as a genome. The genomic RNA has a long open reading frame that encodes for a polyprotein that is processed during and after translation (reviewed in reference 8). This unique open reading frame is flanked by two untranslated regions located at the 5′ and 3′ ends (5′UTR and 3′UTR). In the 5′ end, viral RNA contains a type I cap structure, and in the 3′ end, it lacks the poly(A) track. The 5′UTR and 3′UTR of dengue 4 virus (DEN4) are 101 and 384 nucleotides (nt) in length, respectively. Both regions have been predicted to form stable stem-loop structures that are highly conserved among flaviviruses (5, 7, 26, 37). The presence of conserved secondary structures within both UTRs suggests that they might contain cis-acting elements involved in translation and replication (17, 33). Positive-polarity RNA viruses use the positive-strand genomic 3′UTR [3′UTR (+)] as a promoter for negative-strand RNA synthesis (−). In a similar manner, the 3′UTR present in the minus-polarity RNA could, like the 3′UTR (+), contain promoter sequences for the transcription of positive-polarity RNAs. Besides, it has been revealed that DEN 5′UTR (+) and 3′UTR (+) contain several conserved elements which are involved in viral viability and replication (7, 25, 45). The presence of both regions is required for in vitro replication of DEN, supporting the role of the cis-acting elements in viral growth. Some mutations and deletions within the 5′UTR (+) of DEN4 are lethal to the host or could result in reduced viral replication efficiency (7). Moreover, deletions involving some nucleotides of the conserved sequence CS1 or in the short and long stem-loop structure (3′-SL) present at the 3′UTR (+) generate nonviable viruses, suggesting that these regions may be essential in viral replication (46). All these results support the importance of the sequences present in both UTRs in the replication of DEN.
The replication process in several RNA viruses requires a secondary or tertiary structure located within (+) and (-) 3′ and 5′UTRs and also cellular and viral proteins. The viral nonstructural proteins NS5 and NS3 have been proposed as components of the RNA replicase for several flaviviruses and have been found to be associated with the 3′UTR (+) of Japanese encephalitis virus (JE) (9). For instance, the recombinant NS3 protein from DEN1 could form a ribonucleoprotein complex with the 3′-SL structure within the 3′UTR (+). Since relatively pure preparation of RNA-dependent RNA replicase of DEN and other viruses can efficiently replicate natural or synthetic RNAs in vitro with very little template specificity (43), it has been suggested that cellular factors are necessary for template-specific RNA-dependent RNA synthesis (22).
Some cellular proteins which interact with different positive and negative viral UTRs have been extensively studied and belong to different groups, such as heterogeneous nuclear factors, polymerase III transcript binding proteins, translation factors, and cytoskeletal, chaperone, and miscellaneous proteins (reviewed in reference 22), while other proteins have not been identified yet. Of particular interest is the La autoantigen, which has been found to be associated with the 3′UTR (−) of Sindbis virus (32) and parainfluenza virus (10) in conjunction with glyceraldehyde-3-phosphate dehydrogenase. Some uncharacterized cellular proteins with molecular masses of 42 and 44 kDa, 36 and 38 kDa, and 108, 60, 50, and 42 kDa can stably bind to the 3′UTR (−) of Sindbis virus (31), poliovirus (35), and West Nile virus (38), respectively.
Although replication complexes formed within the 3′UTR (+) and (−) may permit RNA synthesis of negative and positive polarity, respectively, they can also be constituted by different molecules. For example, three cytoplasmic proteins have been found associated with the 3′-SL structure of the rubella virus genome RNA, but only one of them is bound with to the 3′UTR (−) in association with two different additional proteins (27). Furthermore, different cellular proteins bind to the 3′ end of the RNA (−) and to the complementary 5′ end of the RNA genome (+) of mouse hepatitis virus (14). It is possible that the number and type of proteins involved in the interaction with the positive- or negative-strand RNA influence the replication efficiency and polarity of the viral RNA synthesized. Therefore, cellular and viral proteins are not simply bystanders but are active participants in the process of RNA-dependent RNA synthesis. The identification and characterization of viral and cellular proteins involved in DEN replication will help us to understand the pathogenesis, virulence, and tropism of DEN.
In a previous work we reported the binding of some cellular proteins such as La, polypyrimidine tract-binding protein (PTB), and EF-1α bound to the 3′UTR of the DEN genome, suggesting that some of these cellular proteins could form part of the replication complex (12).
The 3′UTR (−) from DEN negative strand was able to form four stable complexes (CI through CIV) with cellular proteins from uninfected and infected S10 extracts from U937 cells. One of the proteins that bound to the region is the La protein, as was demonstrated by supermobility shift and immunoprecipitation assays. Two deletion mutants were generated to determine the region in the 3′UTR (−) that interacts with the La protein. The mutant, called del-96, with a deletion between nucleotides (nt) 96 to 101 within the 3′UTR (−), was unable to interact with the La protein, suggesting that this protein bound efficiently to the 5′ portion of the 3′UTR (−). The CI complex formed with the 3′UTR (−) contains proteins such as calreticulin and protein disulfide isomerase (PDI).
MATERIALS AND METHODS
Cells and virus.
U937 cell (a human monocytic cell line derived from a patient with generalized histiocytic lymphoma) was used in this study. The cells were grown at 37°C in RPMI medium supplemented with 10% fetal calf serum, antibiotics (penicillin and streptomycin), fungizone (Gibco), 5.95 μg of HEPES/ml, and 600 μg of glutamine (pH 7)/ml. Differentiation into macrophage-like cells was induced using 120 mM phorbol myristate acetate (Sigma) for 2 days (30). U937 cells were infected with DEN4 virus strain H-241, generously provided by Goro Kuno (Centers for Disease Control, San Juan, Puerto Rico). Viruses were propagated in suckling mice (16) and in C6/36 cells.
Preparation of S10 extracts.
U937 cells, uninfected and infected (48 h) with DEN4 virus, were scraped in phosphate-buffered saline (PBS) and centrifuged at 1,000 rpm in a Sorvall Tc6 centrifuge. Cells were resuspended in 5 volumes of buffer A (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl) and centrifuged at 3,000 rpm at 4°C. Finally, cells were resuspended in 1 volume of buffer A and proteases inhibitor cocktail (Roche) and broken with 28 strokes in a Dounce homogenizer. The homogenate was centrifuged at 10,000 rpm for 30 min in a Sorvall GSA rotor, and the supernatant (S10 extract) was divided into aliquots and stored at −70°C. The amount of protein was determined with the Bradford assay (Pierce).
Construction of DNA templates by PCR.
Primers for PCR were synthesized by Invitrogen. The following primers were used with the full-length cDNA from DEN4 virus (kindly provided by Chin Yun Lai) (21) as the template: primer A, 5′ AGTTGTTAGTCTGTGTGGACCG 3′; primer B, 5′ TAATACGACTCACTATAGGGGTTTTCCAGAGATCTG 3′; primer C, 5′ AGCTTGCTTAACACAGT 3′; primer D, 5′ TAATACGACTCACTATAGGGGTCCGATTTGGAACTG 3′; primer E, 5′TAATACGACTCACTATAGGGGTTTTCCAGAGATCTGCTCTCTATTCAAACAAACACTGTGTTAAGC 3′; primer F, 5′ TAATACGACTCACTATAGGGGAGAGATCTGCTCTCT 3′. Underlined nucleotides represent the bacteriophage T7 promoter; three G residues were included between the T7 promoter and the DEN-specific sequence to increase the efficiency of transcription by T7 RNA polymerase, as previously described (36).
