Abstract
The transcription of cytomegalovirus (CMV) immediate-early (IE) genes is regulated by a large and complex enhancer containing an array of binding sites for a variety of cellular transcription factors. Previously, using bacterial artificial chromosome recombinants of the virus genome, it was reported that the enhancer region of murine CMV (MCMV) is dispensable but performs a key function for viral multiplication (A. Angulo, M. Messerle, U. H. Koszinowski, and P. Ghazal, J. Virol. 72:8502-8509, 1998). In the present study, we defined, through the reconstitution of infectious enhancerless MCMVs, the growth requirement for the enhancer in tissue culture and explored its significance for steering a productive infection in vivo. A comparison of cis and trans complementation systems for infection of enhancerless virus in permissive fibroblasts revealed a multiplicity-dependent growth phenotype that is severely compromised in the rate of infectious-virus multiplication. The in vivo impact of viruses that have an amputated enhancer was investigated in an extremely sensitive model of MCMV infection, the SCID mouse. Histological examination of spleens, livers, lungs, and salivary glands from animals infected with enhancer-deficient MCMV demonstrated an absence of tissue damage associated with CMV infection. The lack of pathogenic lesions correlated with a defect in replication competence. Enhancerless viruses were not detectable in major target organs harvested from SCID mice. The pathogenesis and growth defect reverted upon restoration of the enhancer. Markedly, while SCID mice infected with 5 PFU of parental MCMV died within 50 days postinfection, all mice infected with enhancerless virus survived for the duration of the experiment (1 year) after infection with 5 × 105 PFU. Together, these results clarify the importance of the enhancer for MCMV growth in cell culture and underscore the in vivo significance of this region for MCMV virulence and pathogenesis.
Following infection of permissive cells, cytomegalovirus (CMV) expresses ∼200 viral products in a temporally regulated fashion, presenting a cascadelike effect in which three phases can be distinguished (for a review, see reference 28). The immediate-early (IE) genes are the first genes to be expressed during infection, and their transcription is not dependent on de novo protein synthesis. The production of IE gene products is necessary for the transition into the early and late phases (14). The genomic region of CMV, by far the most actively transcribed at IE times, is called the major IE (MIE) region. The basic structural organization of the MIE gene locus exhibits a high degree of similarity among the different members of the CMV family studied to date (35). A complex and strong regulatory region, the MIE promoter (MIEP), governs the expression of the MIE genes, encoding the cotransactivator IE1 and the main early gene transactivator IE2 in human CMV (HCMV) and IE3 in murine CMV (MCMV) (15, 16, 26; for an overview, see reference 28). The ie1 genes of both HCMV and MCMV have been disrupted within the context of the viral genome and shown to play nonessential roles in viral replication in tissue culture, although under conditions of low multiplicity of infection (MOI) CMVs defective in ie1 are compromised in the ability to replicate (12, 27, 29). A recent study reported the generation of ie3-deficient mutants and showed that the IE3 protein has an essential regulatory function during the lytic replication cycle of MCMV in tissue culture, being absolutely required for viral replication and lytic gene expression (1). To date, despite the efforts of several groups, it has not been possible to recover an HCMV IE2-defective virus from HCMV genomes with ie2 deleted, indicating that, similar to its murine homolog, IE2 is required for successful HCMV infection in tissue culture (22).
Due to the crucial role played by the MIE proteins during viral infection, regulation of the MIEP has been speculated to be a critical stage in determining CMV permissiveness and switching between latent and lytic infections (18). Transcriptional control of the MIEP comprises a complex interplay between both positive and negative cis-acting regulatory elements (24). The HCMV MIEP consists of several distinct functional units, including a basal promoter, modulator sequences, and a highly potent enhancer containing most of the regulatory elements (reviewed in reference 11). The HCMV enhancer carries reiterated binding sites for a large repertoire of host transcription factors, the majority of which are regulated through signal transduction mechanisms. These cis-acting elements include, among others, binding sites for the transcription factors AP-1, CREB/ATF, NF-κB, retinoic acid receptors, and Sp1. Viral proteins, like the tegument protein pp71 and IE1 (7, 20), have also been reported to regulate the activity of the HCMV MIEP. In addition, cellular transcription factors such as YY1 can repress HCMV enhancer function (21). Analogous MIE enhancers are present in other animal CMV genomes. However, the organization of this region is quite different among CMVs, with variations in the nature, number, and arrangement of the consensus transcription factor sites that they carry (5, 6, 8, 33).
For years, most of our understanding of the significance and regulation of the HCMV-MIEP enhancer has been derived from in vitro transcription and transient-transfection assays. Work with HCMV-MIEP transgenic-mouse models first indicated the importance of the enhancer in conferring cell-type-specific activity in vivo. In these animal models, the expression of the enhancer mimics in part tissues and cell types naturally infected by HCMV (3, 4, 17). More recently, studies with enhancer swap viruses in which the native enhancer of MCMV or rat CMV has been replaced with that of HCMV or MCMV have indicated that the enhancer region is not responsible for species specificity and that the enhancers are paralogous in function (2, 13, 34). Despite some phenotypic differences dependent on the origin of the enhancer, enhancer swap viruses are able to grow in vitro as well as in their respective hosts, suggesting very similar modes of action among enhancer sequences from distinct CMV strains. However, the biological relevance of the MIEP enhancer was not directly addressed until the generation of CMV genomes lacking enhancer sequences. It was previously shown that enhancerless MCMV bacterial artificial chromosome (BAC) genomes, when transfected into murine fibroblasts, led to viral progeny, although substantially reduced compared to that originating from the parental MCMV BAC genome (2). In agreement with these results, HCMV recombinants lacking the MIE distal enhancer region are partially debilitated in their growth on fibroblasts, exhibiting an MOI-dependent effect (23). From these observations, we can infer that for a productive CMV infection of permissive fibroblasts, enhancer elements are not strictly necessary. However, a fundamental question that still remains unanswered is to what extent the MIEP enhancer region contributes to growth and pathogenesis during a natural CMV infection.
To unequivocally define the role of enhancer-mediated transcription control of the MIE locus during viral replication in vitro and to investigate the contribution of this region to MCMV in vivo growth and pathogenesis, we have isolated enhancerless MCMV recombinants with the aid of a complementing cell system. We demonstrate that resection of the enhancer remarkably reduces MCMV replication at low MOIs but not at high MOIs in cultured murine fibroblasts. Studies of severe combined immunodeficient (SCID) mice indicate that enhancer sequences are required for MCMV growth in target organs. Moreover, recombinant viruses with the MIE enhancer deletion exhibit a severely attenuated phenotype. This work provides the long-awaited evidence showing that the MCMV enhancer is absolutely required for efficient replication, pathogenesis, and virulence in its natural host.
(Part of these results was presented at the 25th International Herpesvirus Workshop in Portland, Oregon, July and August 2000.)
MATERIALS AND METHODS
Cells and viruses.
Mouse NIH 3T3 cells (ATCC CRL1658) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% newborn calf serum. Primary mouse embryonic fibroblasts were prepared from BALB/c.ByJ mice and grown in DMEM with 10% fetal calf serum. The parental BAC-derived MCMV strain MW97.01 (36; called MCMV in this study) and MCMVdE::Luc-rev were propagated on NIH 3T3 cells. The enhancerless mutants, MCMVdE and MCMVdE::Luc, were grown on the complementing cell line NIH 3T3-Bam25.
Construction of NIH 3T3-Bam25 cells.
NIH 3T3-Bam25 cells were derived from NIH 3T3 cells by cotransfecting pAMB25 (15; referred to as pBam25 in this publication), a plasmid containing the MCMV ie1 and ie3 genes (nucleotides [nt] 176441 to 187035 of the MCMV genome [32]), and pPUR (Clontech, Palo Alto, Calif.), a plasmid containing the puromycin resistance gene, using the calcium phosphate technique (10) and selecting cells in medium containing puromycin (Sigma) at 5 μg/ml. The cultures were refed every 3 to 5 days. Single colonies were picked using cloning cylinders and analyzed for IE1 expression by indirect immunofluorescence using the monoclonal antibody Croma 101 (kindly provided by Stipan Jonjic). RT PCR using specific primers for ie1 (ie1-R [5′-TAC AGG ACA ACA GAA CGC TC-3] and ie1/3-F [5′-CCT CGA GTC TGG AAC CGA AA-3′]) and ie3 (ie3-R [5′-TGT GAG GCA GTA GTT ATA CC-3′] and ie1/3-F [19]) was carried out to confirm the presence of ie1 and ie3 transcripts in NIH 3T3-Bam25 cells. Several lines were obtained. For the purpose of this study, we primarily used clone 23.
