Abstract
The capsid (CA) domain of the Moloney murine leukemia virus (Mo-MuLV) Gag protein has a unique carboxy terminus with a highly charged arginine-rich sequence. Mutant viruses harboring arginine-to-alanine mutations affecting this region of CA displayed significant defects in virion release, and the few viral particles produced were noninfectious. The interaction between the mutant Gag precursors was affected, as judged by the yeast two-hybrid assay. The results suggest that the unique carboxy terminus of CA in the Mo-MuLV plays an important role in Gag-Gag association during virion production.
The Gag protein of retroviruses directs the assembly and release of the virion particle during the late stages of the viral life cycle (for a review, see references 33 and 37). The Gag precursor of the mammalian gammaretroviruses, such as the Moloney murine leukemia virus (Mo-MuLV), is targeted to the plasma membrane to assemble virion particles that are subsequently released from the infected cells. The Gag protein alone is both necessary and sufficient to mediate its self-assembly and to direct the formation and budding of virus-like particles. Various mutants with alterations in the gag gene have been found to display defects in assembly (14, 31).
During virion maturation, the Gag precursor of Mo-MuLV (Pr65gag) is cleaved by the viral protease into four structural proteins: matrix (MA), p12, capsid (CA), and nucleocapsid (NC). The MA domain is responsible for membrane targeting of the precursor (30). The p12 protein contains an L (late) domain required for the late stage of virion release from cells (39). The NC protein contains a zinc-binding motif required for binding nucleic acids, and point mutations in this region often affect the encapsidation of the genomic RNA (12, 13, 25, 29). In addition, the NC domain includes an I (interaction) domain that is required for multimerization of Gag (16).
The CA domain forms the inner core structure of the virion. Genetic analyses have shown that the CA domain of Mo-MuLV plays essential roles not only in viral entry (1, 9) but also in viral assembly (18, 31). Various experiments have mapped out a number of regions that are required for viral particle assembly (15, 19, 23, 36). The key features include a region called the major homology region that is highly conserved among retroviral CA proteins. The function of this motif remains unclear since deletion of or mutations altering this motif affect virion assembly in certain viruses but not others (8, 24, 32). An amino-terminal fragment of the Mo-MuLV Gag protein with an intact CA domain was incorporated into virion particles (15). The domains of CA proteins required for the assembly of different retroviruses vary in position and size (28), and the exact role of CA in the assembly process remains unclear.
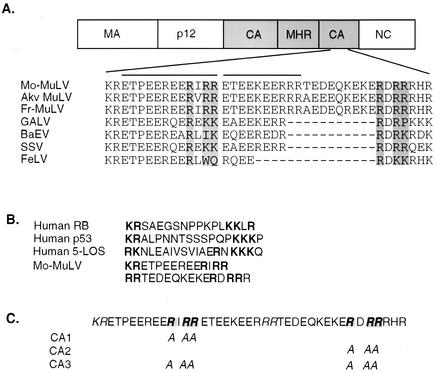
The very carboxy terminus of CA is highly conserved among most mammalian gammaretroviruses but not in more distant retroviruses, such as human immunodeficiency virus type 1 and Rous sarcoma virus (Fig. 1A). Two features of this region have been previously noted. First, it has an 11-amino-acid consensus sequence repeat that is homologous to the protein component of the U1 small nuclear ribonucleoprotein, which is recognized by an autoantibody found in serum samples from systemic lupus erythematosus patients (21, 27). This region is also similar to a murine immunoglobulin E-binding factor (20, 35). Secondly, it contains an extremely hydrophilic sequence with mixed-charge amino acid clusters that have been suggested to harbor two bipartite nuclear localization signals (NLS) (26). The arginine- and lysine-rich region with similarity to bipartite NLS motifs (22) is shown in Fig. 1B.
FIG. 1.
