Abstract
The human immunodeficiency virus type 1 (HIV-1) Rev and human T-cell leukemia virus type 1 (HTLV-1) Rex proteins are essential for the expression of viral structural proteins and productive infection. Both contain a nuclear export signal (NES) in their C-terminal domain and a nuclear localization signal (NLS) in their N-terminal domain. The NES and NLS are necessary for shuttling between nucleus and cytoplasm and are therefore indispensable for the transport of unspliced and singly spliced viral transcripts. HIV-1 Rev function is restricted in A9 cells, a murine fibroblast cell line, whereas HTLV-1 Rex is functional in these cells. Immunofluorescence studies with RevGFP fusion protein demonstrate normal import and export of Rev in A9 cells. To ascertain which domains of Rev are necessary for the restriction of Rev function in A9 cells, we studied a chimeric construct in which the NES domain of Rev was exchanged with Rex C-terminal amino acids 79 to 95, the Rev1-79/Rex79-95 chimera, which restored Rev function in A9 cells. In addition, overexpression of a truncated Rev containing the Rev C-terminal domain in the presence of wild-type Rev, led to restoration of Rev function in A9 cells. These results suggest that the C-terminal domain of HIV-1 Rev plays an important role in restricting Rev function in murine cells.
The human immunodeficiency virus type 1 (HIV-1) Rev function is essential for the cytoplasmic transport of unspliced and singly spliced viral transcripts and thus is required for the expression of viral structural proteins and the establishment of productive infection (12). Rev function appears to be essential in RNA splicing, transport, transcript stabilization, and the translation steps (1, 8, 21). Rev protein binds specifically to the Rev response element (RRE) present in the env RNA region of HIV-1 (23). Rev interacts directly with an RNA loop structure within the RRE through an arginine-rich RNA binding motif (23). This region in the N-terminal end of the Rev protein comprising amino acids 33 to 46 also constitutes a nuclear localization signal (NLS) (6, 20). In addition to the NLS, the Rev protein contains in its C-terminal region a nuclear export signal (NES) encompassing amino acids 73 to 84 (9, 35). The presence of both NLS and NES allows Rev to shuttle between the nucleus and the cytoplasm of HIV-1-infected cells (25). The ability to shuttle between cellular compartments is crucial for HIV-1 Rev function and mutations in the C-terminal NES domain that render the Rev protein unable to shuttle produce a trans-dominant phenotype (20). The trans-dominant mutant Rev protein (RevTD) can multimerize with the wild-type Rev protein and inhibit its nuclear export (34).
The prototypic Rev NES consists of a short stretch of hydrophobic amino acids, primarily leucines (4, 17). Leucine-rich NESs have been identified in several other proteins, including the human T-cell leukemia virus type 1 (HTLV-1) Rex protein (27). Rev and Rex have limited amino acid homology, yet HTLV-1 Rex protein performs functions analogous to those of HIV-1 Rev (13). The Rex protein also shuttles between the nucleus and the cytoplasm of infected cells, and through binding to the Rex response element, it allows expression of the incompletely spliced mRNAs that encode the HTLV-1 Gag, Pol, and Env structural proteins (10). Several cellular proteins specifically interact with the Rev NES, RRE RNA, or the Rev-RRE complex and likely influence HIV-1 Rev function. For example, the Rev NES domain was shown to bind the nuclear export receptor CRM1/exportin 1 (26). Other proteins involved in RNA splicing or processing have been reported to bind to the RRE or the Rev-RRE complex (28).
An important goal in AIDS research has been the development of small animal models for HIV-1 infection that simulate the stages of HIV disease in humans (5). Although several murine models for studies of AIDS pathogenesis have been reported, these models lack long-term, productive viral replication presumably due to blocks at several steps of viral gene expression (24). A goal of this study is to characterize posttranscriptional block in Rev function that prevents productive viral replication in A9 cells, a murine fibroblast cell line. We have shown that in A9 cells, HIV-1 Rev function is restricted, whereas HTLV-1 Rex protein is functional (33). In this study, we rescued Rev function in A9 cells using either a Rev-Rex chimeric protein or expression of the C-terminal domain of Rev in conjunction with the Rev protein. The results suggest that the C-terminal domain of Rev mediates the restriction of Rev function in A9 cells, presumably by its interaction with murine Rev inhibitory factors.
MATERIALS AND METHODS
Plasmids.
pCMVGag2RRE (2) was provided by Frank Maldarelli, National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), Bethesda, Md. pRSVRev1-79/Rex79-95 (17), pRSVRev, and the reporter construct pCMV128 (14) were provided by Thomas J. Hope (University of Illinois at Chicago). pRSVRev1-79 was derived from pRSVRev1-79/Rex79-95 digested with BglII to remove the Rex sequences and religated. pRSVRex79-95 contains amino acids 79 to 95 of Rex, and pRSVCRev61-116 contains amino acids 61 to 116 of Rev protein. The constructs used for indirect immunofluorescence, pcsRevGFP and pcsRevTDGFP, were provided by George N. Pavlakis, National Cancer Institute, Frederick, Md.