Deoxyoligonucleotides (primers) A and B were used to synthesize the cDNA sequence containing the complete 3′UTR (−) from DEN4 virus (from nt 101 to 1) by PCR. Primers B and C were used to synthesize the segment of the 3′UTR (−) from nt 45 to 101, and primers A and D were used for the segment from nt 1 to 44. Primers A and E were used to generate a sequence with a deletion from nt 62 to 68 within the 3′UTR (−), while a sequence with a deletion from nt 96 to 101 within the 3′UTR (−) was generated using primers A and F. Both E and F primers were designed to include the deletion from nt 62 to 68 and nt 96 to 101, respectively. The numbering system employed corresponds to the 5′UTR of the positive-strand RNA.
PCR was performed with a Perkin-Elmer Cetus DNA thermal cycler 480 (40 cycles at 94°C for 30 s, 55°C for 1 min, and 75°C for 30 s) in the presence of Taq DNA polymerase (Roche). Amplification products were purified by electrophoresis in a 3% agarose gel, using the QIAEX II agarose gel extraction protocol (QIAGEN), and used for in vitro transcription.
Transcription of PCR products.
PCR products were used to synthesize 32P-labeled RNA transcripts by in vitro transcription in the presence of [α-32P]UTP (NEN) with T7 RNA polymerase following the conditions described by the manufacturer (New England Biolabs). Transcription reactions were treated with RNase-free DNase (1 U) (Roche) at 37°C for 20 min and later precipitated with ethanol-sodium acetate (3 M; pH 5.8). The purified RNAs were resuspended into diethyl pyrocarbonate-treated water (Sigma).
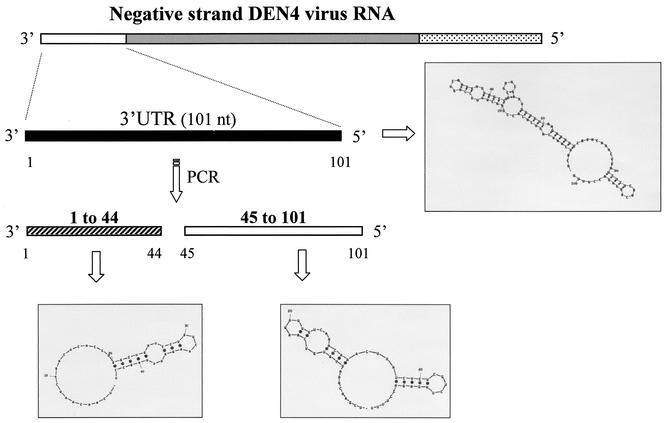
The complete 3′UTR (−) contains 101 nt, the segment of the 3′UTR (−) from nt 45 to 101 contains 57 nt, the segment from nt 1 to 44 contains 44 nt, and the RNA probes containing the deletions (del-62 and del-96) had 95 nt each (Fig. 1).
FIG. 1.
The complete 3′UTR (−) of DEN4 was synthesized, and two shorter RNA probes were generated. The secondary structures for the region between nt 101 and 1 of DEN4 3′UTR (−) RNA and the shorter RNA probes, nt 101 to 44 and nt 44 to 1, were predicted using the mfold2 program (http://mfold2.wustl.edu). The numbers correspond to the 5′UTR of the positive-strand RNA.
Computer prediction of RNA secondary structure.
The secondary structures for the complete, deleted 3′UTR (−) and shorter RNA probes were predicted by mfold2 software (47) (http://mfold2.wustl.edu).
Mobility shift electrophoresis assay.
Three micrograms of S10 extract from U937 cells or 25 ng of recombinant La protein was incubated with 4 μg of yeast tRNA (Sigma) in a buffer containing 10 mM HEPES (pH 7.4), 0.02 mM dithiothreitol, 8 mM MgCl2, 0.1 mM EDTA, 4 mM spermidine, 0.15 mM ATP, 1 mM GTP, and 10% glycerol in a final volume of 10 μl at 4°C for 15 min. Labeled RNA (106 cpm) was added, and the mixture was incubated for 15 min at 4°C for complex formation. Binding reactions were incubated with RNase T1 (20 U) for 20 min at room temperature and loaded gently on a 6% polyacrylamide gel (acrylamide/bisacrylamide, 80:1) prepared in 0.5× Tris-borate-EDTA buffer and electrophoresed at 20 mA for 3 h. Gels were dried and autoradiographed.
For competition experiments, unlabeled RNAs were included in the preincubation reaction. For supermobility shift assay, antibody against La (kindly provided by N. Sonenberg, McGill University, Montreal, Quebec, Canada) was preincubated with the reaction mixture for 15 min before the addition of labeled RNA.
Electroelution of CI complex.
The proteins present in the CI complex from the mobility shift assays were obtained as follows: the mobility shift assay was performed using 10 μg of S10 extract from infected U937 cells, and complexes were separated in the native gel. The CI complex was cut from the gel and electroeluted in a buffer containing 20 mM Tris, 200 mM glycine, and 0.1% sodium dodecyl sulfate (SDS). The electroeluted proteins were concentrated using Microcon ultrafiltration columns (Millipore) and then separated through two-dimensional electrophoresis.
Two-dimensional gel electrophoresis.
The two-dimensional gel electrophoresis was performed essentially using the method of O'Farrel (29) and the conditions suggested by the manufacturer (Miniprotean III; Bio-Rad, Hercules, Calif.). Briefly, the first dimension was run for 3.5 h at 750 V in a 4% polyacrylamide-7 M urea gel tube containing a pH gradient in the range of 4 to 7. The pH gradient was established using isoelectric point (IP) markers under the same conditions. The second dimension was performed using an SDS-10% polyacrylamide gel electrophoresis (PAGE) analysis.
Protein sequencing.
Two proteins with apparent molecular masses of 60 and 62 kDa obtained from CI complex were cut from the two-dimensional gel. Both proteins were reduced and alkylated with iodoacetamide, digested with trypsin, and analyzed by matrix-assisted laser desorption ionization-mass spectrometry using the Protein Chemistry Core Facility in the College of Physicians and Surgeons of Columbia University, New York, N.Y.
Western blot assay.
Samples were fractionated through two-dimensional electrophoresis and transferred to nitrocellulose membranes by using a semidry Transblot apparatus at 16 V for 30 min with a transfer buffer (0.025 M Tris-HCl [pH 9.5], 0.019 M glycine, 20% [vol/vol] methanol). Membranes were blocked for 2 h at room temperature in skimmed milk and then washed three times with washing buffer (PBS-1% Triton [wt/vol]). The first antibody, a goat polyclonal anticalreticulin (Santa Cruz Biotechnology) diluted 1:400 in washing buffer, was incubated with the membrane overnight at 4°C. The second antibody, an anti-goat immunoglobulin G conjugated to alkaline phosphatase (Sigma), diluted 1:1,000 in PBS, was incubated with the polyclonal antibody-bound membrane at room temperature for 1 h. Color was developed with 5-bromo-4-chloro-3-indolylphosphate toluidinium and nitroblue tetrazolium chloride in a buffer containing 100 mM Tris (pH 9.5), 100 mM NaCl, and 5 mM MgCl2.
