Abstract
The induction of the beta interferon (IFN-β) gene constitutes one of the first responses of the cell to virus infection. Its regulation is achieved through an intricate combination of virus-induced binding of transcription factors and local chromatin remodeling. In this work, we demonstrate that transcription factor YY1, known to interact with histone deacetylases (HDAC) and histone acetyltransferases, has a dual activator/repressor role during the regulation of the IFN-β promoter activity. We show that YY1 specifically binds in vitro and in vivo to the murine IFN-β promoter at positions −90 and −122. Overexpression of YY1 strongly repressed the transcriptional capacity of a stably integrated IFN-β promoter fused to a chloramphenicol acetyltransferase reporter gene as well as the endogenous IFN activity of murine L929 cells via an HDAC activity. Stably integrated IFN-β promoters mutated at the −90 site were no longer repressed by YY1, could no longer be activated by trichostatin A, displayed a retarded postinduction turn off, and a reduced virus-induced activity. Introduction of a mutation at the −122 site did not affect YY1-induced repression, but promoters with this mutation displayed a reduced virus-induced activity. Stably integrated full-length promoters (from position −330 to +20) mutated at both YY1-binding sites displayed extremely reduced promoter activities. We conclude that YY1 has a dual activator/repressor role on IFN-β promoter activity depending on its binding site and time after infection.
Beta interferon (IFN-β) plays a key role modulating antiviral response (8, 32). In the absence of external stimuli, the IFN-β gene is maintained in a constitutive transcriptionally silent state while this gene is transiently activated after virus infection (37). As is the case for many other environmentally stimulated genes, the transcriptional regulation of the IFN-β gene is achieved through a complex mechanism during which specific transcription factors as well as chromatin and chromatin-remodeling complexes intervene (1, 28, 36). In a recent work, it was demonstrated that histone deacetylation participates in the establishment of the repressed state of the IFN-β promoter (30). Inhibition of histone deacetylase (HDAC) activity with trichostatin A (TSA) led to the local acetylation of histone H4 tails positioned on the IFN-β promoter region, enhanced the transcriptional capacity of this promoter, and induced an antiviral state to murine fibroblastic L929 cells infected by vesicular stomatitis virus.
Nuclear HDACs deacetylate nucleosomal core histone tails, establishing a locally condensed chromatin structure associated with gene silencing (38). Three classes of nuclear HDACs have been described. The first class includes mammalian HDAC1, HDAC2, and HDAC3, which are highly homologous to the yeast repressor protein Rpd3 (6) and characterized as almost exclusively present in the nucleus. The second class includes mammalian HDAC4, HDAC5, and HDAC6, which are homologous to yeast Hda1 (12) and are able to shuttle between the nucleus and the cytoplasm (23). The third class of HDACs are related to yeast repressor protein SIR2 (18). They differ from the other two classes in that they display NAD-dependent HDAC activity (16) and are often found in the nucleolus. HDACs do not bind directly to DNA but are recruited either directly or indirectly to specific promoters by transcription factors (38) and often function in large multiprotein complexes, such as mSin3A, NuRD (nucleosome remodeling histone deacetylase), or MeCP2 (7, 17, 38).
Protein Yin Yang 1 (YY1) is a transcription factor that binds to DNA through the recognition of a specific consensus sequence and directly interacts with HDACs. YY1 has been shown to bind in vivo to HDAC2 and in vitro to HDAC1, HDAC2, and HDAC3 (6). It is a ubiquitous, Krüppel-like, zinc finger transcription factor (2, 11, 34) known to either repress or activate a high number of genes, among which are c-Myc, c-Fos, β-casein, α-actin, histone H4, IFN-γ, interleukin 5, interleukin 3, adeno-associated virus P5 promoter, human papillomavirus type 16 and 18, Moloney murine leukemia virus, and several other cellular or viral genes (10, 13, 31, 34, 40). The targeted disruption of the mouse YY1 gene is lethal, demonstrating an essential function of this protein during the development of the mouse embryo (9).
A wide variety of transcription factors, such as c-Myc, SP1 (29), and E1A (20), as well as transcriptional corepressors, such as HDACs (as it was mentioned before), or coactivators like CBP, pCAF, and p300 bind to YY1 (34). Such protein-protein interactions can play a key role in establishing YY1 either as a repressor or as an activator, as it has been demonstrated in the case of the human immunodeficiency virus type 1 long terminal repeat, where YY1 acts as a repressor via the recruitment of HDAC1 (5). The intracellular YY1 concentration (4), promoter sequence environment (27), or YY1 posttranslational modifications (39) can also decide whether YY1 acts as a repressor or an activator of transcription.
In this work we show that YY1 specifically binds in vitro as well as in vivo to IFN-β promoter at positions −90 and −122. We demonstrate that YY1 plays a dual repressor/activator role in the transcriptional capacity of the IFN-β promoter depending on its binding site and on the moment after virus infection. Protein YY1 can itself be acetylated and deacetylated by histone acetyltransferases (HAT) and deacetylases (39). The degree of acetylation of YY1 can affect its DNA-binding capacity as well as the capacity of the protein to interact with HDACs. These characteristics of YY1 are discussed in the context of IFN-β gene regulation.
MATERIALS AND METHODS
Gel retardation assays.