Rationale for the design of mutations affecting the CA domain of Mo-MuLV Gag. (A) Alignment of the carboxy terminus of Mo-MuLV CA with those of other gammaretroviruses. The lines on the top of the alignment indicate two tandem repeats homologous to U1 small nuclear ribonucleoproteins (common antigenic epitopes). The arginines that were replaced in the CA mutants and the corresponding amino acids in the proteins of the other viruses are shaded. Akv MuLV, AKV murine leukemia virus; Fr-MuLV, Friend murine leukemia virus; GALV, gibbon ape leukemia virus; BaEV, baboon endogenous virus; SSV, simian sarcoma virus; FeLV, feline leukemia virus. (B) Alignment of two putative bipartite NLS from Mo-MuLV with typical nuclear proteins. The signals are indicated by bold characters. RB, retinoblastoma protein. (C) The structures of the alanine-scanning mutants (CA1 and CA2) and their combination as a double mutant (CA3).
To evaluate the role of this region in the Mo-MuLV replication cycle, we replaced the three arginines in each of the two NLS motifs with alanines to produce mutants CA1 and CA2 (Fig. 1C). Analogous substitutions in other nuclear or shuttling proteins have been shown to inactivate the NLS (17). A third mutant, CA3, was constructed to contain the mutations in both CA1 and CA2 (Fig. 1C). The mutations were generated by two-step overlapping PCR between the unique XhoI and NruI sites with pNCS proviral DNA as a template. The primers to amplify the region were XhoI (forward) (5′-TTTTCCCCTCGAGCGCCCAG-3′) and NruI (reverse) (5′-CACTGGTCGCGATCGAGTTGGGACCTCC-3′). The mutagenic oligodeoxynucleotides used for CA1 were AIAAE5 (5′-GAAGAAAGAGAGGAAGCTATCGCGGCAAGGACAGAGGAAAAA-3′) and AIAAE3 (5′-TTTTTCCTCTGTTTCTGCCGCGATAGCTTCCTCTCTTTCTTC-3′). The primers used for creating CA2 were ADAAR5 (5′-CAGAAAGAGAAAGAAGCAGATGCTGCGAGACATAGAGAGATG-3′) and ADAAR3 (5′-CATCTCTCTATGTCTCGCAGCATCTGC TTCTTTCTCTTTCTG-3′). To combine these two mutations in one construct to generate CA3, a CA1 DNA fragment was used as a PCR template and amplified by XhoI (forward) and the ADAAR5-NruI fragment (reverse). The mutations were all introduced into the wild-type Mo-MuLV provirus, carried on a plasmid vector containing a simian virus 40 origin of replication to allow high-level expression in 293T cells (7).
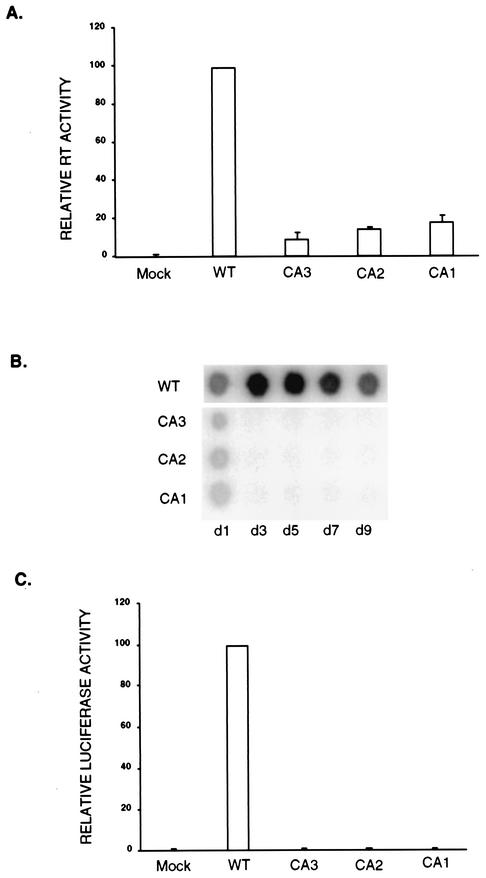
To characterize the effect of the mutations on virion assembly, 293T cells were transiently transfected with the wild-type or mutant provirus DNA constructs by standard procedures (10, 39); a DNA segment encoding β-galactosidase (pCMV-β-gal; Invitrogen) was included as a control for transfection efficiency. The culture supernatants were collected 48 h posttransfection and assayed for their reverse transcriptase (RT) activities (34), which were normalized for transfection efficiency by a β-galactosidase assay of cell lysates. Quantitation of the RT activities by a phosphorimager revealed that mutant CA1 produced approximately 20% of the levels of virion-associated RT of the wild-type virus, while both CA2 and CA3 mutants produced only about 10% of the wild-type levels (Fig. 2A). These results suggest that the arginine-to-alanine mutations dramatically diminish virion production at the stage of viral assembly or release.