UV-induced cross-linking.
Fifty micrograms of S10 extract or 100 ng of recombinant La protein was incubated with 10 mM HEPES, 3 mM MgCl2, 40 mM KCl, and 5% glycerol for 15 min at 4°C. Subsequently, 2 × 106 cpm of each of the 32P-labeled RNAs was added to the mixture and incubated for another 15 min at 4°C. The reaction mixture was then irradiated with 254 nm UV light (UV Ultra-Lum lamp) at 3 cm for 15 min on ice. Samples were analyzed by SDS-10% PAGE. Gels were fixed, dried, and autoradiographed.
Immunoprecipitation with anti-La antibodies.
After the UV cross-linking, the samples were incubated with 10 μl of protein G-agarose beads (Gibco) for 2 h at 4°C and centrifuged at 14,000 × g for 5 min. Supernatants were incubated with monoclonal anti-La antibody in the presence of protein G-agarose beads saturated with bovine serum albumin overnight at 4°C. Unbound materials were removed by five washes with NETS buffer (50 mM Tris-HCl [pH 7.4], 5 mM EDTA, 1 mM dithiothreitol, 100 mM NaCl, 0.05% Nonidet P-40). Bound proteins were analyzed by SDS-10% PAGE followed by autoradiography. Parallel analysis was made, and reactions were carried out with an unrelated antiactin monoclonal antibody (kindly provided by Manuel Hernández, Centro de Investigación y de Estudios Avanzados del IPN, México City, México) as a control.
RESULTS
U937 cellular proteins interact with DEN4 virus 3′UTR (−).
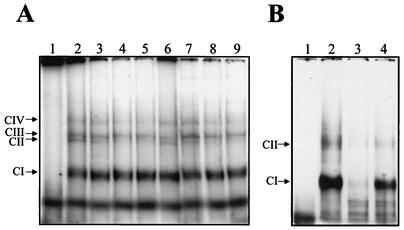
To determine if the 3′UTR (−) of DEN4 bound cellular and/or viral proteins present in U937 cells, mobility shift assays were performed using a 32P-labeled RNA transcript from nt 101 to 1 of the 3′UTR of negative polarity (−) (numbers correspond to the sequence of the 5′UTR of the positive-strand RNA) and S10 extracts from uninfected and DEN4-infected U937 cells. Labeled DEN4 3′UTR (−) formed four RNA-protein complexes with the S10 extract from uninfected (Fig. 2A, lane 2) and infected (Fig. 2A, lane 6) U937 cells. The complex with the fastest migration, which was the most prominent, was referred to as CI, and the additional three minor complexes were designated CII, CIII, and CIV.
FIG. 2.
Mobility shift assay of the 3′UTR (−) RNA of DEN4. (A) 32P-labeled RNA consisting of nt 101 to 1 was incubated without (lane 1) or with 3 μg of S10 extract from uninfected (lanes 2 to 5) or infected (lanes 6 to 9) U937 cells in the presence of 300 mM (lanes 3 and 7), 600 mM (lanes 4 and 8), or 900 mM (lanes 5 and 9) of KCl. (B) Labeled RNA consisting of nt 101 to 1 was incubated without (lane 1) or with 3 μg of S10 extract from infected U937 cells (lanes 2 to 4) in the absence (lane 2) or in presence of a 25-fold molar excess of unlabeled 3′ UTR (−) RNA (lane 3) or a 25-fold molar excess of unlabeled unrelated RNA (nt 111 to 191 of the Norwalk virus genome) (lane 4). The complexes were assayed by electrophoresis through a 6% native polyacrylamide gel. Arrows indicate the migration of the complexes.
In order to determine the stability of the RNA-protein complexes, complex formation was performed in the presence of increasing KCl concentrations. RNA-protein complexes CI, CIII, and CIV were formed with the S10 extracts from both uninfected and infected U937 cells and maintained their migration in the gel up to a 900 mM concentration of KCl (Fig. 2A, lanes 3 to 5 and lanes 7 to 9, respectively), which represents more than seven times the salt concentration under physiological conditions (approximately 120 mM NaCl). This result suggests that the complexes designated CI, CIII, and CIV that formed with both infected and uninfected S10 extracts are stable. However, CIII and CIV complexes were not consistently discernible in subsequent experiments, suggesting that they could represent unstable RNA-protein complexes.
The specificity of the interactions between the DEN4 3′UTR (−) and cellular proteins from infected U937 extracts was determined in competition experiments using a 25-fold-molar excess of homologous and heterologous unlabeled RNA fragments as competitors. A 25-fold molar excess of the complete unlabeled homologous 3′UTR (−) (nt 101 to 1) strongly reduced the formation of the labeled ribonucleoprotein complexes (Fig. 2B, lane 3); however, a 25-fold molar excess of a heterologous RNA (nt 111 to 191 of the Norwalk virus genome) could not compete efficiently (Fig. 2B, lane 4). These data strongly suggest that the proteins present in the S10 extracts of infected U937 cells bound specifically to the 3′UTR (−) RNA of DEN4.
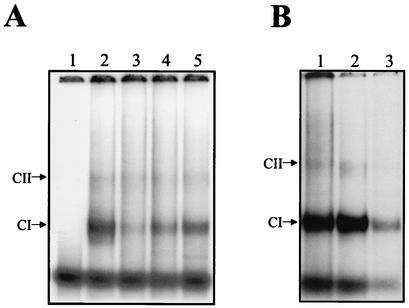
Competition experiments were performed using the 5′UTR (+) (nt 1 to 101) [the complementary sequence of the 3′UTR (−)] and the genomic 3′UTR (+) (nt 10262 to 10644) as competitors. A 25-fold molar excess of the unlabeled 5′UTR (+) was able to induce a slight competition (Fig. 3A, lane 5), while a 25-fold molar excess of the 3′UTR (+) partially competed for the formation of CI and CII (Fig. 3A, lane 4). The homologous RNA added at a similar level of molar excess competed for complex formation (Fig. 3A, lane 3). These results suggest that some of the proteins which bound to the 3′UTR (−) also interacted with the genomic 3′UTR (+) and 5′UTR (+).
FIG. 3.
Mobility shift assay of the complete 3′UTR (−) and shorter RNA probes of DEN4. (A) Labeled RNA consisting of nt 101 to 1 was incubated without (lane 1) or with 3 μg of S10 extract from infected U937 cells (lanes 2 to 5), in the absence (lane 2) or in the presence of a 25-fold molar excess of unlabeled 3′UTR (−) RNA (lane 3) or a 25-fold molar excess of the 3′UTR of the DEN4 genome (lane 4) or a 25-fold excess of the complete 5′UTR of the DEN4 genome (lane 5). (B) Labeled RNAs consisting of nt 101 to 1 (lane 1), nt 101 to 45 (lane 2), and nt 44 to 1 (lane 3) were incubated with 3 μg of S10 extract from infected U937 cells. Arrows indicate the migration of the complexes.