Cell culture and transfection.
HeLa and A9 cells were maintained as described previously (33). Cells were plated at approximately 2 × 105 cells per 35-mm-diameter dish 24 to 48 h before transfection and transfected with 2 to 4 μg of DNA, using Superfect (Qiagen) according to the manufacturer's instructions, or 10 μg of DNA by the calcium phosphate method. Cells were harvested 48 h after transfection and lysed as previously described (33). Protein concentration of the cell lysates was determined by a Bradford assay. For indirect immunofluorescence studies, HeLa and A9 cells were cultured on coverslips and transfected with 1 μg of pcsRevGFP or pcsRevTDGFP in the presence or absence of 1 μg of pCMV128.
CAT assay.
The amount of protein used for chloramphenicol acetyltransferase (CAT) assay was normalized to the transfection efficiency, which in turn was determined by cotransfecting a β-galactosidase expression plasmid, pCMVβ, or a green fluorescent protein (GFP) expression plasmid, pEGFPN1 (Clontech). Fluorescence-activated cell sorter analysis using Cell Quest Program was used to quantify GFP, and a luminometer was used to quantify β-galactosidase enzyme product (Tropix). CAT assays were performed as previously described (33). Results are expressed as fold increase in CAT activity normalized to the control val-ue (pCMV128 reporter construct alone) from three to five independent assays.
Immunoprecipitation and immunoblot analysis.
For immunoprecipitation, A9 cells transfected with 5 μg of pCMV128 alone or cotransfected with both pCMV128 (5 μg) and pRSVRev (10 μg) were washed with phosphate-buffered saline (PBS) and lysed in buffer containing 100 mM KPO4 and 0.2% Triton X-100 (pH 7.8). Cell lysates (1 ml) were preincubated with 100 μl of protein G-Sepharose beads (Bio-Rad) for 1 h and cleared at 12,000 × g for 20 s. Following the preclearing step, the supernatants (500 μl) were incubated with a pool of two mouse anti-Rev monoclonal antibodies, Ab2 and Ab4 (11) (kindly provided by Jonathan Karn, MRC Center, Cambridge, England) for 1 h and precipitated with 50 μl of protein G-Sepharose beads in lysis buffer. The resulting immunoprecipitates were centrifuged for 20 s at 12,000 × g and washed three times with lysis buffer followed by a final wash with 50 mM Tris (pH 8.0). All manipulations were performed at 4°C. The precipitated proteins were separated on a denaturing 4 to 20% gradient acrylamide gel (Bio-Rad) along with prestained protein markers (Invitrogen), transferred to polyvinylidene difluoride membranes (Bio-Rad), and probed for Rev protein (using the same pool of antibodies [at a 1:1,000 dilution] employed in the immunoprecipitation procedure). Purified HIV-1 Rev (NIH AIDS Research and Reference Reagent Program) was used as a positive control.
For Gag immunoblotting, cell lysates were resolved by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and probed with a rabbit polyclonal HIV-1 p25/p24 Gag antiserum (NIH AIDS Research and Reference Reagent Program). The antigen-antibody complexes were detected by chemiluminescence (Amersham).
Export and import of Rev.
Cells were washed with PBS 24 h after transfection and treated with cycloheximide (25 μg/ml) (Sigma) and/or actinomycin D (4 μg/ml) (Sigma) for 3 h at 37°C. The cells were then washed three times in 1× PBS and fixed with cold methanol at 4°C for 5 min. Fixed cells were washed three times with cold 1× PBS and treated with acetone for 4 min at −20°C. The permeabilized cells were then washed with 1× PBS and blocked with 0.3% bovine serum albumin in 1× PBS for 1 h at room temperature. HeLa cells were incubated with a rabbit antinucleolin antibody (16) at a dilution of 1:500 (provided by Raymond Petryshyn, Children's Research Institute, Washington, D.C.), and A9 cells were incubated with a mouse antifibrillarin MAb72B9 antibody (30) at a dilution of 1:500 (provided by Joseph Gall, Carnegie Institution, Baltimore, Md.). After 1 h of incubation with primary antibody at 37°C, cells were washed and incubated with Texas red-conjugated anti-rabbit and anti-mouse secondary antibodies at a dilution of 1:1,000 (Amersham). The immunofluorescence of the stained cells was examined with a Bio-Rad MRC 1024 confocal laser scanning microscope using Bio-Rad Lasersharp MRC 1024 software.
RESULTS
Rev-dependent Gag protein expression is restricted in A9 cells.