Analysis of viral growth in vitro.
Monolayers of NIH 3T3 cells or NIH 3T3-Bam25 cells in 24-well dishes were infected at an MOI of 3 (for single-cycle growth curves) or 0.01 (for multicycle growth curves) PFU/cell with the different MCMV recombinants. After a 1-h adsorption period, the cells were washed three times with phosphate-buffered saline and fed with fresh medium. At different times after infection, the supernatants of three different cultures were harvested, cleared of cellular debris, frozen, and thawed. Viral titers were determined by standard plaque assays (without centrifugal enhancement of infectivity) on NIH 3T3-Bam25 cells. The nature of the enhancerless viruses resulting from growth curves performed at high MOIs on NIH 3T3 cells was checked by parallel titration of the samples on NIH 3T3 cells and the complementing cell line, taking advantage of the different plating efficiencies of enhancerless viruses in these two cell types. Data were plotted as viral titers (PFU/ml) versus time postinfection (in days). After evaluation of the plots, growth constants were determined for the exponential phase of viral growth. Data points from days 1 to 3 after infection were used, except for enhancerless viruses under low-MOI conditions, for which days 3 to 5 after infection were considered for the analysis because a phase shift of exponential growth was observed in these cases. An exponential regression model of type y = b · emx was used to determine the parameters and quality of fit for the untransformed data. Parameter b is the intercept for the curve, and parameter m is the slope and therefore the exponential-growth rate constant. Computations were performed in Excel using the LOGEST function for exponential regression.
Plasmid construction.
Construction of the shuttle plasmids required for introduction of the enhancer deletion and replacement of the enhancer sequences by a stuffer fragment, respectively, in the BAC-cloned MCMV genome has been described previously (2). Briefly, the shuttle plasmid pMBO-dEnh carries an 8.0-kbp fragment comprising the MCMV sequences (32) from nt 178736 to 182940 (SpeI to EspI) and 184086 to 187889 (NdeI to HindIII). Thus, the fragment provides the homologous viral DNA sequences needed to delete the MCMV enhancer sequences from nt 182940 (EspI) to 184086 (NdeI). The shuttle plasmid pST76A-luc carries the same MCMV DNA sequences and in addition a 770-bp fragment of the luciferase gene (referred to as the luciferase stuffer) inserted between the EspI (nt 182940) and NdeI (nt 184086) sites, replacing the MCMV enhancer sequences. For the construction of a revertant virus genome, we decided to tag the MCMV enhancer sequences in order to allow differentiation between the genomes of the revertant and the parental viruses. A 22-bp oligonucleotide linker (forward, 5′-GGC AAG CTT GCG ATG CCT GGA T-3′; reverse, 5′-CCA GGC ATC GCA AGC TTG CCA T-3′) providing a HindIII site (underlined) was inserted into the unique PacI site (MCMV nucleotide position 183155) of the plasmid pUC-L (2). Then, a 5.3-kbp ApaI/SfiI fragment was transferred to the ApaI/SfiI-digested plasmid pST76A-luc, resulting in the shuttle plasmid pST76A-RevH1.
BAC mutagenesis and reconstitution of MCMV mutants.
Recombination between the BAC plasmid pSM3fr containing the complete MCMV genome (referred to as C3X in this publication) (36) and the shuttle plasmids pMBO-dEnh and pST76A-luc was performed by a two-step replacement procedure in the Escherichia coli strain CBTS as described previously (2, 36) in order to generate the mutant MCMV BAC plasmids C3XdE and C3XdE::Luc, respectively. The shuttle plasmid pST76A-RevH1 and the BAC plasmid C3XdE::Luc were used to generate the BAC plasmid C3XdE::Luc-rev, which contains the repaired enhancer sequences. Recombinant viruses were reconstituted by transfection of the BAC plasmids into NIH 3T3 cells or the complementing cell lines using the calcium phosphate transfection method.
Viral nucleic acid isolation and analysis.
Preparation of total DNA from infected cells, restriction enzyme analysis, and gel electrophoresis were done essentially as described previously (2). BAC plasmids were isolated from 400-ml E. coli cultures by using Nucleobond PC 100 columns (Machery-Nagel, Düren, Germany) according to the instructions of the manufacturer. To analyze whether appropriate excision of the BAC vector sequences from the recombinant MCMV genomes had occurred, PCRs were carried out as described previously (36). Primers b (5′-GCC CGC CTG ATG AAT GCT C-3′) and g (5′-GGA TAC TCA GCG GCA GTT TGC-3′), which bind in the BAC vector sequences and in the EcoRI g fragment, respectively, were used to detect a 1,950-bp PCR fragment within the BAC-containing genomes. Primers f (5′-GGT TAC TGG ATG GGT ACG AG-3′) and g, which bind to viral sequences flanking the excision site, were used to amplify a 590-bp product after successful excision of the BAC vector from the viral genomes. The template used in the PCR was either genomic DNA from cells infected with the recombinant viruses or viral genomes from extracellular virus (105 PFU) that was heated at 95°C for 5 min before being added to the reaction mixture. PCRs were performed under the following conditions: 1 cycle at 94°C for 3 min; 30 cycles of 1 min at 94°C, 1 min at 51°C, and 1 min at 72°C; and 1 cycle at 72°C for 10 min.
Infection of immunocompromised mice.
Female CB17 SCID mice (6 weeks old) were obtained from Taconic Farms and housed in the vivarium (The Scripps Research Institute) under specified pathogen-free conditions. The animals were injected intraperitoneally with 6 × 105 PFU of tissue culture-derived MCMV recombinants. On various days after infection, mice were sacrificed and organs were removed, weighed, and harvested as a 10% (wt/vol) tissue homogenate, and virus titers were determined on NIH 3T3-Bam25 cells by standard plaque assay. To maximize sensitivity, samples from animals infected with enhancerless virus and in which infectious virus could not be detected by standard plaque assays were also titrated using centrifugal enhancement of infectivity. To assess levels of virulence, the lethalities in CB17 SCID mice of recombinant and parental MCMV viruses were compared. Animals were infected intraperitoneally with various amounts of tissue culture-passaged recombinant or parental wild-type virus, and their survival was monitored daily for at least 150 days after infection.
Histopathology.
Six-week-old female CB17 SCID mice were injected intraperitoneally with 6 × 105 PFU of tissue-culture-derived MCMV recombinants. At 14 days after infection, the mice were sacrificed and selected organs were removed and fixed in Bouin's solution. The fixed tissues were subsequently processed and embedded in paraffin by standard procedures. The organs and tissues included the spleen, liver, lung, and salivary gland. Sections (3 μm thick) were cut with a rotary microtome and stained with hematoxylin and eosin for examination. In addition to routine examination and qualitative assessment of selected tissues, liver sections were examined using the 10× objective to determine the number of inflammatory foci (>10 inflammatory cells per focus) per unit area.
RESULTS
Generation of enhancerless MCMV recombinants.