To determine the major protein binding site within the 3′UTR (−), mobility shift assays were performed using two RNA probes, one for nt 101 to 45 and the other for nt 44 to 1 (Fig. 1). Labeled RNAs from nt 101 to 1, 101 to 45, and 44 to 1 were incubated with S10 extracts from DEN4-infected U937 cells. All the RNA probes were able to form the CI complex (Fig. 3B, lanes 1, 2, and 3, respectively), while the CII complex was observed only with the complete 3′UTR (−) RNA and with the transcript from nt 45 to 101 (Fig. 3B, lanes 1 and 2, respectively). The CI complex formed with the RNA probe from nt 44 to 1 showed a smaller amount of radioactivity than the CI complex formed with the other two probes. This result indicates that the two shorter RNAs (nt 45 to 101 and 44 to 1) contain sequences involved in RNA-protein complex formation. Since all interactions were treated with RNases, free RNA probes had the same migration as expected.
Cellular proteins that bind the DEN4 virus 3′UTR (−).
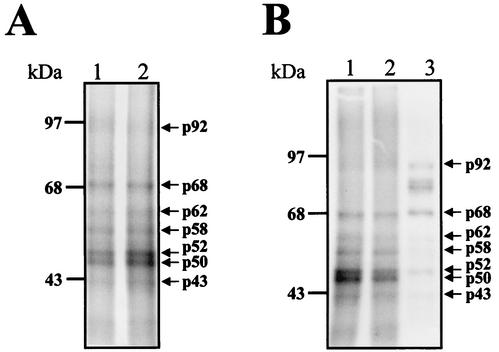
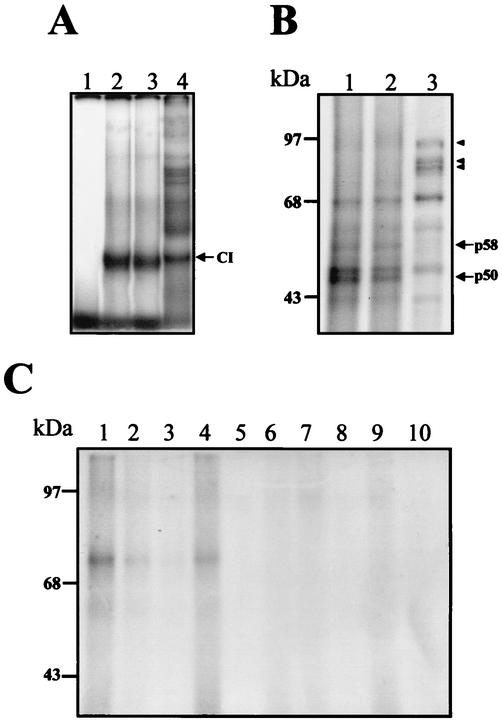
To identify the proteins that bind to the 3′UTR (−) of DEN4 virus, labeled RNA consisting of nt 101 to 1 was incubated with S10 extracts from uninfected and infected U937 cells, followed by UV-induced cross-linking. The 3′UTR (−) cross-linked with seven major proteins with molecular masses of 92, 68, 62, 58, 52, 50, and 43 kDa in both uninfected and infected S10 extracts (Fig. 4A, lanes 1 and 2, respectively).
FIG. 4.
UV-induced cross-linking assay of the complete 3′UTR (−) and shorter RNA probes of DEN4 virus. (A) Labeled 3′UTR (−) RNA consisting of nt 101 to 1 was UV cross-linked with protein present in uninfected (lane 1) or infected (lane 2) U937 cell extracts. (B) Labeled RNA probes from the 3′UTR (−) RNA consisting of nt 101 to 1 (lane 1), nt 101 to 45 (lane 2), or nt 44 to 1 (lane 3) was incubated with 50 μg of infected S10 extract, and the UV-induced cross-linking assay was performed. Proteins were separated by SDS-10% PAGE and detected by autoradiography. Molecular masses in kilodaltons are indicated on the left. Arrows indicate cross-linked proteins, with the molecular masses given in kilodaltons.
To determine the proteins that bind to the different segments of the 3′UTR (−) of DEN4, the RNA probes containing nt 101 to 45 and 44 to 1 were used to perform a UV cross-linking assay. The RNA probe which includes nt 101 to 45 cross-linked with proteins similar to those with which the complete region cross-linked (Fig. 4B, lane 2), but with lower intensity than the complete RNA probe (Fig. 4B, lane 1), suggesting that it contains most of the binding motifs. However, the RNA probe consisting of nt 44 to 1 cross-linked with the same proteins as did the complete 3′UTR (−) with the exception of the 50-, 58-, and 62-kDa proteins, which bound mainly to the complete 3′UTR (−) (Fig. 4B, lane 3), suggesting that the three protein molecules require sequences present in the probe (consisting of nt 101 to 45) in order to bind to the 3′UTR (−). Furthermore, the RNA probe from nt 44 to 1 cross-linked with two additional proteins with molecular masses of approximately 68 and 92 kDa, suggesting that the secondary structure present in this RNA sequence could permit the interaction with some cellular proteins.
La protein binds to the 3′UTR (−).
Since a cross-linked protein has a molecular mass similar to that of La (50 kDa), which could bind to other UTRs of different RNA viruses, we analyzed its presence in the complexes. To determine if La was one of the proteins present in the RNA-protein complexes formed with the 3′UTR (−), a supermobility shift assay was performed using a polyclonal anti-La antibody. The addition of the anti-La antibody to the binding reaction induced the supershifting of the complexes (Fig. 5A, lane 2), while antiactin polyclonal antibody did not alter complex formation (Fig. 5A, lane 3), suggesting that the La protein binds to the 3′UTR (−). It is interesting that the del-96 probe cross-linked with two additional proteins of molecular masses between 58 and 92 kDa as well as in the cross-linking assay using the 44 to 1 probe (Fig. 4B, lane 3).
FIG. 5.
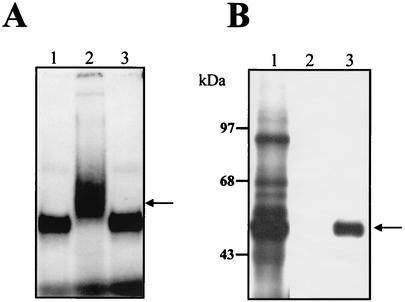
Supermobility shift and immunoprecipitation assays of the La protein. (A) Three micrograms of S10 extract from infected U937 cells was incubated without (lane 1) or with an anti-La (lane 2) or antiactin (lane 3) monoclonal antibodies before the incubation with labeled RNA consisting of nt 101 to 1. The complex was assayed by electrophoresis through a 6% native polyacrylamide gel. The arrow indicates the supershifted complex migration. (B) Immunoprecipitation of the cross-linked La protein. UV cross-linked proteins (lane 1) were immunoprecipitated with an anti-La (lane 3) or antiactin (lane 2) monoclonal antibody. Molecular masses in kilodaltons are indicated on the left. The arrow indicates the immunoprecipitated La protein.
To corroborate the presence of the La protein in the ribonucleoprotein complexes formed with the 3′UTR (−), an immunoprecipitation assay was performed. After UV-induced cross-linking (Fig. 5B, lane 1), the cross-linked proteins were immunoprecipitated with anti-La or antiactin monoclonal antibodies. The anti-La antibody was able to immunoprecipitate a 50-kDa radiolabeled protein (Fig. 5B, lane 3). Expectedly, the antiactin antibody does not immunoprecipitate any radiolabeled protein as expected (Fig. 5B, lane 1). These results demonstrate that the La protein interacts directly with the complete 3′UTR (−) of DEN4.