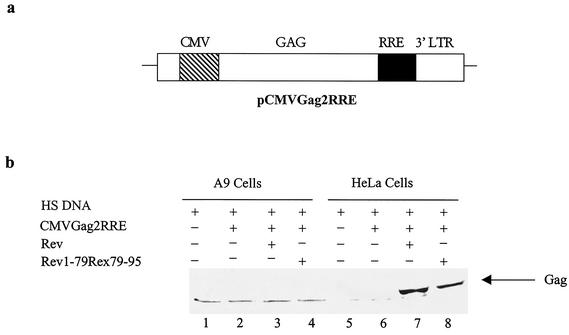
In an earlier report, using the HIV-1 Env-CAT reporter, pCMV128, we showed that A9 cells did not support Rev function, although HTLV-1 Rex was functional in these cells (33). To determine whether the Rev-dependent expression of both intron-containing and intron-lacking transcripts was restricted in A9 cells, HeLa and A9 cells were transfected with a gag expression vector, pCMVGag2RRE, and evaluated for the cytoplasmic expression of Gag protein in response to Rev or the Rev1-79/Rex79-95 chimera. The Gag expression vector consists of HIV-1 gag p17, p24, and a portion of p9 coding sequences, linked to the RRE sequence (2). Expression of Gag protein in this reporter system is dependent upon binding of Rev to the RRE in the unspliced transcript. The Rev function was monitored by detecting the presence of 44-kDa unprocessed Gag protein in extracts from A9 and HeLa cells. As expected, in HeLa cells in the absence of gag expression plasmid or the reporter alone, there was no detectable Gag expression. However, in the presence of Rev or the Rev1-79/Rex79-95 chimera, a 44-kDa unprocessed Gag protein was detected in HeLa cell extracts. On the other hand, A9 cells showed no Gag protein expression with Rev or the Rev 1-79/Rex79-95 chimera (Fig. 1b, compare lanes 3 and 4 to lanes 7 and 8). Thus, we conclude that the restriction of Rev function in A9 cells is not limited to the HIV-1 env reporter (pCMV128). Importantly, substitution of C-terminal Rev with amino acids 79 to 95 of Rex did not rescue the inhibition of Gag expression in A9 cells.
FIG. 1.
HIV-1 Rev-dependent Gag expression in HeLa and A9 cells. (a) Diagram depicting pCMVGag2RRE. CMV, cytomegalovirus major immediate-early promoter; GAG, HIV-1 gag p17, p24, and part of p9 open reading frame (protease gene not included); RRE, HIV-1 RRE; 3′ LTR, the HIV-1 long terminal repeat polyadenylation signal. (b) Gag expression in HeLa and A9 cells. A9 and HeLa cells were transfected with herring sperm DNA (HS DNA) (lanes 1 and 5), pCMVGag2RRE (2 μg) alone (lanes 2 and 6) or with pRSVRev (1 μg) (lanes 3 and 7) or pRSVRev1-79Rex79-95 (1 μg) (lanes 4 and 8). The position of the Gag protein is indicated by the arrow.
Rev-Rex fusion protein rescues Rev function in A9 cells.
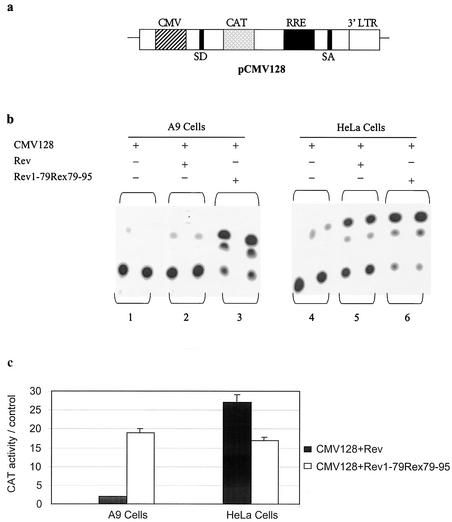
We showed earlier that nuclear proteins from HeLa cells formed specific ribonucleoprotein complexes with RRE RNA, whereas nuclear proteins from A9 cells lacked this ability (32). We also reported that Rev-mediated expression of the reporter construct pCMV128 was restricted in A9 cells (33). The reporter pCMV128 is derived from the second intron of HIV-1 expressed under the cytomegalovirus major immediate-early promoter with the CAT gene inserted within the intron upstream of the RRE (14). Since HIV-1 Rev and HTLV-1 Rex proteins serve analogous functions (13), we attempted to clarify which domain of the Rev protein is required to restrict Rev function in A9 cells, utilizing a Rev-Rex chimeric construct in which amino acids 1 to 79 of Rev were fused in frame with Rex amino acids 79 to 95, pRSVRev1-79/Rex79-95.
A9 and HeLa cells were transfected with either the pCMV128 reporter alone or in the presence of the pRSVRev or pRSVRev1-79/Rex79-95 expression vector. HeLa cells cotransfected with pCMV128 and pRSVRev yielded on average a 27-fold increase in CAT activity compared to cells transfected with the pCMV128 reporter plasmid alone (Fig. 2b, lanes 4 and 5, and c). Cotransfection of pCMV128 and the Rev-Rex chimera (pRSVRev1-79/Rex79-95) in HeLa cells resulted in, on average, a 17-fold increase in CAT activity (Fig. 2b, lane 6, and c). On the other hand, A9 cells cotransfected with pCMV128 and pRSVRev showed no significant increase in CAT activity compared to cells transfected with pCMV128 alone (Fig. 2b, lanes 1 and 2, and c), confirming our previous observation (33). However, when A9 cells were cotransfected with pCMV128 and pRSVRev1-79/Rex79-95, a 19-fold increase in CAT activity was observed (Fig. 2b, lane 3, and c).