FIG. 2.
Viral assembly, particle release, and viral infectivity analyzed by RT assay. Proviruses containing the mutations were transiently transfected into 293T cells, and the culture medium was collected 48 h posttransfection. (A) Phosphorimager quantitation of relative RT activities in the culture medium. The means of results from three independent experiments are displayed. The amount of RT activity in wild-type (WT) virus is set as 100. Standard deviations of the results are shown by error bars. (B) Equal volumes of culture medium were used to infect Rat2-2 cells, and the culture medium was assayed for RT activity on the indicated days postinfection. (C) Single-round infection. Rat2-2 cell lysates were prepared 36 h after infection with equal numbers of luciferase reporter viruses packaged by mutant viruses serving as helpers. Quantitation of the relative luciferase activities detected in three independent experiments is displayed. The amount of luciferase activity in wild-type virus is set as 100.
To investigate whether the few virions produced were capable of completing the replication cycle, the mutant viral particles were assayed for their infectivities. 293T cells were transiently transfected with either wild-type virus or the individual mutant proviruses as before. Culture medium was collected 48 h posttransfection, and equal volumes were used to infect Rat2-2 cells by standard procedures (10). Viral spread was monitored by assaying the RT activity of the culture medium on successive days (Fig. 2B). The wild-type virus exhibited high progeny yields within 2 to 3 days, as expected. In contrast, all three of the CA mutant viruses failed to display any RT activity even after 9 days in culture.
To further evaluate the infectivity, the mutants were tested as helpers for single-round infections with a firefly luciferase reporter virus. 293T cells were transiently transfected with either wild-type virus or the individual mutants together with a murine retrovirus vector, SRαLluc (a gift from Dong-Sung An) (3). The supernatants were collected, and equal numbers of wild-type and mutant reporter viruses, as judged by RT activities, were used to infect Rat2-2 cells. The infectivity was assessed by measuring the luciferase activity (Promega) in lysates of the infected cells prepared 36 h postinfection (Fig. 2C) with an EG&G Berthold luminometer (Lumat model LB9507). Whereas high levels of luciferase were detected in the cells infected with the wild-type helper virus, no luc expression above background was present in the cells infected with the three CA mutant viruses. These data suggest that the virions produced by these mutants were not infectious.
Mutations in CA may affect the synthesis and stability of the Gag precursor protein or its function during assembly. To examine Gag levels, cells were transfected with the viral DNAs and the Gag proteins in intracellular lysates and in the released virions were analyzed by Western blots (10, 38) probed with anti-CA serum (79S-107) from the National Cancer Institute. The L domain p12 mutant DPY, an example of a mutant defective in viral assembly and release (39), was used as a control. The wild-type virus produced Pr65gag and some processing intermediates, but the major intracellular species was the mature CA protein (Fig. 3A). Cells expressing the DPY mutant retained higher levels of unprocessed Pr65gag and showed a lower portion of the total Gag being processed to the mature CA. Cells expressing the CA mutants accumulated even higher levels of the Pr65gag precursor and displayed aberrant processing compared to those expressing the wild-type virus. The mutants accumulated higher levels of intermediate bands than DPY (Fig. 3A); a major cleavage product of about 55 kDa was particularly prominent.
FIG. 3.