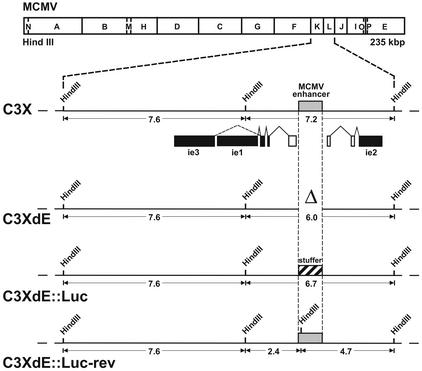
Previously, the construction of MCMV genomes lacking enhancer sequences, based on the parental MCMV BAC plasmid pSM3, was reported (2). Viral progeny reconstituted from this parental BAC construct following transfection of permissive cells exhibits an attenuated phenotype in vivo due to a deletion of viral sequences and insertion of the BAC vector (36). Recently, a BAC plasmid, pSM3fr (36; called C3X in this study), containing the full-length MCMV genome was generated. Virus reconstituted from this new construct after transfection into permissive cells shows biological properties in vivo comparable to those of wild-type MCMV. Since the purpose of this study was to analyze the relevance of the enhancer region during an acute CMV infection, we decided to generate new enhancerless MCMV genomes based on the C3X backbone. A schematic representation of the various BAC recombinant genomes constructed is shown in Fig. 1. In order to remove the MCMV enhancer sequences, a 1.1-kbp EspI-NdeI deletion was introduced within the HindIII L fragment of the parental MCMV genome C3X by using recombination techniques in E. coli as described in Materials and Methods. The resulting BAC recombinant genome, C3XdE, contained a deletion that encompasses nucleotide positions −48 to −1191 of the MCMV MIEP. In this recombinant BAC plasmid, deletion of the 1.1-kbp segment places nucleotide position −183 relative to the ie2 transcription start site immediately adjacent to nucleotide position −48 of the MIEP. It is possible that in this recombinant the remains of the MIEP could be influenced by the juxtaposition of the ie2 promoter-proximal control elements, that the junction point introduces a novel regulatory sequence, or that the relative positional distance between the ie2 and MIEP TATA boxes interferes with or modulates their activities. For these reasons we sought to maintain spatial integrity of the ie1-ie3 and ie2 promoters flanking the MIEP enhancer, and we constructed a second BAC plasmid, C3XdE::Luc, that contains a stuffer fragment replacing the deleted enhancer segment. In C3XdE::Luc, a nonregulatory 770-bp fragment from the firefly luciferase reporter gene was introduced within nucleotides −48 to −1191 of the enhancer region of MCMV. Finally, to determine whether the phenotype of the enhancer-deficient genome C3XdE::Luc was caused solely by deletion of the enhancer region, a revertant genome designated C3XdE::Luc-rev was generated. In this recombinant BAC construct, the luciferase stuffer present in C3XdE::Luc was replaced by the 1.1-kbp segment of the natural MCMV enhancer (spanning nucleotide positions −48 to −1191 of the MCMV MIEP). To distinguish this new MCMV BAC plasmid from the parental C3X genome, a new HindIII restriction site was introduced within the HindIII L fragment (described in more detail in Materials and Methods).
FIG. 1.
Construction of enhancerless MCMV BAC genomes. The HindIII map of the MCMV genome is shown at the top. The HindIII K and L fragments from the parental MCMV BAC genome, C3X, are expanded below to show the major IE gene region. The structures of the ie1, ie2, and ie3 transcripts are indicated below the expanded map. Coding exons are shown solid, and the first noncoding exons of the ie1-ie3 and ie2 transcription units are depicted as open rectangles. The shaded box marks the MCMV enhancer ie1-ie3 promoter. The recombinant MCMV BAC plasmids C3XdE, C3XdE::Luc, and C3XdE::Luc-rev, shown below the C3X genome, were generated by successive rounds of homologous recombination in E. coli starting from the parental BAC plasmid C3X (for C3XdE and C3XdE::Luc) or C3XdE::Luc (for C3XdE::Luc-rev), as indicated in Materials and Methods. The MCMV BAC plasmid C3XdE contains a 1.1-kbp deletion within the HindIII L fragment extending from nucleotide position −48 to −1191 relative to the ie1-ie3 MCMV transcription start site. C3XdE::Luc contains a 770-bp fragment from the luciferase gene replacing nucleotide sequences from −48 to −1191 of the MCMV MIEP enhancer region. C3XdE::Luc-rev is a revertant of C3XdE:: Luc in which the enhancer sequences were reconstituted. The 1.1-kbp deletion in the enhancer region is marked Δ. The crosshatched box represents the luciferase stuffer region. The sizes of the natural and new HindIII K and L fragments are indicated. The illustration is not drawn to scale.
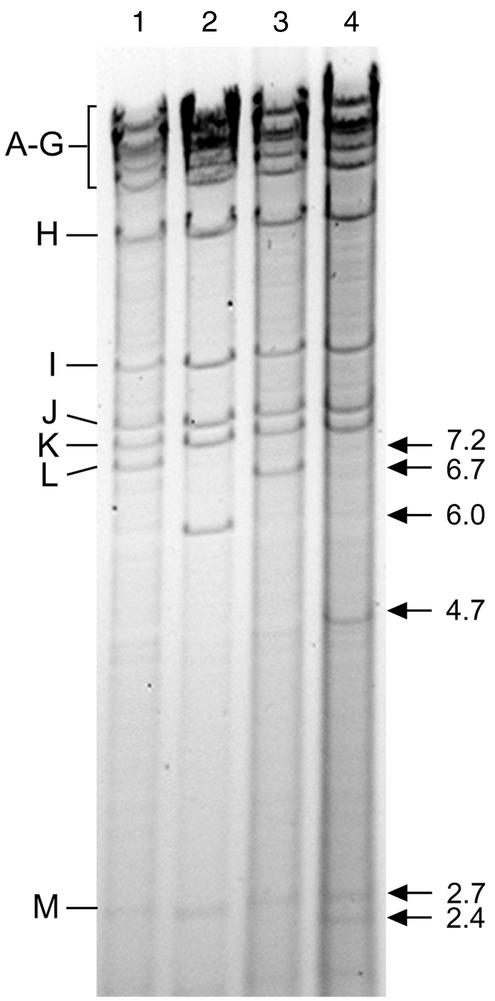
To verify the genomic structure of the recombinant BAC plasmids generated, their HindIII restriction patterns were analyzed. As shown in Fig. 2, the 7.2-kbp HindIII L fragment of the parental BAC plasmid C3X was replaced by a 6.0-kbp fragment in C3XdE (Fig. 2, compare lanes 1 and 2). Insertion of the stuffer sequences into the BAC plasmid C3XdE::Luc led to a shift of the 7.2-kbp HindIII L fragment to a novel fragment of 6.7-kbp (Fig. 2, lanes 1 and 3). Restoration of the enhancer sequences in the revertant BAC plasmid C3XdE::Luc-rev resulted in the appearance of two new fragments of 2.4 and 4.7 kbp (Fig. 2, compare lanes 1 and 4). These findings confirmed the successful deletion of the enhancer in C3XdE and C3XdE::Luc and the correct introduction of the stuffer and enhancer segments into the BAC plasmids C3XdE::Luc and C3XdE::Luc-rev, respectively. Additional restriction enzyme digest analyses (with EcoRI) indicated that the recombinant BAC plasmids generated were as expected and that no deletions or rearrangements had occurred elsewhere in the genome (data not shown).
FIG. 2.
Structural analysis of the enhancerless MCMV BAC genomes. DNAs of the HindIII-digested BAC plasmids C3X (lane 1), C3XdE (lane 2), C3XdE::Luc (lane 3), and C3XdE::Luc-rev (lane 4) after separation on a 0.7% agarose gel and ethidium bromide staining. The HindIII fragment names (9) are shown at the left. The sizes of the natural HindIII L and M fragments and the new HindIII L fragment for each BAC plasmid are indicated with arrows at the right.
We next sought to reconstitute and isolate viral progeny from the recombinant BAC constructs. Because in our previous experiments it became clear that enhancerless MCMV BAC genomes produce significantly fewer plaques after transfection into murine fibroblasts than wild type genomes, we examined whether reconstitution of enhancerless viruses from the recombinant BAC plasmids would be facilitated by the use of a complementing cell line expressing the MCMV IE proteins IE1 and IE3. To this end, NIH 3T3 cells were transfected with a recombinant plasmid, pBam25, which carries the MCMV ie1 and ie3 genes, and stable NIH 3T3-Bam25 cell lines were isolated as described in Materials and Methods. The parental and revertant BAC plasmids C3X and C3XdE::Luc-rev were transfected into NIH 3T3 cells, while the enhancerless BAC plasmids C3XdE and C3XdE::Luc were transfected into one of the NIH 3T3-Bam25 cell lines. In all four cases, numerous plaques developed ∼5 to 6 days posttransfection, and the infection spread rapidly throughout the cultures, reaching a complete cytopathic effect ∼2 to 3 days later. These results are in agreement with our previous studies, indicating that the growth defect of enhancerless virus is due to a lack of transacting IE protein activity. The supernatants derived from these cultures were used to infect new cell monolayers, the corresponding viruses were subsequently purified by three rounds of plaque purification, and viral stocks were generated. For enhancerless MCMV recombinants, NIH 3T3-Bam25 cells were used throughout the amplification and purification steps. The viruses generated were named MCMV (derived from C3X), MCMVdE (derived from C3XdE), MCMVdE::Luc (derived from C3XdE::Luc), and MCMVdE::Luc-rev (derived from C3XdE::Luc-rev).