FIG. 2.
Rev-Rex chimera rescues CAT reporter activity in A9 cells. (a) Diagram depicting pCMV128. CMV, cytomegalovirus major immediate-early promoter; SD, HIV-1 splice donor site; SA, HIV-1 splice acceptor site; 3′LTR, HIV-1 3′ long terminal repeat. (b) HIV-1 Rev-dependent CAT activity in A9 and HeLa cells. Both cell lines were transfected with 1 μg of reporter construct pCMV128 alone (lanes 1 and 4) or cotransfected with 500 ng of pRSVRev (lanes 2 and 5) or 500 ng of pRSVRev1-79/Rex79-95 (lanes 3 and 6). CAT enzyme activity was determined as described in Materials and Methods. (c) Quantification of CAT assay results. Graphed results are expressed as fold increase in the pCMV128 reporter CAT activity in cells cotransfected with pRSVRev or pRSVRev1-79/Rex79-95 compared to cells transfected with pCMV128 alone. Values are averages ± standard errors (error bars) from five independent transfections.
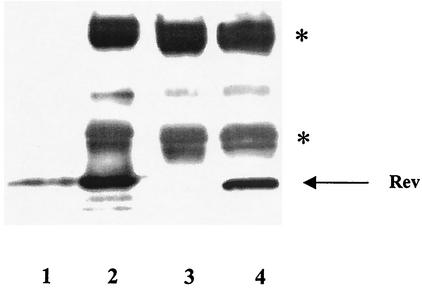
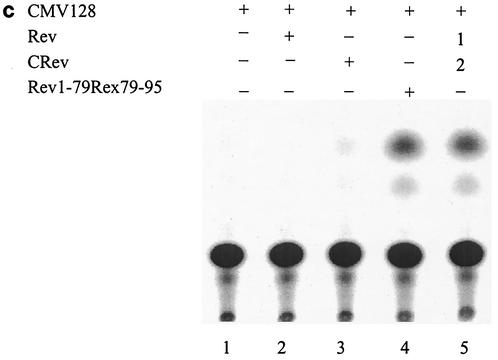
The production of Rev protein in A9 and HeLa cells was confirmed by dot blot analysis of extracts prepared from cells transfected with 1, 2, or 5 μg of Rev expression plasmids (results not shown) and by immunoprecipitation followed by Western blot analysis (Fig. 3). The results shown in Fig. 3 make two important points that HIV-1 Rev is expressed and stable in A9 cells, thus excluding the possibility that restriction of Rev function in A9 cells is due to a block in the production or the instability of Rev protein in these cells. Furthermore, the block in Rev-mediated expression of pCMV128 (the env reporter) can be rescued in A9 cells by the Rev/Rex (Rev1-79/Rex79-95) chimera, which is not the case with the Gag reporter (pCMVGag2RRE), arguing that the nucleocytoplasmic transport of Env and Gag mRNA may be differentially regulated in murine cells.
FIG. 3.
HIV-1 Rev expression in A9 cells. A9 cells were cotransfected with pCMV128 and pRSVRev expression constructs and lysed 48 h posttransfection, as described in Materials and Methods. Lane 1, purified wild-type Rev protein (2 μg); lane 2, purified wild-type Rev protein (2 μg) immunoprecipitated as described in Materials and Methods; lane 3, A9 cells transfected with 5 μg of pCMV128; lane 4, A9 cells cotransfected with 5 μg of pCMV128 and 10 μg of pRSVRev. The positions of Rev protein (arrow) and heavy and light immunoglobulin G chains (asterisks) are indicated.
Restriction of Rev function in A9 cells is not due to impaired localization or shuttling of Rev protein.
HIV-1 Rev function requires proper utilization of Rev's NLS and NES (25). To verify whether an impaired nucleocytoplasmic shuttling of HIV-1 Rev in A9 cells is responsible for the restriction of Rev function, fusion constructs of either wild-type Rev or trans-dominant mutant Rev and GFP (pcsRevGFP and pcsRevTDGFP, respectively) were employed. In order to characterize the intracellular transport of Rev, cells were transfected with RevGFP or the RevTDGFP plasmids in the presence or absence of the reporter, pCMV128, and with or without incubation with the translation and transcription inhibitors cycloheximide (25 μg/ml) and actinomycin D (4 μg/ml). In A9 and HeLa cells, both RevTDGFP and RevGFP localized primarily to the nucleolus. Upon drug treatment, RevTDGFP remained in the nucleolus in both cell types. On the other hand, in both A9 and HeLa cells, the wild-type RevGFP was able to exit the cell nucleus (data not shown). Even though RevGFP does not support Rev function in A9 cells, its intracellular localization is similar to that in HeLa cells, suggesting that the mechanism of restriction of Rev function in A9 cells is not related to impaired localization or shuttling of Rev protein.