Localization of the La binding site at the 3′UTR (−).
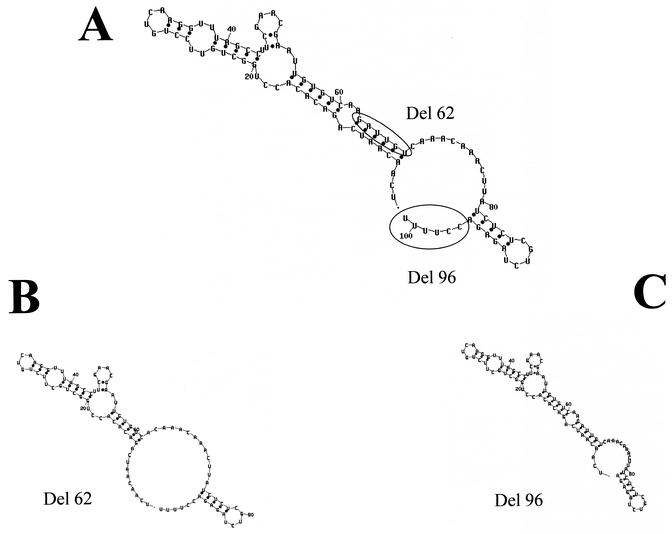
It has been reported that the La protein binds mainly to the UUUOH terminal motif of the nascent transcripts of RNA polymerase III (41). The La protein binds to tRNA precursors, other nuclear RNAs and the cytoplasmic YRNAs (41), the internal ribosome entry site, and other elements in viral and cellular RNAs (reviewed in reference 23). Since the sequence present in the 5′ region of our 3′UTR RNA contains a U-rich track, we performed a deletion of 6 nt, from nt 101 to 96 (CCUUUU), later referred to as del-96 (Fig. 6C). In the del-96, two C's were eliminated. The YRNAs U tail is also preceded by one or two C'sR. This sequence is located in a single-strand region beside the initiation codon (Fig. 6A). As a control, a second mutant, called del-62, was also created (Fig. 6B). This mutant contains a 6-nt deletion (from nt 62 to 68), thereby eliminating a stem present in the secondary structure predicted from the complete 3′UTR (−) (Fig. 6A). The mutants and wild-type 3′UTR (−) RNA probes were first used in a mobility shift assay to determine if the interactions were modified in the presence of the different deletions. The del-62 mutant was able to form RNA-protein complexes similar to those formed by the complete 3′UTR (−) (Fig. 7A, lanes 3 and 2, respectively), while the deletion present in the del-96 mutant caused an important reduction in CI formation (Fig. 7A, lane 4). Furthermore, the del-96 mutant was able to form additional RNA-protein complexes with lower migration in the native gel. This result clearly indicates that the deletion in nt 96 to 101 modified the interaction with cellular proteins present in the U937 extract. To determine which of the proteins modified the interaction with the del-96 mutant, a UV cross-linking assay was performed. The binding ability of most of the proteins was conserved when the complete region and the del-62 mutant were assayed (Fig. 7B, lanes 1 and 2, respectively), while the 50- and 58-kDa proteins were not able to cross-link with the del-96 probe (Fig. 7B, lane 3). The del-96 probe cross-linked with two other proteins with a molecular mass of approximately 80 kDa. The 92-kDa protein which cross-linked with the del-96 mutant showed a larger amount of radioactivity than the one that cross-linked with the complete and del-62 regions. The inability of the 50-kDa protein to cross-link with the del-96 mutant suggests that La could be one of the proteins that interacts with the region located within nt 96 to 101.
FIG. 6.
The complete 3′UTR (−) of DEN4 was used as a template to produce two deletions, del-62 and del-96 (A). The secondary structures for the two deleted regions del-62 (B) and del-96 (C) were predicted using the mfold2 program (http://mfold2.wustl.edu).
FIG. 7.
Mobility shift and UV-induced cross-linking assay of the 3′UTR (−) and deleted RNA probes of DEN4. (A) Labeled RNA consisting of nt 101 to 1 (lanes 1 and 2), del-62 (lane 3), or del-96 (lane 4) was incubated without (lane 1) or with (lanes 2 to 4) 3 μg of S10 extract from infected U937 cells. Complex was assayed by electrophoresis through a 6% native polyacrylamide gel. (B) Labeled 3′UTR (−) RNA consisting of nt 101 to 1 (lane 1), del-62 (lane 2), or del-96 (lane 3) was UV cross-linked with protein present in 50 μg of infected U937 cell extracts. Proteins were separated by SDS-10% PAGE and detected by autoradiography. (C) Labeled 3′UTR (−) RNA consisting of nt 101 to 1 (lanes 1 and 6), del-62 (lanes 4 and 9), del-96 (lanes 5 and 10), or RNA consisting of nt 101 to 45 (lanes 2 and 7) or nt 44 to 1 (lanes 3 and 8) was UV cross-linked with 100 ng of recombinant GST-La protein (lanes 1 to 5) or with GST recombinant protein (lanes 6 to 10). Proteins were separated by SDS-10% PAGE and detected by autoradiography. Molecular masses in kilodaltons are indicated on the left.
To corroborate that the deletion mutant del-96 has lost the La protein binding site, a UV-induced cross-linking assay was performed using a recombinant glutathione S-transferase (GST)-La protein (12). The recombinant GST-La, with a molecular mass of approximately 76 kDa, was able to cross-link with the complete 3′UTR (−), the RNA probe from nt 101 to 45, and the del-62 mutant (Fig. 7C, lane 1, 2, and 4, respectively) but was not with the RNA probe from nt 44 to 1 and the del-96 mutant (Fig. 7C, lanes 3 and 5, respectively). The GST protein, included as a control, was unable to cross-link with either of the RNA probes (Fig. 7C, lanes 6 to 10). This result demonstrate that the sequence from nt 96 to 101 present in the 3′UTR (−) is necessary for the interaction with the La protein from U937-infected cells as well as the recombinant human La protein.
Calreticulin binds to the 3′UTR (−).