The complementing cell line contains sequences from nt 176441 to 187035 of the MCMV genome, and the deletion of the enhancer in the enhancerless genomes spans nt −48 to −1191. Therefore, there are sequences in the NIH 3T3-Bam25 cell line homologous to regions directly flanking the enhancer segment. Homologous recombination between the shared sequences could potentially result in the generation of revertant viruses due to the restoration of the enhancer segment. In order to analyze the genomic structure of the reconstituted mutants, and to confirm that during the replication process in the complementing cell line the enhancerless viruses did not acquire the missing enhancer sequences, total cell DNA was isolated from infected cells and subjected to HindIII digestion. In all four cases, the DNA patterns of the viral genomes correctly matched the predicted ones and proved to be identical to those of the corresponding BAC plasmids (data not shown). Thus, with the help of a complementing cell system we were able to isolate and produce stocks of mutant MCMV lacking the enhancer with which to analyze the biological phenotype.
Growth rates of enhancerless MCMV recombinants in tissue culture: an evaluation in cis and trans complementation systems (in a functional context).
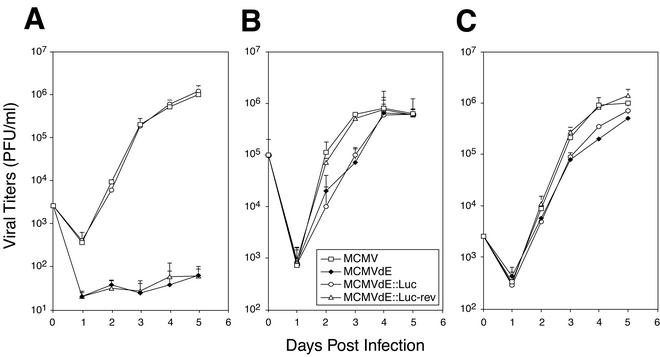
The fact that we could propagate and obtain relatively high titers of enhancerless mutants with the aid of the complementing cell line allowed us to characterize in more detail the growth properties of the enhancerless mutants in tissue culture. To this end, viral replication was examined after both high- and low-MOI infections of NIH 3T3 cells and compared to that exhibited by the parental or revertant virus. First, NIH 3T3 cells were infected with recombinant MCMVs at an MOI of 0.01, and viral titers were determined by standard plaque assays at various times postinfection. As shown in Fig. 3A, the amounts of virus that could be detected in the supernatants of the cultures infected with MCMVdE or MCMVdE::Luc were very small. Viral titers of ∼8 × 101 to 1 × 102 PFU/ml were obtained by day 5 postinfection. These results were consistent with previous data that documented a reduced efficacy after transfection of enhancerless BAC genomes into permissive fibroblasts compared to parental or revertant plasmids (2). In contrast, cultures that were infected with viruses that contained the enhancer region displayed a rapid increase in viral titers. Viral yields of ∼106 PFU/ml were reached in these cultures 5 days after infection. The growth kinetics of the revertant virus MCMVdE::Luc-rev were indistinguishable from those of the parental virus, demonstrating that reinsertion of the enhancer region led to complete rescue of the growth phenotype. These results ruled out the possibility that other regions outside the enhancer were responsible for the replication defect associated with the enhancer-deficient viruses, MCMVdE and MCMVdE::Luc, under low-MOI conditions. To document further the replication deficiency associated with enhancerless viruses under these conditions, we calculated from the growth kinetics the exponential-growth rate constants for the four recombinant viruses as described in Materials and Methods. While the exponential-growth rate constants were 3.17 days−1 and 3.05 days−1 for parental and revertant MCMV, respectively, the corresponding values for enhancerless MCMVs proved to be 0.47 days−1 for MCMVdE and 0.40 days−1 for MCMVdE::Luc. Thus, a marked reduction of almost 10-fold in the rate constant of exponential growth was observed after resection of the enhancer segment of MCMV, resulting in >104-fold difference in the production of infectious viral particles per milliliter. Altogether, therefore, these data indicate that the enhancer region makes a vital contribution to accelerating productive infection when a low input of virus is used.
FIG. 3.
Growth curve analysis of enhancerless MCMV mutants in tissue culture. NIH 3T3 cells (A and B) or NIH 3T3-Bam25 cells (C) were infected at an MOI of 0.01 (A and C) or 3 (B) PFU/cell with parental MCMV, MCMVdE, MCMVdE::Luc, and MCMVdE::Luc-rev. At the indicated time points after infection, supernatants from the infected cultures were harvested, and titers were determined on monolayers of NIH 3T3-Bam25 cells. The error bars indicate the standard deviation of the viral titers from three separate cultures.
To examine whether the inefficient growth displayed by enhancerless viruses could be overcome by increasing the virus dose, NIH 3T3 cells were infected at an MOI of 3. In these single-step growth analyses, both MCMVdE and MCMVdE::Luc presented slightly lower titers than enhancer-containing viruses (<1-log-unit difference in this experiment) by day 2 or 3 after infection (Fig. 3B). The differences in growth levels between enhancer-deficient and enhancer-containing viruses were statistically significant (P < 0.01, as determined by Student's test) at the time point where the maximum difference was observed. However, the final viral titers reached in cultures infected with enhancerless MCMV were similar to those obtained after infection with wild-type or revertant MCMV (Fig. 3B). In agreement with these observations, the exponential-growth rate constants calculated under these high-MOI conditions were 3.37 days−1 for parental MCMV, 3.15 days−1 for MCMVdE::Luc-rev, 2.23 days−1 for MCMVdE, and 2.44 days−1 for MCMVdE::Luc. Thus, when high input doses of enhancerless viruses were used, growth constants at the exponential phase were very close (1.4- to 1.5-fold difference) to the ones exhibited by enhancer-containing MCMVs. These findings suggest that enhancer sequences from nt −48 to −1191 are dispensable for replication in cultured murine fibroblasts under high-MOI conditions.
We next carried out growth analysis on the NIH 3T3-Bam25 cell line to examine the extent to which the IE protein-complementing system could restore replication of the enhancerless mutants under low-MOI conditions. When NIH 3T3-Bam25 cells were infected at an MOI of 0.01 PFU/cell, MCMVdE and MCMVdE::Luc grew with kinetics that, although not identical, were comparable to those exhibited by wild-type and revertant MCMVs (Fig. 3C). Altogether, these data clearly indicate that the enhancerless virus phenotype is multiplicity dependent and that the rate constant defect and impaired growth exhibited under low-MOI conditions can be mostly rescued by trans-complementation with IE protein expression.
Absence of MCMV-induced pathogenicity in SCID mice infected with enhancer-deficient MCMV.