Overexpression of the Rev C-terminal domain rescues Rev function in A9 cells.
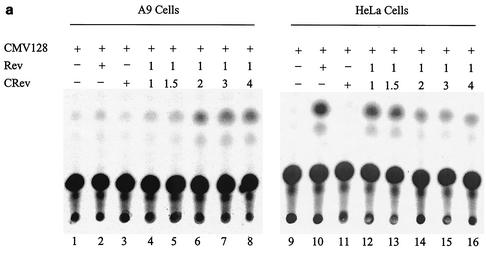
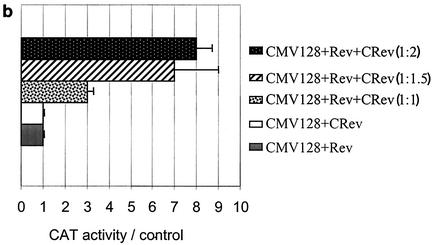
The above data suggest that the restriction of HIV-1 Rev function in A9 cells involves a host cell factor(s) which requires the C-terminal domain of Rev. Such factor(s) may bind the C-terminal domain of Rev and either fail to mediate Rev function or actively inhibit Rev function. In the latter case, overexpression of Rev C-terminal domain sequences might squelch the host factor(s) responsible for inhibiting Rev function in A9 cells. To test the hypothesis, HeLa and A9 cells were transfected with Rev expression plasmid (pRSVCRev), comprising the C-terminal amino acids 61 to 116 of the Rev protein along with pCMV128 reporter plus the Rev expression vector, pRSVRev. The ratio of C-terminal Rev (CRev)/Rev expression vectors used ranged from 0.5:1 to 4:1 (Fig. 4).
FIG. 4.
Overexpression of amino acids 61 to 116 of Rev C-terminal domain restores Rev function in A9 cells. (a) HIV-1 Rev-dependent CAT activity in A9 and HeLa cells. A9 and HeLa cells were transfected with 1 μg of the reporter construct pCMV128 alone (CMV128) or cotransfected with 500 ng of pRSVRev (Rev) or 1 μg of pRSVCRev (CRev) alone or with 500 ng of pRSVRev and increasing amounts of C-Rev (numbers represent ratios of plasmid amounts). (b) Quantification of CAT assay results for A9 cells. Graphed results are expressed as fold increase in the pCMV128 reporter CAT activity in response to cotransfected constructs. The Rev/CRev ratios are shown in parentheses in the symbol key. (c) Comparison of the level of rescue of Rev function in A9 cells. A9 cells were transfected with pCMV128 alone (lane 1) or cotransfected with pRSVRev (lane 2), pRSVCRev (lane 3), pRSVRev1-79Rex79-95 (lane 4) or pRSVCRev and pRSVRev at a 2:1 ratio.
HeLa cells cotransfected with the reporter (pCMV128) and the C-terminal Rev expression plasmid (pRSVCRev) showed no increase in CAT activity over background levels (Fig. 4a, lanes 9 and 11), whereas cotransfection of the reporter and the Rev expression plasmid (pRSVRev) yielded on average 15-fold increase in CAT activity over background levels (Fig. 4a, lane 10). When both Rev and increasing concentrations of CRev were transfected in HeLa cells along with the reporter plasmid, a decrease in CAT activity was observed (Fig. 4a, lanes 12 to 16), presumably due to the titration of host cell Rev cofactors that interact with the carboxyl end of the Rev protein.
On the other hand, at CRev/Rev ratios of 1.5:1 and greater, a significant increase in CAT activity was observed in A9 cells compared to cells with the reporter alone (Fig. 4a, lanes 4 to 8, and b). The results suggest a squelching of Rev inhibitory factor(s) in A9 cells with affinity for the Rev carboxyl-terminal sequences. In other experiments, A9 cells cotransfected with the reporter and increasing amounts of either Rev or CRev expression plasmid (ranging from 1 to 6 μg) showed no increase in CAT activity above background levels, suggesting that the restriction of Rev function in A9 cells is not dependent on the amount of Rev and is not rescued by increasing amounts of CRev without the presence of full-length Rev protein (data not shown).
DISCUSSION
A greater understanding of immune dysfunction during HIV-1 infection is compromised by the absence of a suitable animal model system (24). Mice provide an attractive model for studying HIV-1 pathogenesis given the unparalleled understanding of their immune function, the ease with which they can be genetically manipulated, and the expanding repertoire of mice with genetically defined immunological defects (15). However, previous efforts to develop a transgenic murine model for HIV-1 infection have been hindered by specific blocks to virus replication in murine cells (24).