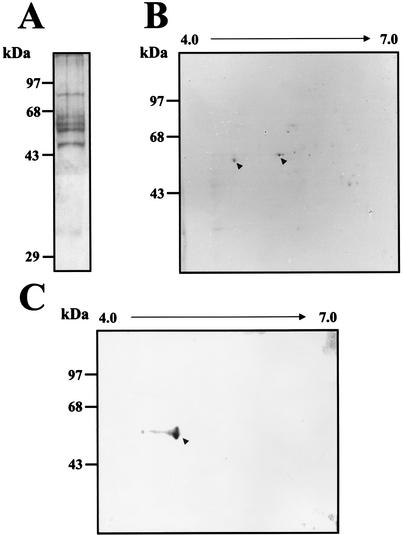
Using different antibodies, we were unable to demonstrate the presence of the other proteins in the complexes formed with the 3′UTR (−) and U937 cell extracts. Therefore, as an alternative strategy, the CI (which is the most specific and intense complex) was isolated from native gel and eluted. The proteins obtained were subjected to SDS-10% PAGE and Coomassie blue staining. Six proteins with apparent molecular masses of 88, 66, 62, 60, 50, and 35 kDa were observed when infected S10 extract was used (Fig. 8A). Some of the proteins detected by Coomassie blue staining in the CI have molecular masses similar to those of the ones detected in the UV-induced cross-linking assay. To further identify the proteins present in the CI, the proteins were separated through two-dimensional electrophoresis. Two of the proteins, with molecular masses of 60 and 62 kDa, were clearly observed (Fig. 8B) and were sequenced using the Protein Core Lab facilities at Columbia University. The procedure used with both proteins included a tryptic digestion, separation, and sequencing of the tryptic peptides. The sequence of the analysis revealed that the 60-kDa protein was calreticulin, while the 62-kDa protein was identified as the PDI. This is an important finding because calreticulin and PDI interact with each other in the endoplasmic reticulum for regulation of cellular functions.
FIG. 8.
Proteins present in the CI complex. (A) After a mobility shift assay, the CI from infected S10 extract was cut from the native gel, electroeluted, and then subjected to SDS-10% PAGE. The proteins were stained using Coomassie blue. (B) The proteins obtained from the CI were separated through two-dimensional electrophoresis. The proteins indicated with arrowheads were cut and sequenced. (C) Western blot assay of the proteins present in the CI complex using an anticalreticulin polyclonal antibody. Molecular mass markers are indicated on the left. The arrowhead indicates the calreticulin protein.
The presence of calreticulin in the ribonucleoprotein CI formed with the complete 3′UTR (−) RNA was corroborated by a Western blot assay of the electroeluted proteins present in CI using an anticalreticulin polyclonal antibody. The anticalreticulin antibody was able to detect the 60-kDa protein present in the two- dimensional electrophoresis from CI (Fig. 8C).
DISCUSSION
The mechanism followed by most of the RNA viruses to replicate their genomes is poorly understood. Nevertheless, in general it has been proposed that the first step for the replication of positive-strand RNA viruses requires the interaction between cellular and/or viral proteins (which constitute trans-acting factors) with some cis elements present in the 3′UTR, 5′UTR, and/or the coding regions (45) of the sequence to replicate. Among the members of flavivirus family, two viral proteins, NS5 and NS3, have been proposed as the viral components involved in the RNA replicase complex (1, 9). However, the interaction of the RNA replicase complex with the cis elements and their function in the replication process require the presence of additional cellular and viral proteins.
Recently it has been suggested that the regulatory sequences involved in viral replication reside in both ends of the flavivirus genome (44). The promoter sequence for the negative strands resides in the 3′ end of the positive-polarity RNA, while the promoter for the synthesis of the positive strands resides on the 3′ end of the negative-strand RNA. Although the UTRs contain the sequences needed for replication (7, 25, 34, 46), the presence of cis- and trans-acting elements is essential in regulating the functions of the replication sequences.
Some cellular and viral proteins, such as NS3, NS5, EF1-α, La, PTB, and some uncharacterized proteins with molecular masses of 84 and 105 kDa (9, 12), have been shown to bind to the 3′UTR of DEN, West Nile virus, and JE virus, while proteins of 105 kDa have been observed to bind to the 3′UTR of the negative-strand RNA from West Nile virus (38). The replication complexes involved in the negative- and positive-polarity RNA synthesis from some viruses are not the same. Apparently, some proteins can be shared for the two replication complexes, but others are specific for the replication of one specific strand (4).
To initially characterize components that could be involved in DEN replication, we attempted to first identify some cellular proteins that interact with the 3′UTR (−) of DEN4 RNA. The interactions were analyzed using mobility shift and UV-induced cross-linking assays with uninfected and infected U937 cell extracts and different segments of the 3′UTR (−) from DEN4.
The complete 3′UTR (−) from DEN4 was able to form stable and specific RNA-protein complexes with cellular factors in uninfected and infected U937 S10 extracts. However, we were unable to differentiate between the various complexes formed with the extracts, since the same proteins were observed to also cross-link with the 3′UTR (−) using uninfected and infected cell extracts. Similar results have been obtained with West Nile virus, another member of the flavivirus family, in which no differences were found between the RNA-protein complexes or the cross-linked proteins which interact with the 3′UTR (−) from uninfected or infected cell extracts (38). This observation could be explained in two different ways: the viral RNA might have interacted first with cellular factors to form ribonucleoprotein complexes which later can interact with viral proteins, probably in the presence of membranes in the form of membrane-associated proteins or the 5′UTR (44, 45). The second possibility is that viral RNA does not directly interact with viral proteins. However, the 3′UTR of JE virus interacted with different proteins in uninfected and infected BHK cell extracts, through the viral proteins NS3 and NS5 detected in infected cell extracts (9). The inability to detect viral proteins in our assays could be related to the use of S10 extracts, which contain only cytoplasmic protein and not membrane-associated proteins that are present in total cell extracts, which were used in the assay with JE virus. In addition, our experiments were performed with U937 proteins obtained at 48 h after infection at a multiplicity of infection of 0.1, while the infected cell extract from BHK cells used in the JE virus experiments was obtained 24 h after infection at a multiplicity of infection of 5. It is possible that a smaller amount of NS3 and NS5 protein was present in the S10 extract.
The cellular proteins bound specifically to the 3′UTR (−) because the interaction was competed using homologous 3′UTR sequence but not with an heterologous RNA. Furthermore, the interaction between the 3′UTR (−) and the cellular protein was partially competed with the genomic 3′UTR, suggesting that both 3′UTRs (positive and negative) share some proteins. This phenomenon has also been observed with other RNA viruses, such as West Nile virus, where the protein with a molecular weight of 105 binds to both 3′UTR (positive and negative polarity) RNAs. The presence of the 5′UTR induced only a light reduction in the interaction between the cellular proteins and the 3′UTR (−).
Seven proteins specifically bound to the complete 3′UTR (−). One of them, with a molecular mass of 50 kDa, has molecular mass similar to those of the two cellular factors that have been described interacting with other viral RNAs, the EF1-α and La proteins. Using mobility supershift, immunoprecipitation, and cross-linking assays in the presence of a monoclonal anti-La antibody, we were able to demonstrate the presence of the La protein from U937 cells in the ribonucleoprotein complexes. The La protein is a nuclear protein involved in transcription initiation and termination of RNA polymerase III (15). It has also been demonstrated to be involved in protein transport from the nucleus to the cytoplasm (24). For instance, La is relocated from the nucleus to the cytoplasm in poliovirus-infected cells (24). Although the La protein could be playing an essential role in viral translation of poliovirus (24, 42) and rhinovirus (36), etc., its role in replication has not been fully described. As we described previously, the La protein interacts with the genomic 3′UTR (12), which could make it an important protein in DEN replication. La could be playing different roles in replication, such as an RNA chaperone to maintain RNA structure or, as has been described for hepatitis C, La can protect the RNA from rapid degradation. Since DEN RNA does not contain a poly(A) tail (40), both functions could be exploited by the virus to enhance the replication. Additionally, La has an ATP-dependent helicase activity that unwinds double-strand RNA substrates (18), which is also required for viral replication.