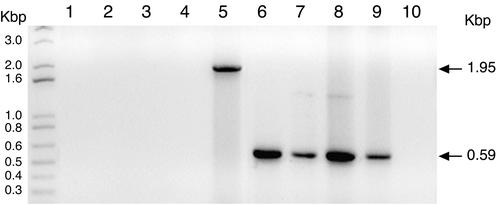
In the next set of experiments, we sought to address the role and biological significance of the enhancer in vivo. The parental MCMV BAC plasmid, C3X, used for the generation of the mutants described in this study contains the full-length MCMV genome. In addition, the MCMV BAC plasmid contains an insertion of BAC vector sequences flanked by short identical viral sequences that serve as a substrate for homologous recombination during virus reconstitution in cells, thus allowing the elimination of all nonviral sequences. Excision of the BAC vector via the duplicated sequences can be achieved in an accurate and reproducible manner after five virus passages (36). Although the BAC sequences within the MCMV genome have been shown not to alter viral replication in tissue culture, they might have some unpredictable effects during the course of an infection in vivo. We therefore examined whether the viral stocks generated, after the cloning and amplification steps to which they were subjected, had appropriately excised the BAC vector sequences. For this purpose, PCRs were performed using two different primer sets with the recombinant viral genomes as templates (36; see Materials and Methods for further details). First, reactions were carried out with a primer that binds in the BAC vector sequences and a second primer that anneals in the MCMV EcoRI fragment. This primer pair should yield a 1,950-bp PCR fragment if the BAC vector segment was present within the viral genomes or fail to amplify a product in the absence of these BAC sequences. As expected, a specific PCR-amplified product was not obtained with this primer set when the DNA from the recombinant viral stocks was used as a template (Fig. 4, lines 1 to 4). In contrast, a 1,950-bp fragment could be detected in a DNA preparation isolated from an early MCMVdE::Luc passage, examined before all the cloning and amplification steps (Fig. 4, line 5). Thus, the data suggested that BAC-containing genomes were not present in the recombinant viral stocks. Next, reactions were performed with a set of primers that bind to viral sequences flanking the BAC excision site. This second primer set should either fail to detect a product in the recombinant viral genomes if the BAC sequences were present or yield a 590-bp product in cases where the BAC vector had been appropriately excised. As shown in Fig. 4, when DNA isolated from any of the four viral recombinant stocks was used as a template in the reactions, the expected 590-bp fragment was obtained (lanes 6 to 9), while no specific amplified product was detected when DNA from an early MCMVdE::Luc passage was used (lane 10). Together, these results confirmed that our fibroblast-passaged recombinant MCMV stocks were homogeneous and their genomes were free of BAC vector sequences.
FIG. 4.
Excision of the BAC vector sequences from enhancerless MCMV genomes. PCRs were performed using as templates genomic DNAs isolated from viral stocks of MCMV (lanes 1 and 6), MCMVdE (lanes 2 and 7), MCMVdE::Luc (lanes 3 and 8), and MCMVdE::Luc-rev (lanes 4 and 9) or from an early MCMVdE::Luc passage (lanes 5 and 10). Two different primer sets were used to examine the correct excision of the BAC vector sequences within the viral genomes. The first primer set contains a primer that anneals with the BAC sequence and a primer located with the EcoRI g fragment (lanes 1 to 5), and the second primer set contains two primers that bind to viral sequences flanking the excision site (lanes 6 to 10). The amplified products were separated on a 1% agarose gel and visualized by ethidium bromide staining. Shown are the products obtained in the PCR. The positions of the size markers are shown at the left. The sizes of the amplified products are indicated by arrows.
In initial experiments in which BALB/c mice were infected with enhancerless viruses, we failed to detect any symptoms of MCMV-associated disease or to recover any enhancerless infectious virus (data not shown). To determine the extent to which deletion of the enhancer affects viral pathogenesis and replication in vivo, we turned to an extremely susceptible model for MCMV infection, CB17 SCID mice. The selection of such a system was based on the defective phenotype displayed by enhancerless MCMVs in vitro under low-MOI conditions and the fact that SCID mice are highly sensitive to low levels of infectious virus. CB17 SCID mice, which are devoid of Ly49-NK cells and functional T and B lymphocytes, are incapable of efficiently controlling an MCMV infection, succumbing to as few as 1 PFU (30, 31). We first studied the pathogenicities of the enhancerless MCMV recombinants in these immunodeficient animals. For this pupose, histological examination of selected key organs following infection with enhancerless viruses was carried out. SCID mice were injected intraperitoneally with 6 × 105 PFU of enhancerless, parental, or revertant MCMV. Fourteen days postinfection, the mice were sacrificed, and their livers, spleens, lungs, and salivary glands were harvested. Tissue sections were prepared and stained with hematoxylin and eosin for morphological examination.
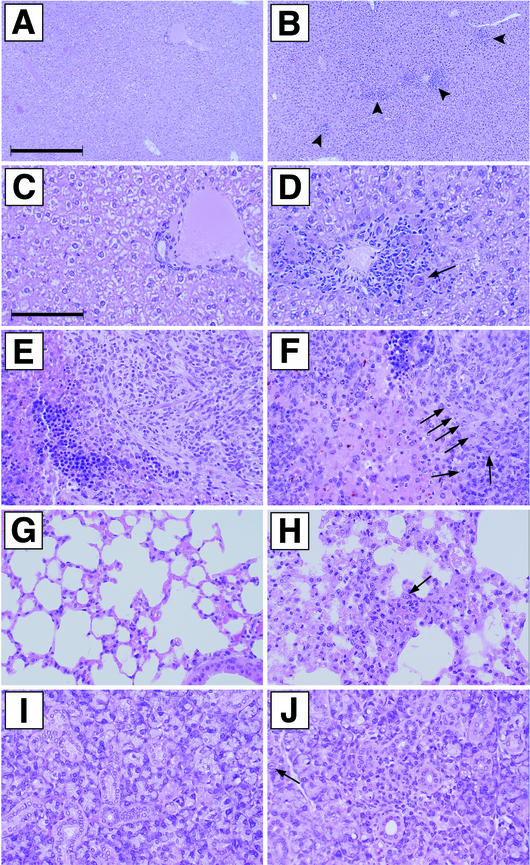
Sections of the livers were analyzed first. As expected, CMV infection-associated lesions were detected in the livers of wild-type and revertant MCMV recombinant-infected mice (Fig. 5B and D [revertant virus] and data not shown [wild-type virus]). The lesions generally were characterized by the presence of multifocal neutrophilic infiltrates, occasionally associated with large cells with eosinophilic intranuclear Cowdry type A inclusions, as well as eosinophilic intracytoplasmic inclusions (cytomegalic cells [Fig. 5B]). In addition, in the livers of affected animals, the inflammation was often correlated with individual hepatocellular necrosis (Fig. 5D). In marked contrast, the livers of mice infected with enhancerless virus were essentially normal (Fig. 5A and C [MCMVdE::Luc] and data not shown [MCMVdE]), and signs of CMV infection were not detected.
FIG.5.
Histological examination of livers, spleens, lungs, and salivary glands from mice infected with enhancerless MCMV. Shown are sections from livers (A to D), spleens (E and F), lungs (G and H), and salivary glands (I and J) of CB17 SCID mice 14 days after infection with 6 × 105 PFU of tissue culture-passaged MCMVdE::Luc (A, C, E, G, and I) or MCMVdE::Luc-rev (B, D, F, H, and J). (A, C, E, G, and I) No lesions present. (B) Multifocal inflammatory foci (marked by arrowheads). (D) Note cytomegalic cell with inclusion (arrow). The number of inflammatory foci present in liver sections from MCMVdE::Luc-rev-infected animals was 23 ± 10 per mm2. (A and B) Overview; (C and D) details of portions of A and B, respectively. (F, H, and J) Note the cytomegalic cells (indicated by arrows). (F) Neutrophilic infiltrate in splenic white pulp. (H) Lung alveolar septal thickening with fibrin and inflammatory cells. (J) Salivary gland focal necrosis with inflammation. Bars, 400 (for A and B) and 100 (for C through J) μm. Magnification, ×25 (A and B) and ×100 (C through J).
When spleens, lungs, and salivary glands of mice infected with parental and revertant viruses were analyzed, damage induced by CMV infection was detected in the three organs (Fig. 5F, H, and J [revertant MCMV] and data not shown [parental MCMV]). Again, lesions were associated with the presence of multifocal neutrophilic infiltrates, occasionally correlated with large cells with eosinophilic intranuclear Cowdry type A inclusions, as well as cytomegalic cells. Both the white pulp and the red pulp of the spleens from these animals were affected (Fig. 5F). The lungs were characterized by multifocal interstitial and intra-alveolar neutrophilic infiltrates and focally increased pulmonary macrophage numbers (Fig. 5H). In the salivary glands, inflammation was associated with focal epithelial necrosis, while cytomegalic cells at times were located away from the inflammatory infiltrates (Fig. 5J). In contrast, signs of CMV infection in these three organs were not detected in mice infected with enhancerless virus (Fig. 5E, G, and I [MCMVdE::Luc] and data not shown [MCMVdE]). Consistent with these results, we were unable to detect damage induced by CMV after examination of the livers, spleens, lungs, and salivary glands of mice infected for 7 days with enhancerless MCMV (data not shown). From all these data together we conclude that the enhancer region significantly contributes to MCMV pathogenesis.