Several possible reasons for the restriction of HIV-1 Rev function in murine cells have been proposed. One would expect that a combination of one or more of these mechanisms may contribute to the loss of Rev function in different murine cells. For example, our previous results suggested a lack of cellular factors in A9 cells that interacted with RRE RNA (32). Mariani and colleagues (24) reported inefficient processing of Gag protein in murine cells. Bieniasz and Cullen (3) reported reduced levels of the unspliced and genomic RNA transcripts in several rodent cells, but the results varied considerably among different rat and mouse cell lines. Yet another suggested mechanism for the lack of Rev function in murine cells (22) envisions a more efficient splicing of viral transcripts in murine cells than in human cells. While these results indicate wide variations in Rev function in different murine and rodent cells, they underscore the fact that the Rev function is restricted in murine cells whose mechanisms may vary depending upon the cell type used in the study. In this study, the results suggest that the murine cellular environment may restrict HIV-1 Rev function due to the presence of an inhibitor of Rev function or due to the modification of a positive cofactor of Rev.
Rev function block in A9 cells.
Murine A9 cells do not support Rev function in transient-transfection assays or form RRE-host protein ribonucleoprotein complexes in vitro (32, 33). However, rescue of Rev function in A9 cells can be achieved using human and mouse somatic cell hybrids containing human chromosomes 6 and 11 (33). It is noted that human chromosome 12 did not rescue Rev function in these somatic cell hybrids (33). These results suggested that multiple host factors are required for optimal Rev function. In A9 cells, Rev cofactors may be posttranslationally modified, allowing them to act as Rev inhibitors or preventing their function as Rev cofactors, unlike their role in human cells. Alternatively, restriction of HIV-1 Rev function in A9 cells may be due to the presence of specific inhibitor(s) that interact with the Rev C-terminal domain.
Block in Rev function in A9 cells is not due to impaired localization or shuttling of the Rev protein.
Yet another possibility is that the nucleocytoplasmic shuttling of Rev is impaired in these cells, as has been described for astrocytes (19). Astrocytes show impaired Rev function that is attributable to a predominantly cytoplasmic localization of the Rev protein (19). Other examples of inhibition of Rev function through changes in its localization include the trans-dominant mutant Rev protein (RevTD) (34) and a Sam68 C-terminal deletion mutant (29). RevTD has been shown to inhibit Rev function by multimerization with the wild-type Rev protein and trapping it in the cell nucleus, whereas the C-terminal deletion mutant of Sam68 has been shown to inhibit Rev function by trapping it in the cell cytoplasm (23, 29). In this study, proper nucleolar localization of both Rev and RevTD were confirmed in both A9 and HeLa cells using antibodies against nucleolus-specific proteins and Rev. Export of Rev-GFP was observed in both HeLa and A9 cells (data not shown), suggesting that the restriction of Rev function in A9 cells is not due to a defect in the nucleocytoplasmic shuttling of Rev protein.
Rev function in A9 cells can be rescued for singly spliced transcripts but not for unspliced transcripts.
In contrast to HIV-1, HTLV-1 can replicate efficiently in murine cells (7; R. Feng, A. Kabayama, K. Uchida, H. Hoshino, and M. Miwa, Abstr. 2000 Meet. Retroviruses Cold Spring Harbor Lab., abstr. 171, 2000). Our results show that in A9 cells HTLV-1 Rex protein is functional, whereas HIV-1 Rev function is restricted (33). The region of the Rex protein which is necessary to complement Rev function has been mapped to a core activation domain represented by residues 79 to 95 of Rex protein (13, 17). We showed that Rev/Rex fusion protein was able to restore Rev function in A9 cells. Interestingly, the Rev/Rex chimera was insufficient to rescue Gag expression in A9 cells. The results suggest two important points: first, the restriction of Rev function with respect to singly spliced env transcripts and nonspliced gag transcript in murine cells is specific to Rev, and second, the transport of singly spliced env and unspliced gag transcripts may be independently regulated. Possible explanations for the difference may be the presence of host factors that interact with the cis-inhibitory sequences in gag coding sequence which are not present in the env/CAT reporter construct (31). A recent report demonstrated that murine NIH 3T3 fibroblasts stably expressing human CD4, human CCR5, and human cyclin T1 produced unspliced HIV-1 mRNA transcripts albeit at reduced levels but presented a dramatic block to virus assembly (24). The researchers suggested that even though the murine cells produced large amounts of Gag and Gag-Pol, the Gag protein was inefficiently processed.
Determining the nature of the block in Rev function in A9 cells.
The rescue of Env-CAT reporter expression in A9 cells using a chimeric construct containing the first 79 amino acids of Rev fused to the C-terminal 79 to 95 amino acids of Rex suggested that the restriction of Rev function in A9 cells was associated with the carboxyl-terminal domain of the Rev protein. This is further supported by the fact that Rev inhibition can be titrated by coexpression of Rev and the C-terminal domain of HIV-1 Rev (amino acids 61 to 116 of Rev).