By generating two different mutants, we were able to observe that the sequence from nt 96 to 101 is part of the La protein binding site. This sequence is present in a putative single-strand conformation and is close to the AUG codon in the complementary sequence. Cahour et al. (7) induced a lethal mutant in a deletion involving three of the six nucleotides that we eliminated in the del-96 mutant, indicating that this region is important for the viability of DEN. An additional role of the La protein could be to facilitate the interaction of viral RNA with other cellular or viral proteins in order to initiate replication of the negative strand. Further analyses, including functional studies, are necessary to demonstrate the role of this protein in DEN virus replication.
Two other proteins were found to interact with the 3′UTR (−), both of which were present in CI. Calreticulin, a ubiquitous protein which is the major intracellular calcium binding protein, is highly conserved, with more than 90% amino acid identity between rat, human, and rabbit. Calreticulin, apart from being a simple calcium storage protein, also plays an important role in modulating calcium signals. It also functions as a chaperone in the endoplasmic reticulum and modulates the expression of hormonally regulated genes. In addition, calreticulin has been found to bind to an SLSL structure present within the 3′UTR (+) of rubella virus through its aminoterminal sequence (2, 39). During rubella virus infection, calreticulin is hyperphosphorylated and its binding affinity for the 3′SL region increases temporally with the onset of the negative-strand synthesis (39). The presence of calreticulin in the ribonucleoprotein CI formed with the 3′UTR (−) RNA could suggest that this protein plays a role in replication of DEN. Nevertheless, additional experiments need to be performed to elaborate on these observations.
The PDI can interact in vivo with calreticulin. This interaction could be Zn dependent and requires the N-terminal and P domains of calreticulin (3). The PDI protein is an abundant protein in the lumen of the endoplasmic reticulum and catalyzes the in vivo folding of a variety of proteins by assisting in isomerization of intramolecular disulfide bridges (13, 28). Calreticulin and PDI have both been proposed to be multifunctional proteins capable of interaction with a variety of proteins (13, 20, 28) and have been implicated in modulating gene expression (6, 11, 19). Although the interaction of both proteins with a cis-acting element present in an RNA virus has not been described for other RNA viruses, it is possible that the complex of both calreticulin and PDI present in the lumen of the endoplasmic reticulum could bind to the 3′UTR of DEN4 through the ability of calreticulin to interact with RNA sequences. It is possible that calreticulin and PDI could be involved in the assembly of the replication complex within the 3′UTR (−) of DEN4. Further analyses directed at determining the role of both molecules in DEN replication, translation, and/or assembly are currently being done in this laboratory.
The data presented in this paper indicate that the La, calreticulin, and PDI proteins from a human monocytic cell line bind to the 3′UTR (−) of DEN4. The La protein has been reported to interact with the genomic 3′UTR of DEN4 (12). Part of the sequence involved in the interaction with the La protein is present between nt 96 to 101, which is an unstructured sequence close to the complementary sequence of the AUG codon. Calreticulin and PDI are important components of the endoplasmic reticulum, an important compartment for viral translation, replication, and encapsidation. Calreticulin and PDI are able to interact with each other in vivo and have been found regulate several functions. The three proteins may have a chaperone activity; therefore, it is possible to speculate that they could play a role in viral replication, translation, or encapsidation. Further analyses, including functional assays, are required to demonstrate the role of each of the proteins in DEN replication.
Acknowledgments
We thank Jaime Escobar and Salvador Chavarría for their technical assistance. We also acknowledge with thanks the assistance of Lorena Gutiérrez and Saheed S. Baba in their critical comments on the manuscript.
This work was supported by a grant from Consejo Nacional de Ciencia y Tecnología. Rosa Martha E. Yocupicio-Monroy received a scholarship from Consejo Nacional de Ciencia y Tecnología.
REFERENCES
- 1.Ackermann, M., and R. Padmanabhan. 2001. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 276:39926-39937. [DOI] [PubMed] [Google Scholar]
- 2.Atreya, C. D., N. K. Singh, and H. L. Nakhasi. 1995. The rubella virus RNA binding activity of human calreticulin is localized to the N-terminal domain. J. Virol. 69:3848-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baksh, S., K. Burns, C. Andrin, and M. Michalak. 1995. Interaction of calreticulin with protein disulfide isomerase. J. Biol. Chem. 270:31338-31344. [DOI] [PubMed] [Google Scholar]
- 4.Barrera, I., D. Schuppli, J. M. Sogo, and H. Weber. 1993. Different mechanisms of recognition of bacteriophage Q beta plus and minus strand RNAs by Q beta replicase. J. Mol. Biol. 232:512-521. [DOI] [PubMed] [Google Scholar]
- 5.Brinton, M., and J. Dispoto. 1988. Sequence and secondary structure analysis of the 5′-terminal region of flavivirus genome RNA. Virology 162:290-299. [DOI] [PubMed] [Google Scholar]
- 6.Burns, K., B. Duggan, E. A. Atkinson, K. S. Famulski, M. Nemer, R. C. Bleackley, and M. Michalak. 1994. Modulation of gene expression by calreticulin binding to the glucocorticoid receptor. Nature 367:476-480. [DOI] [PubMed] [Google Scholar]
- 7.Cahour, A., A. Pletnev, M. Vazeille-Falcoz, L. Rosen, and C. Lai. 1995. Growth-restricted dengue virus mutants containing deletions in the 5′ noncoding region of the RNA genome. Virology 207:68-76. [DOI] [PubMed] [Google Scholar]
- 8.Chambers, T., C. Hahn, R. Galler, and C. Rice. 1990. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44:649-688. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C., M. Kuo, L. Chien, S. Hsu, Y. Wang, and J. Lin. 1997. RNA-protein interactions: involvement of NS3, NS5, and 3′ noncoding regions of Japanese encephalitis virus genomic RNA. J. Virol. 71:3466-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De, B. P., S. Gupta, H. Zhao, J. A. Drazba, and A. K. Banerjee. 1996. Specific interaction in vitro and in vivo of glyceraldehyde 3-phosphate dehydrogenase and La protein with cis-acting RNAs of human parainfluenza virus type 3. J. Biol. Chem. 271:24728-24735. [DOI] [PubMed] [Google Scholar]
- 11.Dedhar, S., P. S. Rennie, M. Shago, C. Y. Hagesteijn, H. Yang, J. Filmus, R. G. Hawley, N. Bruchovsky, H. Cheng, and R. J. Matusik. 1994. Inhibition of nuclear hormone receptor activity by calreticulin. Nature 367:480-483. [DOI] [PubMed] [Google Scholar]
- 12.De Nova Ocampo, M., N. Villegas Sepúlveda, and R. M. del Angel. 2002. Translation elongation factor 1α, La, and PTB interact with the 3′ untranslated region of dengue 4 virus RNA. Virology 295:337-347. [DOI] [PubMed] [Google Scholar]
- 13.Freedman, R. B., T. R. Hirst, and M. F. Tuite. 1994. Protein disulphide isomerase: building bridges in protein folding. Trends Biochem. Sci. 19:331-336. [DOI] [PubMed] [Google Scholar]
- 14.Furuya, T., and M. Lai. 1993. Three different cellular proteins bind to complementary sites on the 5′-end-positive and 3′-end-negative strands of mouse hepatitis virus RNA. J. Virol. 67:7215-7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottlieb, E., and J. A. Stelitz. 1989. Function of the mammalian La protein: evidence for its action in transcription termination by RNA polymerase III. EMBO J. 8:851-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gould, E., and S. C. J. Clegg. 1991. Growth, titration and purification of alphaviruses and flaviviruses, p. 43-78. In B. W. J. Mahy (ed.), Virology: a practical approach. IRL Press, Oxford, United Kingdom.