Loss of pathogenicity of enhancerless MCMV recombinants is correlated with an inability to establish a productive infection in vivo.
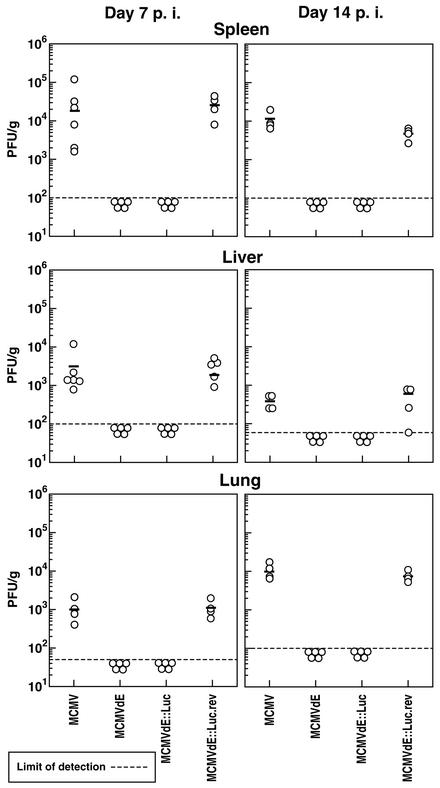
We next sought to analyze whether the lack of virus-induced damage after infection by enhancerless MCMV was due to the inability of these viruses to replicate in CB17 SCID mice. For these experiments, animals were injected intraperitoneally with 6 × 105 PFU of enhancerless, revertant, or parental MCMV. At 7 days postinfection, spleens, livers, and lungs were harvested, and virus titers in these three target organs for MCMV replication were determined on NIH 3T3-Bam25 cells. As shown in Fig. 6, under these conditions of infection, the parental MCMV exhibited titers of ∼2 × 104, 3 × 103, and 1 × 103 PFU/g in the spleen, liver, and lungs, respectively. Conversely, replication of the enhancerless viruses, MCMVdE and MCMVdE::Luc, could not been detected in these three organs (Fig. 6). As expected, the revertant virus MCMVdE::Luc-rev was found to replicate at levels similar to those of the parental MCMV in the three organs, confirming that the enhancerless phenotype was indeed due to the absence of the enhancer sequences. Titers of revertant virus in the spleens, livers, and lungs of the SCID mice were ∼3 × 104, 2 × 103, and 1 × 103 PFU/g, respectively (Fig. 6). To further extend this analysis and to examine whether a delayed infection was responsible for the defective growth observed for enhancerless virus, we determined viral titers in spleens, livers, and lungs 14 days after infection of SCID mice with the recombinant viruses. At this time point, similar to what was observed at day 7 postinfection, the titers of enhancerless MCMV recombinants were below the levels of detection, while parental MCMV exhibited significant viral titers in these three organs (Fig. 6). Again, the growth properties of the revertant virus MCMVdE::Luc-rev were very similar to those of the parental MCMV. Consistent with the histological analysis, we were unable to detect infectious virus in the salivary glands of the immunocompromised mice 14 days after infection with MCMVdE or MCMVdE::Luc (data not shown). Therefore, these results indicate that the lack of MCMV-induced pathology correlates with lack of replication, revealing that the enhancer region of MCMV is a major determinant of viral replication in vivo, at least in the target organs analyzed.
FIG. 6.
Growth of enhancerless MCMV mutants in the spleens, livers, and lungs of SCID mice. Groups of SCID mice (CB17; four to six per group) were inoculated intraperitoneally with 6 × 105 PFU of tissue culture-propagated MCMV, MCMVdE, MCMVdE::Luc, and MCMVdE::Luc-rev. The mice were sacrificed on day 7 or 14 postinfection (p. i.), and their spleens, livers, and lungs were harvested for determination of viral titers. The organs were weighed and sonicated as a 10% (wt/vol) tissue homogenate in DMEM. The viral titers of the resulting homogenates were determined on NIH 3T3-Bam25 cells. Titers corresponding to the spleen, liver, and lungs from each individual mouse within a group are shown. The dashed lines represent the limits of detection. The horizontal bars indicate the mean values.
Enhancerless MCMV recombinants are severely attenuated.
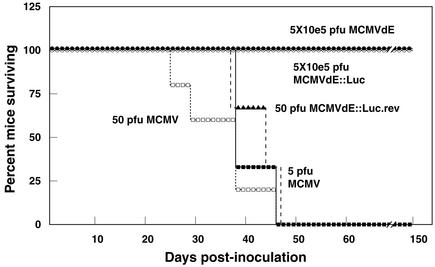
The experiments detailed above indicated that recombinant MCMVs defective in the enhancer region are severely impaired in the ability to grow in at least four of the major organs targeted during CMV infection. Because the enhancer determines the efficiency of replication, we could not rule out from these experiments the existence of enhancerless virus that could be either replicating in sites other than the spleen, liver, lungs, and salivary glands or present at very low levels in these organs (below the limit of detection of the plaque assays) and that could eventually lead to CMV disease in the host. We therefore sought to examine the extent to which enhancerless MCMVs were attenuated. For this purpose, the virulence of enhancerless MCMV was compared to that of parental or revertant MCMV by monitoring the kinetics of lethality in SCID mice. Since it has been documented that 1 PFU of MCMV is sufficient to kill a SCID mouse, we assessed the virulence of enhancerless viruses by infecting these animals with increasing concentrations of the recombinant viruses. Mice were intraperitoneally infected with 5 × 100, 5 × 101, 5 × 102, 5 × 103, 5 × 104, and 5 × 105 PFU of enhancerless MCMVs, and survival was monitored daily after infection. The mice in control groups were infected with 5 or 5 × 101 PFU of parental MCMV. As shown in Fig. 7, 100% of the mice infected with the parental MCMV died within <50 days after infection. However, all the animals infected with enhancerless viruses, with or without the stuffer sequences, survived the infection for at least 150 days, the time at which the experiment was terminated for most of the groups (Fig. 7). Animals infected with 5 × 105 PFU of MCMVdE and MCMVdE:Luc were further monitored for a total period of 365 days. During this time, all infected mice remained alive, and infectious enhancerless virus could not be recovered from their main target organs (spleen, lung, liver, and salivary glands) at the time the animals were finally sacrificed (1 year after infection; data not shown). Therefore, even after inoculation with 5 × 105 PFU of MCMVdE or MCMVdE::Luc, a dose 105-fold higher than the dose of wild-type MCMV that killed 100% of the mice, none of the animals succumbed to the infection. Restoration of the enhancer region in the revertant virus led to the rescue of the virulence phenotype. Fifty PFU of MCMVdE::Luc-rev was sufficient to kill all the SCID mice within <47 days (Fig. 7). Thus, this analysis provides strong evidence that enhancerless viruses are severely attenuated (>10,000-fold difference) in the SCID mouse model. Based on these experiments, we conclude that the enhancer is indispensable for establishing a productive infection in vivo.
FIG. 7.
Attenuation of enhancerless MCMV mutants in SCID mice. Groups of SCID mice (CB17; three to four mice per group) were inoculated intraperitoneally with the indicated amounts of either parental MCMV, MCMVdE, MCMVdE::Luc, or MCMVdE::Luc-rev and monitored daily for survival. One hundred percent of the mice that received 5 × 100, 5 × 101, 5 × 102, 5 × 103, or 5 × 104 PFU of MCMVdE or MCMVdE::Luc survived the infection for the time the experiment lasted (150 days; data not shown).
DISCUSSION
To precisely define the role that the CMV enhancer plays in viral growth and pathogenesis, we constructed two recombinant viruses with a deletion of the complete enhancer region of the MIEP but leaving intact the TATA box region and associated downstream promoter-proximal regulatory sequences. In this report, we show that the enhancer plays a critical role in determining the rate of virus multiplication. Enhancerless viruses exhibit an MOI-dependent phenotype on permissive NIH 3T3 cells. Under low-MOI conditions, there is a strong requirement for viral replication, while at high MOI the enhancer is dispensable for growth. Remarkably we found that genetic resection of the enhancer results in a CMV that is completely disabled in vivo. Enhancerless viruses are nonpathogenic and highly attenuated in immunocompromised SCID mice. The results presented define the role of the enhancer in viral multiplication and pathogenesis, contributing to a more complete understanding of the key regulatory mechanisms that operate during CMV growth.