The data are consistent with a model which suggests the existence of an inhibitor of Rev function in A9 cells which can be squelched by overexpression of the C terminus of Rev. The results however do not exclude the possibility that a murine homologue of a Rev cofactor that recognizes the C-terminal domain of Rev may be unable to function, as is the case with murine cyclin T1 in Tat transactivation (18). The impairment in Rev function in A9 cells is not coupled to impaired shuttling of the Rev protein in this cell line. In addition, our results indicate that the translation of Rev protein and its stability is not impaired in A9 cells (Fig. 3). Determining the nature of the host factor(s) responsible for restriction of Rev function in A9 cells will help determine the mechanism of restriction of HIV-1 replication in murine cells and therefore aid in the development of an animal model for studying HIV-1 pathogenesis.
Acknowledgments
We thank Wilhelm E. Woolery, Mamatha Garige, and Mazer R. Ally for assistance and Silvio Urcuqui-Inchima, Anamaris Colberg-Poley, Frank Maldarelli, Kathleen Boris-Lawrie, Nick Somia, Sally Moody, Sergei Nekhai, and Susan Ceryak for critical comments on the manuscript. We are grateful to Robyn Rufner from the Center for Microscopy and Image Analysis (C.M.I.A.) of George Washington University for expert assistance with confocal microscopy.
This work was supported in part by NIH grants CA72147 (A.K.) and AI42491 (R.R.S.), and a fellowship from the PROGRAMA PRAXIS XXI of the Portuguese Fundação para a Ciência e Tecnologia (to S.M.P.M.).
REFERENCES
- 1.Arrigo, S. J., and I. S. Chen. 1991. Rev is necessary for translation but not cytoplasmic accumulation of HIV-1 vif, vpr, and env/vpu2 RNAs. Genes Dev. 5:808-819. [DOI] [PubMed] [Google Scholar]
- 2.Berthold, E., and F. Maldarelli. 1996. cis-acting elements in human immunodeficiency virus type 1 RNA direct viral transcripts to distinct intranuclear locations. J. Virol. 70:4667-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bieniasz, P. D., and B. R. Cullen. 2000. Multiple blocks to human immunodeficiency virus type 1 replication in rodent cells. J. Virol. 74:9868-9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogerd, H. P., R. A. Fridell, R. E. Benson, J. Hua, and B. R. Cullen. 1996. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol. Cell. Biol. 16:4207-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyle, M. J., M. Connors, M. E. Flanigan, S. P. Geiger, H. Ford, Jr., M. Baseler, J. Adelsberger, R. T. Davey, Jr., and H. C. Lane. 1995. The human HIV/peripheral blood lymphocyte (PBL)-SCID mouse. A modified human PBL-SCID model for the study of HIV pathogenesis and therapy. J. Immunol. 154:6612-6623. [PubMed] [Google Scholar]
- 6.Cochrane, A. W., A. Perkins, and C. A. Rosen. 1990. Identification of sequences important in the nucleolar localization of human immunodeficiency virus Rev: relevance of nucleolar localization to function. J. Virol. 64:881-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang, J., S. Kushida, R. Feng, M. Tanaka, T. Kawamura, H. Abe, N. Maeda, M. Onobori, M. Hori, K. Uchida, and M. Miwa. 1998. Transmission of human T-cell leukemia virus type 1 to mice. J. Virol. 72:3952-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felber, B. K., M. Hadzopoulou-Cladaras, C. Cladaras, T. Copeland, and G. N. Pavlakis. 1989. Rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl. Acad. Sci. USA 86:1495-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer, U., J. Huber, W. C. Boelens, I. W. Mattaj, and R. Luhrmann. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82:475-483. [DOI] [PubMed] [Google Scholar]
- 10.Green, P. L., and I. S. Chen. 1990. Regulation of human T cell leukemia virus expression. FASEB J. 4:169-175. [DOI] [PubMed] [Google Scholar]
- 11.Henderson, B. R., and P. Percipalle. 1997. Interactions between HIV Rev and nuclear import and export factors: the Rev nuclear localisation signal mediates specific binding to human importin-beta. J. Mol. Biol. 274:693-707. [DOI] [PubMed] [Google Scholar]
- 12.Hope, T. J. 1999. The ins and outs of HIV Rev. Arch. Biochem. Biophys. 365:186-191. [DOI] [PubMed] [Google Scholar]
- 13.Hope, T. J., B. L. Bond, D. McDonald, N. P. Klein, and T. G. Parslow. 1991. Effector domains of human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type I Rex are functionally interchangeable and share an essential peptide motif. J. Virol. 65:6001-6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hope, T. J., X. J. Huang, D. McDonald, and T. G. Parslow. 1990. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping cryptic functions of the arginine-rich motif. Proc. Natl. Acad. Sci. USA 87:7787-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jamieson, B. D., and J. A. Zack. 1999. Murine models for HIV disease. AIDS 13(Suppl. A):5-11. [PubMed] [Google Scholar]