Analysis of gag gene products in lysates and virions of transfected 293T cells. (A) Lysates of transfected 293T cells were analyzed by Western blotting by using anti-CA antibody. The positions of the precursor Pr65gag and two of its cleavage products, Pr55 and CA, are marked. WT, wild type. (B) The corresponding virions were lysed and subjected to Western blot analysis probing with anti-CA antiserum. The position of CA is marked. (C) Transfection efficiency control. Luciferase activities in lysates are indicated relative to that in the wild-type control. (D) Pulse-chase study. Transiently transfected 293T cells were pulse-labeled with [35S]methionine for 20 min, and the cultures were either harvested immediately or chased with unlabeled methionine for 4 h. Proteins in cell lysates or in virion particles were immunoprecipitated with anti-CA serum and analyzed on an SDS-PAGE gel. Lanes 1 to 5, 20-min pulse, no chase; lanes 6 to 10, 4-h chase. Lanes 1 and 6, mock; 2 and 7, wild type; 3 and 8, CA1 mutant; 4 and 9, CA2 mutant; 5 and 10, CA3 mutant.
Very high yields of CA were found in the wild-type virions released from the cells (Fig. 3B). Lower amounts of virion-associated Gag proteins were detected in the DPY mutant, with only partial processing, as previously reported (39). Only exceedingly low levels of released virion-associated Gag proteins could be detected in the CA mutants (Fig. 3B). The results of these experiments were consistent with those of the RT assay and together suggest a strong block for these mutants at the stage of viral assembly and release. We note that the precursor Pr65gag and cleavage products Pr55 and mature CA in the mutants migrated more slowly on the sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel than did the corresponding wild-type products, probably due to the change of positively charged arginines to neutral amino acid alanines. To confirm that there were no problems with the transfection efficiency or toxicity in these experiments, DNA encoding luciferase was included in the transfections and luciferase activity in the cell lysates was measured (Fig. 3C).
To further examine Gag production, pulse-chase experiments were performed. Cells expressing proviral DNAs and maintained on duplicate 10-cm plates were starved for 20 min in 3 ml of methionine-free Dulbecco modified Eagle medium. A pulse of [35S]methionine (200 μCi) was added for 20 min, the medium was removed, and the cells were washed with phosphate-buffered saline. The cells in one plate were lysed immediately with 3 ml of ice-cold radioimmunoprecipitation assay buffer. Chasing medium containing unlabeled methionine was added to the remaining plate, and the cells were incubated at 37°C for a 4-h chase period. The medium was harvested for analysis of virion proteins, and the cells were lysed. Virion particles were purified by sedimentation through a 25% sucrose cushion. The Gag proteins in the cell lysate and virions were immunoprecipitated and analyzed by electrophoresis followed by fluorography (Fig. 3D). The results showed that the mutants directed the synthesis of Gag at rates approximately half that of the wild type. Upon chasing, the intracellular levels of wild-type Gag dropped while those of the mutant proteins remained high. The label chased into CA in the extracellular virions efficiently in the wild-type virus but only inefficiently in mutant CA1 and even less efficiently in mutants CA2 and CA3. As in the Western blots, the mutant Gag and CA proteins migrated more slowly than the corresponding wild-type proteins.
Because the mutants showed defects in virion production and Gag processing, we tested whether the oligomerization of Gag was affected in these mutants by using the yeast two-hybrid system (11). DNA segments encoding the complete Mo-MuLV Gag with CA mutations were recovered by PCR and moved into yeast vectors pSH2-1 and pGADNOT (2). The Gag sequences were fused to either the C terminus of the DNA binding domain of LexA (in pSH2-1) or the activation domain of Gal4 (in pGADNOT). Saccharomyces cerevisiae strain CTY10-5d (the gift of R. Sternglanz) carrying an integrated lacZ reporter gene under the control of the lexA operator was transformed with the DNA segments, and the interactions between the Gag precursors were first assessed by staining yeast colonies for β-galactosidase activity with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (Table 1). The wild-type Mo-MuLV Gag interacted with itself very strongly. The CA1 mutant protein interacted with itself only weakly, and CA2 and CA3 proteins did not interact with themselves to induce detectable levels of the reporter. To control for the specificity of the effects on Gag-Gag interactions, we examined the ability of the mutant proteins to interact with another partner. We have found that the Mo-MuLV Gag protein interacts strongly in the yeast system with endophilin, a cellular protein involved in endocytosis and membrane vesicular transport, largely through contacts in the MA domain (unpublished data). The Gag proteins of the mutants CA1, CA2, and CA3 all continued to interact strongly with endophilin, as judged by the staining of yeast colonies for β-galactosidase activity, indicating that the mutations did not cause a global defect in the synthesis, folding, or nuclear transport of the fusion proteins (Table 1).