Previously, the generation of enhancerless BAC genomes and the possibility of recovering viral progeny after transfection into permissive cells were reported (2). However, the recombinant genomes constructed contained BAC vector sequences replacing an array of genes within the HindIII fragment E′ (open reading frames m151 to m158), shown to be dispensable for viral replication on murine fibroblasts but required for appropriate in vivo CMV growth (2, 36). Because a major goal of this study was the analysis of the enhancer region during infection in the natural host, we used a full-length MCMV BAC genome (C3X) to construct two new recombinant BAC genomes (MCMVdE and MCMVdE::Luc) with a deletion of the enhancer region (base positions −48 to −1191 with respect to the +1 start site of MIE RNAs). In addition, since our earlier work indicated an important function of this region during lytic infection, we generated a stable cell line expressing the MCMV IE proteins IE1 and IE3 and used it for the isolation of the enhancer-deficient mutants. A relevant feature of the enhancerless recombinant viruses obtained after transfection in the stable cell line is an MOI dependence for growth on permissive NIH 3T3 cells (Fig. 3A and B). These viruses grow poorly in cells infected with single virus particles, but high doses of virus can compensate for the most part for the growth defect caused by the absent sequences at low MOI. The facts that two different types of enhancerless virus exhibit identical behavior and that restoration of the enhancer sequences in the revertant virus MCMVdE::Luc-rev rescues its ability to grow in vivo indicate that this phenotype is the result of the engineered enhancer deletion. In addition, the enhancerless phenotype cannot be attributed to the loss of the putative open reading frames m124, m124.1, and m125, contained in the deleted enhancer sequences, since a hybrid virus in which the native MCMV enhancer is replaced by the HCMV enhancer exhibits infectious kinetics identical to those of the parental wild-type MCMV in cultured fibroblasts and is permissive in vivo (2, 13; A. Angulo and P. Ghazal, unpublished data).
The behavior displayed by enhancer-deficient viruses is remarkably similar to the growth phenotype reported for other herpesvirus mutants carrying lesions within the MIE loci. For CMV, these include mutant viruses unable to express IE1 (12, 27) or containing deletions in the distal MIEP HCMV enhancer (23). This MOI-dependent growth phenotype is in agreement with the model of IE protein function which suggests that the levels of IE proteins determine the outcome of a productive CMV infection in permissive cells. In this connection, the MCMV IE3 transactivator protein has been shown to be essential for progression to the early and late phases of infection (1). Therefore IE3 is required to be expressed at sufficient levels in order to push the MCMV replication cycle forward. Of the many potential roles that the CMV enhancer can play, it is clear that one of the most prominent functions is to augment viral-gene expression in order to achieve IE1 and IE3 levels sufficient to guarantee efficient entry into the lytic cycle. Thus, under low-MOI conditions, the enhancing effect of the enhancer is most likely revealed because MIE gene expression levels are below the threshold required to get replication started. However, this control region might be completely dispensable when increasing concentrations of virus are present because under these conditions sufficient amounts of MIE products are synthesized. This notion is supported in our system by the results obtained in the complementing cell line, in which expression of IE1 and IE3 in trans reverses the enhancerless replication deficit to a significant degree (Fig. 3C). Along this line, recent work by Meier et al. has shown that UV-inactivated HCMV virions at a high particle/cell ratio can overcome the defects exhibited by a virus with the distal enhancer deleted when grown at a low MOI (25). In this case, virion-associated components most likely augment MIE enhancer promoter activity, increasing MIE gene expression above threshold levels and compensating for the missing distal enhancer sequences. On the other hand, studies with the rat CMV enhancer swap have revealed a lack of correlation between IE expression levels and viral growth in permissive cells, reflecting the complexity and multivalency of this region and manifesting distinct characteristics of the rat CMV enhancer (33). Indeed, it is possible that the enhancer could play a more direct role in promoting the rate of firing of the origin of replication and in this capacity may be uncoupled from the control of lytic gene expression. Clarification of the mode of action of the enhancer will require further studies.
The impact of the enhancer region on the ability of CMV to cause disease and eventually death was primarily explored in SCID mice. In this model system, MCMV causes a systemic infection replicating to high viral titers and inducing damage in a variety of organs that include the lungs, liver, spleen, adrenal glands, and kidneys. In this setting, viral replication is not subjected to tight control and animals eventually succumb to as little as 1 PFU of MCMV. Thus, the selection of this mouse model system to evaluate the activity of enhancerless viruses was based on the high degree of sensitivity exhibited by the SCID mouse to infection by MCMV. In initial experiments, we failed to detect infectious virus in a variety of organs of immunocompetent BALB/c mice after infection with enhancerless viruses (data not shown). Three lines of evidence firmly support the notion that the enhancer region is indispensable during the course of acute MCMV infection. First, histological examination of a variety of organs of MCMVdE- and MCMVdE::Luc-infected animals resulted in the absence of detectable MCMV-induced associated damage (Fig. 5). Second, enhancerless viruses failed to replicate to detectable levels in the major target organs for CMV replication (Fig. 6). And finally, MCMVdE and MCMVdE::Luc were unable to kill SCID mice even when the animals were infected at a dose 105 times higher than the dose of parental MCMV required to cause death (Fig. 7). The finding that two different enhancerless viruses, MCMVdE and MCMdE::Luc, exhibited identical behavior, together with the fact that repairing this region in MCMVdE::Luc-rev restored virus-induced damage and virulence to the wild-type level, indicate that this attenuated phenotype can be attributed to the deletion of the enhancer.
Due to the robust growth of enhancerless virus in cultured cells at high MOI, the absolute requirement for the enhancer for CMV replication in immunocompromised mice was not anticipated. Our in vivo results using high viral doses do not even parallel events occurring in cells infected with single virus particles. Thus, the residual activity of enhancerless viruses exhibited in permissive NIH 3T3 cells and mouse embryonic fibroblasts (data not shown) under low-MOI conditions does not seem to be sufficient to establish or maintain low-level replication in vivo. It is noteworthy that while the SCID mice are deficient in T, B, and NK (Ly49) cells, they retain a certain degree of innate immunity. Thus, one possible corollary from these observations is that the enhancer plays a critical role in driving an infection past the initial innate response of SCID mice. Alternatively, it is possible that enhancerless virus may manifest a more cell-type-specific effect, such as replication deficiency in macrophages, that severely compromises dissemination and the establishment of productive infection. However, when injected intravenously into SCID mice, enhancerless viruses failed to replicate to detectable levels in the spleen and liver during a 3-day period (data not shown). Nevertheless, in our studies, we cannot rule out the presence of low levels of enhancerless virus replication (below our levels of detection) in certain cell types in SCID mice, which would be kept in check by T- and B-lymphocyte- and NK cell-independent mechanisms. In this sense, the use of alternative and more sensitive techniques to detect infected cells in mouse tissues, such as the one described by Grzimek and coworkers (13), might be of great help in addressing this issue. Finally, one has to consider the limitations of the animal model used in this study and the possibility that enhancerless viruses would replicate to some extent in the complete absence of immune functions.
Due to the strict species specificity associated with CMV, the study of MCMV in its natural host has been extensively used to address many aspects of HCMV pathogenesis. Whether the observations reported here in the murine model might be extended to the HCMV enhancer remains an open question. While different at the primary sequence level, MCMV and HCMV enhancers share many of the same regulatory elements (such as NF-κB, retinoic acid receptors, AP-1, and CREB/ATF). At the in vitro level, MCMV and HCMV containing deletions in the enhancer region exhibit very similar phenotypes (23). Furthermore, the fact that the human enhancer can replace the natural MCMV enhancer-promoter in vitro and in vivo suggests a functional conservation of this control region in the two species (2, 13). These findings, in concert with the results presented here demonstrating the lack of residual ability of enhancer-deficient genomes to replicate in vivo, establish the basis for the use of enhancer swap strains as tools for the identification of functional HCMV enhancer elements in the context of mouse infection. Viral mutants containing subtle mutations in the transcription binding sites within this control region can now be engineered in order to dissect the contributions that the different signaling pathways may make to CMV pathogenesis.