- 17.Hahn, C. S., Y. S. Hahn, C. M. Rice, E. Lee, L. Dalgarno, E. G. Strauss, and J. H. Strauss. 1987. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 198:33-41. [DOI] [PubMed] [Google Scholar]
- 18.Hühn, P., G. J. M. Pruijn, W. J. Van Venrooij, and M. Bachmann. 1997. Characterization of the autoantigen La (SS-B) as a dsRNA unwinding enzyme. Nucleic Acids Res. 25:410-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, E., W. Henzel, and A. Deisseroth. 1992. An isoform of protein disulfide isomerase isolated from chronic myelogenous leukemia cells alters complex formation between nuclear proteins and regulatory regions of interferon-inducible genes. J. Biol. Chem. 267:14412-14417. [PubMed] [Google Scholar]
- 20.Krause, K. H., and M. Michalak. 1997. Calreticulin. Cell 88:439-443. [DOI] [PubMed] [Google Scholar]
- 21.Lai, C., B. Zhao, H. Hori, and M. Bray. 1991. Infectious RNA transcribed from stably cloned full-length cDNA of dengue type 4 virus. Proc. Natl. Acad. Sci. USA 88:5139-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai, M. 1998. Cellular factors in the transcription and replication of viral RNA genomas: a parallel to DNA-dependent RNA transcription. Virology 244:1-12. [DOI] [PubMed] [Google Scholar]
- 23.Maraia, R., and R. V. A. Intine. 2001. Recognition of nascent RNA by the human La antigen: conserved and divergent features of structure and function. Mol. Cell. Biol. 21:367-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meerovitch, K., Y. V. Svitkin, H. S. Lee, F. Lejbkowics, D. J. Kenan, E. K. L. Chan, V. I. Agol, J. D. Keene, and N. Sonenberg. 1993. La autoantigen enhances and corrects aberrant translation of poliovirus RNA and reticulocyte lysate. J. Virol. 67:3798-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Men, R., M. Bray, D. Clark, R. Chanock, and C. Lai. 1996. Dengue type 4 virus mutants containing deletions in the 3′ noncoding region of the RNA genome: analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J. Virol. 70:3930-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohan, P. M., and R. Padmanabhan. 1991. Detection of stable secondary structure at the 3′ terminus of dengue virus type 2 RNA. Gene 108:185-191. [DOI] [PubMed] [Google Scholar]
- 27.Nakhashi, H. L., X. Q. Cao, T. A. Rouault, and T. Y. Liu. 1991. Specific binding of host cell proteins to the 3′-terminal stem-loop structure of rubella virus negative-strand RNA. J. Virol. 65:5961-5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noiva, R., and W. J. Lennarz. 1992. Protein disulfide isomerase. A multifunctional protein resident in the lumen of the endoplasmic reticulum. J. Biol. Chem. 267:3553-3556. [PubMed] [Google Scholar]
- 29.O'Farrel, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 30.O'Sullivan, M., and H. Killen. 1994. The differentiation state of monocytic cells affects their susceptibility to infection and the effects of infection by dengue virus. J. Gen. Virol. 75:2387-2392. [DOI] [PubMed] [Google Scholar]
- 31.Pardigon, N., and J. Strauss. 1992. Cellular proteins bind to the 3′ end of Sindbis virus minus-strand RNA. J. Virol. 66:1007-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pardigon, N., and J. Strauss. 1996. Mosquito homolog of the La autoantigen binds to Sindbis virus RNA. J. Virol. 70:1173-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Proutski, V., E. Gould, and E. Holmes. 1997. Secondary structure of the 3′ untranslated region of flaviviruses: similarities and differences. Nucleic Acids Res. 25:1194-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Proutski, V., T. S. Gritsun, E. A. Gould, and E. C. Holmes. 1999. Biological consequences of deletions within the 3′-untranslated region of flaviviruses may be due to rearrangements of RNA secondary structure. Virus Res. 64:107-123. [DOI] [PubMed] [Google Scholar]
- 35.Roehl, H., and B. Semler. 1995. Poliovirus infection enhances the formation of two ribonucleoprotein complexes at the 3′ end of viral negative-strand RNA. J. Virol. 69:2954-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rojas-Eisenring, I. A., M. Cajero-Juarez, and R. M. del Angel. 1995. Cell proteins bind to a linear polypyrimidine-rich sequence within the 5′-untranslated region of rhinovirus 14 RNA. J. Virol. 69:6819-6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi, P., M. Brinton, J. Veal, Y. Zhong, and W. Wilson. 1996. Evidence for the existence of a pseudoknot structure at the 3′ terminus of the flavivirus genomic RNA. Biochemistry 35:4222-4230. [DOI] [PubMed] [Google Scholar]
- 38.Shi, P., W. Li, and M. Brinton. 1996. Cell proteins bind specifically to West Nile virus minus-strand 3′ stem-loop RNA. J. Virol. 70:6278-6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sing, H. N., C. D. Atreya, and H. L. Nakhasi. 1994. Identification of calreticulin as a rubella virus RNA binding protein. Proc. Natl. Acad. Sci. USA 91:12770-12774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spangberg, K., L. Wiklund, and S. Schwartz. 2001. Binding of the La autoantigen to the hepatitis C virus 3′ untranslated region protects the RNA from rapid degradation in vitro. J. Gen. Virol. 82:113-120. [DOI] [PubMed] [Google Scholar]
- 41.Stefano, J. E. 1984. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell 36:145-154. [DOI] [PubMed] [Google Scholar]
- 42.Svitkin, Y., V. K. Meerovitch, H. S. Lee, J. N. Dholakia, D. J. Kenan, V. I. Agol, and N. Sonenberg. 1994. Internal initiation on poliovirus RNA: further characterization of La function in poliovirus translation in vitro. J. Virol. 68:1544-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan, B. H., J. Fu, R. J. Sugrue, E. H. Yap, Y. C. Chan, and Y. H. Tan. 1996. Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology 216:317-325. [DOI] [PubMed] [Google Scholar]
- 44.You, S., B. Falgout, L. Markoff, and R. Padmanabhan. 2001. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J. Biol. Chem. 276:15581-15591. [DOI] [PubMed] [Google Scholar]
- 45.You, S., and R. Padmanabhan. 1999. A novel in vitro replication system for dengue virus. J. Biol. Chem. 274:33714-33722. [DOI] [PubMed] [Google Scholar]
- 46.Zeng, L., B. Falgout, and L. Markoff. 1998. Identification of specific nucleotide sequences within the conserved 3′-SL in the dengue type 2 virus genome required for replication. J. Virol. 72:7510-7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zuker, M. 1989. Computer prediction of RNA structure. RNA processing. Methods Enzymol. 180:262-288. [DOI] [PubMed] [Google Scholar]