TABLE 1.
Gag-Gag and Gag-endophilin interactions in the yeast two-hybrid systema
| DNA binding partner | Test results with activation partner(s)
|
||
|---|---|---|---|
| Gal4AD | Corresponding Gal4AD-Gag | Gal4AD-endophilin 2 | |
| LexADB | − | − | − |
| LexADB-WTb | − | ++ | +++ |
| LexADB-CA3 | − | − | +++ |
| LexADB-CA2 | − | − | ++ |
| LexADB-CA1 | − | + | ++ |
LacZ levels as judged by X-Gal staining of yeast colonies. −, white; +, blue in 2 h; ++, blue in 1 h; +++, blue in 30 min.
WT, wild-type.
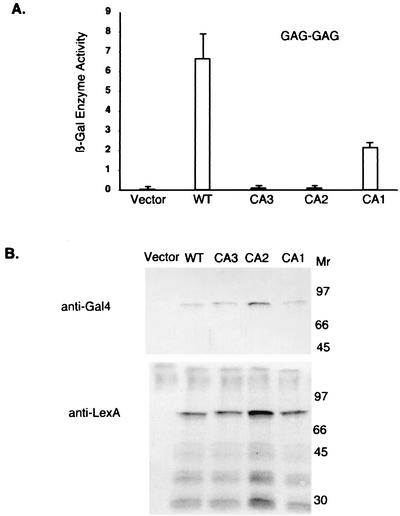
To provide a more quantitative measure of the ability of Gag proteins to form homodimers, the levels of β-galactosidase were measured in lysates of yeast cells (Fig. 4A). The level of the reporter responsive to the interaction of the mutant Gag-Gag precursor was reduced threefold in CA1 and virtually eliminated in both CA2 and CA3. To monitor the levels of expression of the mutant fusion proteins, Western blotting was performed on yeast lysates (Fig. 4B). There were no significant effects of the mutations on the levels of production of the Gal4 or LexA fusion proteins, and all proteins were available at levels that should have allowed interaction. These results suggest that the mutation in CA1 has a modest but specific effect on the homomeric interaction between the Gag precursors and that the CA2 mutation in the very C-terminal region of the CA domain can virtually eliminate Gag oligomerization.
FIG. 4.
Homomeric interactions in the two-hybrid system between Gag proteins with CA mutations. (A) β-galactosidase assay. Yeast strain CTY10-5d was transformed with plasmids encoding LexADB-Gag fusions and Gal4AD-Gag fusions. The y axis presents units of β-galactosidase enzyme activity. Results are the averages from four separate yeast clones, and the control represents the interaction between the empty vector and fusion proteins. The error bars represent the standard deviations. The low levels in CA2 and CA3 were almost below detection. WT, wild type. (B) Yeast cell lysates expressing pairs of fusions were analyzed by Western blotting by probing with anti-Gal4 or anti-LexA antibodies. Molecular weights are given in thousands.
The analysis of the mutations described here suggests that the highly charged C-terminal region of CA is crucial for Gag-Gag interactions and for virion production. The experiments suggest that Gag-Gag interactions were directly affected by the mutations and support the notion that this region of Gag is likely to be important for assembly. This notion is consistent with other mapping data. A single amino acid change in this region, L477P, very near to the carboxy terminus of CA, also strongly disrupted virion assembly (4). Recent results suggest that the downstream NC region of the Gag precursor also plays an important role in Gag-Gag interactions, perhaps through RNA bridges between the proteins (5, 6, 29). It is not clear if the basic residues of CA similarly use RNA to promote their association.
These CA mutations are also likely to affect the trafficking and intracellular localization of the Gag precursor in virus-expressing cells. We have examined the localization of epitope-tagged versions of Gag by immunofluorescence and found that the intracellular localization of the Gag precursor was profoundly affected by the mutations (data not shown). None of the constructs, wild type or mutant, showed significant localization to the nucleus. Thus, the mutations were unlikely to be affecting NLS functions but did alter the distribution of the proteins in the membrane. Gag-Gag association and the intracellular localization of Gag may both be coordinately dependent on the CA domain. Determining which of these effects is primary and which secondary will be important for further understanding of this region of the molecule.