- 16.Kibbey, M. C., B. Johnson, R. Petryshyn, M. Jucker, and H. K. Kleinman. 1995. A 110-kD nuclear shuttling protein, nucleolin, binds to the neurite-promoting IKVAV site of laminin-1. J. Neurosci. Res. 42:314-322. [DOI] [PubMed] [Google Scholar]
- 17.Kim, F. J., A. A. Beeche, J. J. Hunter, D. J. Chin, and T. J. Hope. 1996. Characterization of the nuclear export signal of human T-cell lymphotropic virus type 1 Rex reveals that nuclear export is mediated by position-variable hydrophobic interactions. Mol. Cell. Biol. 16:5147-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwak, Y. T., D. Ivanov, J. Guo, E. Nee, and R. B. Gaynor. 1999. Role of the human and murine cyclin T proteins in regulating HIV-1 tat-activation. J. Mol. Biol. 288:57-69. [DOI] [PubMed] [Google Scholar]
- 19.Ludwig, E., F. Ceccherini-Silberstein, J. vanEmpel, V. Erfle, M. Neumann, and R. Brack-Werner. 1999. Diminished Rev-mediated stimulation of human immunodeficiency virus type 1 protein synthesis is a hallmark of human astrocytes. J. Virol. 73:8279-8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malim, M. H., S. Bohnlein, J. Hauber, and B. R. Cullen. 1989. Functional dissection of the HIV-1 Rev trans-activator-derivation of a trans-dominant repressor of Rev function. Cell 58:205-214. [DOI] [PubMed] [Google Scholar]
- 21.Malim, M. H., and B. R. Cullen. 1993. Rev and the fate of pre-mRNA in the nucleus: implications for the regulation of RNA processing in eukaryotes. Mol. Cell. Biol. 13:6180-6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malim, M. H., and B. R. Cullen. 1991. HIV-1 structural gene expression requires the binding of multiple Rev monomers to the viral RRE: implications for HIV-1 latency. Cell 65:241-248. [DOI] [PubMed] [Google Scholar]
- 23.Malim, M. H., J. Hauber, S. Y. Le, J. V. Maizel, and B. R. Cullen. 1989. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338:254-257. [DOI] [PubMed] [Google Scholar]
- 24.Mariani, R., G. Rutter, M. E. Harris, T. J. Hope, H. G. Krausslich, and N. R. Landau. 2000. A block to human immunodeficiency virus type 1 assembly in murine cells. J. Virol. 74:3859-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer, B. E., and M. H. Malim. 1994. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev. 8:1538-1547. [DOI] [PubMed] [Google Scholar]
- 26.Neville, M., F. Stutz, L. Lee, L. I. Davis, and M. Rosbash. 1997. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr. Biol. 7:767-775. [DOI] [PubMed] [Google Scholar]
- 27.Palmeri, D., and M. H. Malim. 1996. The human T-cell leukemia virus type 1 posttranscriptional trans-activator Rex contains a nuclear export signal. J. Virol. 70:6442-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Powell, D. M., M. C. Amaral, J. Y. Wu, T. Maniatis, and W. C. Greene. 1997. HIV-1 Rev-dependent binding of SF2/ASF to the Rev response element: possible role in Rev-mediated inhibition of HIV-1 splicing. Proc. Natl. Acad. Sci. USA 94:973-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reddy, T. R., W. Xu, J. K. Mau, C. D. Goodwin, M. Suhasini, H. Tang, K. Frimpong, D. W. Rose, and F. Wong-Staal. 1999. Inhibition of HIV replication by dominant negative mutants of Sam68, a functional homolog of HIV-1 Rev. Nat. Med. 5:635-642. [DOI] [PubMed] [Google Scholar]
- 30.Reimer, G., K. M. Pollard, C. A. Penning, R. L. Ochs, M. A. Lischwe, H. Busch, and E. M. Tan. 1987. Monoclonal autoantibody from a (New Zealand black × New Zealand white)F1 mouse and some human scleroderma sera target an Mr 34,000 nucleolar protein of the U3 RNP particle. Arthritis Rheum. 30:793-800. [DOI] [PubMed] [Google Scholar]
- 31.Schneider, R., M. Campbell, G. Nasioulas, B. K. Felber, and G. N. Pavlakis. 1997. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J. Virol. 71:4892-4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shukla, R. R., P. L. Kimmel, and A. Kumar. 1994. Human immunodeficiency virus type 1 Rev-responsive element RNA binds to host cell-specific proteins. J. Virol. 68:2224-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shukla, R. R., S. M. Marques, P. L. Kimmel, and A. Kumar. 1996. Human chromosome 6- and 11-encoded factors support human immunodeficiency virus type 1 Rev function in A9 cells. J. Virol. 70:9064-9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stauber, R., G. A. Gaitanaris, and G. N. Pavlakis. 1995. Analysis of trafficking of Rev and transdominant Rev proteins in living cells using green fluorescent protein fusions: transdominant Rev blocks the export of Rev from the nucleus to the cytoplasm. Virology 213:439-449. [DOI] [PubMed] [Google Scholar]
- 35.Wen, W., J. L. Meinkoth, R. Y. Tsien, and S. S. Taylor. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell 82:463-473. [DOI] [PubMed] [Google Scholar]