In summary, our results clearly document an absolutely essential role of the MCMV enhancer during acute infection. These findings indicate that this regulatory region might be a good target for antiviral therapy, as blocking enhancer activity should eliminate or lessen the burden of CMV infection and disease. The severe attenuated phenotype suggests novel vaccine approaches or gene therapy agents. Further studies will be required to thoroughly understand how this complicated control region operates.
Acknowledgments
We thank Michelle Griffiths and Andrea Reus for excellent technical assistance. We also thank Thorsten Forster for advice on the determination of viral growth constants.
This work was supported in part by grants AI-30627 (P.G.) and AI-44851 (A.A.) from the National Institutes of Health; by Sonderforschungsbereich 455, project A2 (M.M.); and by the Wellcome Trust (P.G.). A.A. is a fellow from the Ramón y Cajal Research Program.
REFERENCES
- 1.Angulo, A., P. Ghazal, and M. Messerle. 2000. The major immediate-early gene ie3 of mouse cytomegalovirus is essential for viral growth. J. Virol. 74:11129-11136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angulo, A., M. Messerle, U. H. Koszinowski, and P. Ghazal. 1998. Enhancer requirement for murine cytomegalovirus growth and genetic complementation by the human cytomegalovirus enhancer. J. Virol. 72:8502-8509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baskar, J. F., P. P. Smith, G. S. Climent, S. Hoffman, C. Tucker, D. J. Tenney, A. M. Colberg-Poley, J. A. Nelson, and P. Ghazal. 1996. Developmental analysis of the cytomegalovirus enhancer in transgenic animals. J. Virol. 70:3215-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baskar, J. F., P. P. Smith, G. Nilaver, R. A. Jupp, S. Hoffmann, N. J. Peffer, D. J. Tenney, A. M. Colberg-Poley, P. Ghazal, and J. A. Nelson. 1996. The enhancer domain of the human cytomegalovirus major immediate-early promoter determines cell type-specific expression in transgenic mice. J. Virol. 70:3207-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boshart, M., F. Weber, G. Jahn, K. Dorsch-Hasler, B. Fleckenstein, and W. Schaffner. 1985. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell 41:521-530. [DOI] [PubMed] [Google Scholar]
- 6.Chang, Y. N., K. T. Jeang, T. Lietman, and G. S. Hayward. 1995. Structural organization of the spliced immediate-early gene complex that encodes the major acidic nuclear (IE1) and trans-activator (IE2) proteins of African green monkey cytomegalovirus. J. Biomed. Sci. 2:105-130. [DOI] [PubMed] [Google Scholar]
- 7.Cherrington, J. M., and E. S. Mocarski. 1989. Human cytomegalovirus ie1 transactivates the promoter-enhancer via an 18-base-pair repeat element. J. Virol. 63:1435-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorsch-Hasler, K., G. M. Keil, F. Weber, M. Jasin, W. Schaffner, and U. H. Koszinowski. 1985. A long and complex enhancer activates transcription of the gene coding for the highly abundant immediate early mRNA in murine cytomegalovirus. Proc. Natl. Acad. Sci. USA 82:8325-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ebeling, A., G. M. Keil, E. Knust, and U. H. Koszinowski. 1983. Molecular cloning and physical mapping of murine cytomegalovirus DNA. J. Virol. 47:421-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghazal, P., and J. A. Nelson. 1991. Enhancement of RNA polymerase II initiation complexes by a novel DNA control domain downstream from the cap site of the cytomegalovirus major immediate-early promoter. J. Virol. 65:2299-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghazal, P., and J. A. Nelson. 1993. Transcription factors and viral regulatory proteins as potential mediators of human cytomegalovirus pathogenesis, p. 360-383. In Y. Becker, G. Darai, and E.-S. Huang (ed.), Molecular aspects of human cytomegalovirus diseases. Springer-Verlag, Heidelberg, Germany.
- 12.Greaves, R. F., and E. S. Mocarski. 1998. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low multiplicity of infection by a human cytomegalovirus ie1 mutant. J. Virol. 72:366-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grzimek, N. K. A., J. Podlech, H.-P. Steffens, R. Holtappels, S. Schmalz, and M. J. Reddehase. 1999. In vivo replication of recombinant murine cytomegalovirus driven by the paralogous major immediate-early promoter-enhancer of human cytomegalovirus. J. Virol. 73:5043-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honess, R. W., and B. Roizman. 1975. Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc. Natl. Acad. Sci. USA 72:1276-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keil, G. M., K. A. Ebeling, and U. H. Koszinowski. 1984. Temporal regulation of murine cytomegalovirus transcription and mapping of viral RNA synthesized at immediate early times after infection. J. Virol. 50:784-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keil, G. M., K. A. Ebeling, and U. H. Koszinowski. 1987. Sequence and structural organization of murine cytomegalovirus immediate-early gene 1. J. Virol. 61:1901-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koedood, M., A. Fichtel, P. Meier, and P. J. Mitchell. 1995. Human cytomegalovirus (HCMV) immediate-early enhancer/promoter specificity during embryogenesis defines target tissues of congenital HCMV infection. J. Virol. 69:2194-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurz, S. K., and M. J. Reddehase. 1999. Patchwork pattern of transcriptional reactivation in the lungs indicates sequential checkpoints in the transition from murine cytomegalovirus latency to recurrence. J. Virol. 73:8612-8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurz, S. K., M. Rapp, H.-P. Steffens, N. K. A. Grzimek, S. Schmalz, and M. J. Reddehase. 1999. Focal transcription activity of murine cytomegalovirus during latency in the lungs. J. Virol. 73:482-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, B., and M. F. Stinski. 1992. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1 cis-acting elements. J. Virol. 66:4434-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, R., J. Baillie, J. G. Sissons, and J. H. Sinclair. 1994. The transcription factor YY1 binds to negative regulatory elements in the human cytomegalovirus major immediate early enhancer/promoter and mediates repression in nonpermissive cells. Nucleic Acids Res. 22:2453-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchini, A., H. Liu, and H. Zhu. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meier, J. L., and J. A. Pruessner. 2000. The human cytomegalovirus major immediate-early distal enhancer region is required for efficient viral replication and immediate-early gene expression. J. Virol. 74:1602-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meier, J. L., and M. F. Stinski. 1996. Regulation of cytomegalovirus immediate early genes. Intervirology 39:331-342. [DOI] [PubMed] [Google Scholar]
- 25.Meier, J. L., M. J. Keller, and J. J. McCoy. 2002. Requirement of multiple cis-acting elements in the human cytomegalovirus major immediate-early distal enhancer for viral gene expression and replication. J. Virol. 76:313-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Messerle, M., B. Buhler, G. M. Keil, and U. H. Koszinowski. 1992. Structural organization, expression, and functional characterization of the murine cytomegalovirus immediate-early gene 3. J. Virol. 66:27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 29.Mocarski, E. S., G. W. Kemble, J. M. Lyle, and R. F. Greaves. 1996. A deletion mutant in the human cytomegalovirus gene encoding IE1491aa is replication defective due to a failure in autoregulation. Proc. Natl. Acad. Sci. USA 93:11321-11326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okada, M., and Y. Minamishima. 1987. The efficacy of biological response modifiers against murine cytomegalovirus in normal and immunodeficient mice. Microbiol. Immunol. 31:45-57. [DOI] [PubMed] [Google Scholar]
- 31.Pollock, J. L., and H. W. Virgin III. 1995. Latency, without persistence, of murine cytomegalovirus in the spleen and kidney. J. Virol. 69:1762-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandford, G. R., and W. H. Burns. 1996. Rat cytomegalovirus has a unique immediate early gene enhancer. Virology 222:310-317. [DOI] [PubMed] [Google Scholar]
- 34.Sandford, G. R., L. E. Brock, S. Voight, C. M. Forester, and W. H. Burns. 2001. Rat cytomegalovirus major immediate-early enhancer switching results in altered growth characteristics. J. Virol. 75:5076-5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stenberg, R. M. 1996. The human cytomegalovirus major immediate-early gene. Intervirology 39:343-349. [DOI] [PubMed] [Google Scholar]
- 36.Wagner, M., S. Jonjic, U. H. Koszinowski, and M. Messerle. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]