Acknowledgments
We thank Jeremy Luban, Gregg Gundersen, Guangxia Gao, Gilda Tachedjian, Theodora Hatziioannou, Dag Helland, and Masha Orlova for helpful discussions and Kenia de los Santos for laboratory assistance.
This work was supported by PHS grant CA 30488 from the National Cancer Institute. S.P.G. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Alin, K., and S. P. Goff. 1996. Amino acid substitutions in the CA protein of Moloney murine leukemia virus that block early events in infection. Virology 222:339-351. [DOI] [PubMed] [Google Scholar]
- 2.Alin, K., and S. P. Goff. 1996. Mutational analysis of interactions between the Gag precursor proteins of murine leukemia viruses. Virology 216:418-424. [DOI] [PubMed] [Google Scholar]
- 3.An, D. S., Y. Koyanagi, J. Q. Zhao, R. Akkina, G. Bristol, N. Yamamoto, J. A. Zack, and I. S. Chen. 1997. High-efficiency transduction of human lymphoid progenitor cells and expression in differentiated T cells. J. Virol. 71:1397-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bacharach, E., J. Gonsky, K. Alin, M. Orlova, and S. P. Goff. 2000. The carboxy-terminal fragment of nucleolin interacts with the nucleocapsid domain of retroviral Gag proteins and inhibits virion assembly. J. Virol. 74:11027-11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burniston, M. T., A. Cimarelli, J. Colgan, S. P. Curtis, and J. Luban. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 73:8527-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cimarelli, A., S. Sandin, S. Hoglund, and J. Luban. 2000. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 74:3046-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colicelli, J., and S. P. Goff. 1988. Sequence and spacing requirements of a retrovirus integration site. J. Mol. Biol. 199:47-59. [DOI] [PubMed] [Google Scholar]
- 8.Craven, R. C., A. E. Leure-duPree, R. A. Weldon, Jr., and J. W. Wills. 1995. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J. Virol. 69:4213-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fassati, A., and S. P. Goff. 1999. Characterization of intracellular reverse transcription complexes of Moloney murine leukemia virus. J. Virol. 73:8919-8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao, G., and S. P. Goff. 1999. Somatic cell mutants resistant to retrovirus replication: intracellular blocks during the early stages of infection. Mol. Biol. Cell 10:1705-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goff, S. P. 2001. Retroviridae: the retroviruses and their replication, p. 1871-1939. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins Publishers, Philadelphia, Pa.
- 12.Gorelick, R. J., D. J. Chabot, D. E. Ott, T. D. Gagliardi, A. Rein, L. E. Henderson, and L. O. Arthur. 1996. Genetic analysis of the zinc finger in the Moloney murine leukemia virus nucleocapsid domain: replacement of zinc-coordinating residues with other zinc-coordinating residues yields noninfectious particles containing genomic RNA. J. Virol. 70:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorelick, R. J., L. E. Henderson, J. P. Hanser, and A. Rein. 1988. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc. Natl. Acad. Sci. USA 85:8420-8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Granowitz, C., and S. P. Goff. 1994. Substitution mutations affecting a small region of the Moloney murine leukemia virus MA gag protein block assembly and release of virion particles. Virology 205:336-344. [DOI] [PubMed] [Google Scholar]
- 15.Hansen, M., L. Jelinek, S. Whiting, and E. Barklis. 1990. Transport and assembly of gag proteins into Moloney murine leukemia virus. J. Virol. 64:5306-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen, M. S., and E. Barklis. 1995. Structural interactions between retroviral Gag proteins examined by cysteine cross-linking. J. Virol. 69:1150-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Healy, A. M., M. Peters-Golden, J. P. Yao, and T. G. Brock. 1999. Identification of a bipartite nuclear localization sequence necessary for nuclear import of 5-lipoxygenase. J. Biol. Chem. 274:29812-29818. [DOI] [PubMed] [Google Scholar]
- 18.Hsu, H. W., P. Schwartzberg, and S. P. Goff. 1985. Point mutations in the P30 domain of the gag gene of Moloney murine leukemia virus. Virology 142:211-214. [DOI] [PubMed] [Google Scholar]
- 19.Jones, T. A., G. Blaug, M. Hansen, and E. Barklis. 1990. Assembly of gag-β-galactosidase proteins into retrovirus particles. J. Virol. 64:2265-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuff, E. L., J. A. Mietz, M. L. Trounstine, K. W. Moore, and C. L. Martens. 1986. cDNA clones encoding murine IgE-binding factors represent multiple structural variants of intracisternal A-particle genes. Proc. Natl. Acad. Sci. USA 83:6583-6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lerner, M. R., and J. A. Steitz. 1979. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc. Natl. Acad. Sci. USA 76:5495-5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang, S. H., and M. F. Clarke. 1999. A bipartite nuclear localization signal is required for p53 nuclear import regulated by a carboxyl-terminal domain. J. Biol. Chem. 274:32699-32703. [DOI] [PubMed] [Google Scholar]
- 23.Lobel, L. I., and S. P. Goff. 1984. Construction of mutants of Moloney murine leukemia virus by suppressor-linker insertional mutagenesis: positions of viable insertion mutations. Proc. Natl. Acad. Sci. USA 81:4149-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mammano, F., A. Ohagen, S. Hoglund, and H. G. Gottlinger. 1994. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J. Virol. 68:4927-4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meric, C., and S. P. Goff. 1989. Characterization of Moloney murine leukemia virus mutants with single-amino-acid substitutions in the Cys-His box of the nucleocapsid protein. J. Virol. 63:1558-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nash, M. A., M. K. Meyer, G. L. Decker, and R. B. Arlinghaus. 1993. A subset of Pr65gag is nucleus associated in murine leukemia virus-infected cells. J. Virol. 67:1350-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Query, C. C., and J. D. Keene. 1987. A human autoimmune protein associated with U1 RNA contains a region of homology that is cross-reactive with retroviral p30gag antigen. Cell 51:211-220. [DOI] [PubMed] [Google Scholar]
- 28.Rayne, F., F. Bouamr, J. Lalanne, and R. Z. Mamoun. 2001. The NH2-terminal domain of the human T-cell leukemia virus type 1 capsid protein is involved in particle formation. J. Virol. 75:5277-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rein, A., D. P. Harvin, J. Mirro, S. M. Ernst, and R. J. Gorelick. 1994. Evidence that a central domain of nucleocapsid protein is required for RNA packaging in murine leukemia virus. J. Virol. 68:6124-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rein, A., M. R. McClure, N. R. Rice, R. B. Luftig, and A. M. Schultz. 1986. Myristylation site in Pr65gag is essential for virus particle formation by Moloney murine leukemia virus. Proc. Natl. Acad. Sci. USA 83:7246-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwartzberg, P., J. Colicelli, M. L. Gordon, and S. P. Goff. 1984. Mutations in the gag gene of Moloney murine leukemia virus: effects on production of virions and reverse transcriptase. J. Virol. 49:918-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strambio-de-Castillia, C., and E. Hunter. 1992. Mutational analysis of the major homology region of Mason-Pfizer monkey virus by use of saturation mutagenesis. J. Virol. 66:7021-7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 34.Telesnitsky, A., S. Blain, and S. P. Goff. 1995. Assays for retroviral reverse transcriptase. Methods Enzymol. 262:347-362. [DOI] [PubMed] [Google Scholar]
- 35.Toh, H., M. Ono, and T. Miyata. 1985. Retroviral gag and DNA endonuclease coding sequences in IgE-binding factor gene. Nature 318:388-389. [DOI] [PubMed] [Google Scholar]
- 36.Wang, C. T., and E. Barklis. 1993. Assembly, processing, and infectivity of human immunodeficiency virus type 1 gag mutants. J. Virol. 67:4264-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wills, J. W., and R. C. Craven. 1991. Form, function, and use of retroviral gag proteins. AIDS 5:639-654. [DOI] [PubMed] [Google Scholar]
- 38.Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 74:7250-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan, B., X. Li, and S. P. Goff. 1999. Mutations altering the moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18:4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]