Nuclear extracts of murine L929 cells were prepared by microextraction as described by Therrien and Drouin (33). Five micrograms of nuclear extracts was incubated with 1.5 μg of poly(dI-dC) in 20 μl (final volume) of 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 5 mM EDTA, 10% glycerol, and 5 mM dithiothreitol for 10 min at room temperature before adding the corresponding 5′ 32P-labeled probes (0.05 pmol). After adding the labeled probe, the mixture was incubated at room temperature for 15 min. Electrophoresis was carried out in an 8% polyacrylamide gel in 0.25× Tris-borate-EDTA. During competition experiments, the corresponding unlabeled DNA probes were added at the same time as the labeled probes. When indicated, 1 μg of anti-YY1 monoclonal H-10 antibodies (Santa Cruz no. 7341X) were added to the nuclear extracts prior to the addition of the labeled probes. Nuclear extracts and antibodies were incubated in ice for 40 min in 20 μl of 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 5 mM EDTA, 10% glycerol, 5 mM dithiothreitol in the presence of 1.5 μg of poly(dI-dC). After adding labeled probes, the samples were further incubated for 15 min at room temperature. Gel retardation assays carried out with recombinant HMGI protein were done as previously described (3).
Gel retardation assays followed by Western blotting.
Gel retardation assays were carried out as described above, except that 25 μg of nuclear extracts (instead of 5 μg) and 5 pmol of unlabeled DNA probe (instead of 0.05 pmol) were used. After migration, the gel was soaked in transfer buffer (50 mM Tris-HCl, 40 mM Gly, 1 mM sodium dodecyl sulfate [SDS], 20% methanol) for 40 min at room temperature before being submitted to Western blotting with Santa Cruz rabbit polyclonal anti-YY1 antibody (H-414) as the primary antibody.
Chromatin immunoprecipitation.
L929 wt330, mut122, and mut90 cells were fixed with 1% formaldehyde added to the medium for 10 min, scraped, and collected by centrifugation. Cells were resuspended in 0.1 ml of lysis buffer [5 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES, pH 8.0), 85 mM KCl, 0.5% NP-40] with 0.5 mM phenylmethylsulfonyl fluoride, 1 μg of pepstatin A/ml, and 1 μg of leupeptin/ml. Cells were pelleted by centrifugation and resuspended in 200 μl of 1% SDS, 10 mM EDTA, and 50 mM Tris-HCl (pH 8.0) containing protease inhibitors. After incubation on ice for 10 min, cells were sonicated 10 times for 10 s. Lysates were then cleared by centrifugation, and the concentration of DNA was determined. DNA was diluted 10-fold in dilution buffer (0.01% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], 150 mM NaCl). The chromatin solution was precleared for 45 min at 4°C on protein A-Sepharose 4B beads preadsorbed with sonicated single-stranded DNA (1 ml of a 50% suspension of protein A-Sepharose 4B beads plus 8 μl of sonicated 10-mg/ml single-stranded DNA). Corresponding aliquots of chromatin solution were then incubated with 5 μl of anti-YY1 (Santa Cruz no. 4703) or anti-HMGI (Santa Cruz no. 8982) antibodies overnight at 4°C. Immune complexes were collected on protein A beads preadsorbed with sonicated single-stranded DNA. Beads were washed sequentially in TSE (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1]) with 150 mM NaCl, TSE with 500 mM NaCl, buffer A (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl [pH 8.1]), and three times with Tris-EDTA and then extracted three times with 1% SDS and 0.1 M NaHCO3. Cross-links were reversed by heating at 65°C for 4 h, and DNA was precipitated with ethanol. Precipitates were resuspended in 20 μl of Tris-EDTA, digested with proteinase K (50 μg/ml for 1 h at 37°C), extracted with phenol-chloroform (1:1), and finally ethanol precipitated. PCR analysis of immunoprecipitated DNA was performed with the oligonucleotide F-40 (5′-GTT TTC CCA GTC ACG AC-3′, specific for the pBLCAT3 vector) as the 5′ primer and oligonucleotide CAT (5′-CCA TTT TAG CTT CCT TAG-3′, specific for the chloramphenicol acetyltransferase [CAT] reporter gene) as the 3′ primer. PCR conditions were as follows: 1 cycle of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min; 1 cycle of 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min; 1 cycle of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min; 20 cycles of 94°C for 1 min, 53°C for 1 min, and 72°C for 1 min; and 1 cycle of 72°C for 10 min.
Plasmid construction.
Plasmid constructions were carried out by double PCR as previously described (3) by using plasmid pBLCAT3-muIFN-β wt330 as a template. This plasmid carries the wild-type murine IFN-β (muIFN-β) promoter fragment from −330 to +20 cloned in front of the CAT reporter gene of plasmid pBLCAT3 (3). For primers, we used the corresponding mutated oligonucleotides described in Table 1 as well as primer −40 (5′-GTTTTCCCAGTCACGAC-3′) and primer CAT (5′-CCATTTTAGCTTCCTTAG-3′). Plasmid mut122 carries a mutation at the −122 site, and plasmid mut90 carries a mutation at the −90 site. Plasmid mut122 was used as a template for the construction of plasmid mut122/90, which carried mutations at both the −122 and −90 sites.
TABLE 1.
Sequences of oligonucleotides containing wild-type or mutated YY1 DNA-binding sitesa
| Name | Sequence (5′ to 3′) | Promoter |
|---|---|---|
| Consensus | GA(C/g/a)(G/t)(C/a/t)CATN(T/a)(T/g/c) | |
| c-Fos | GCAGAGGGGACCATCTCCGAAA | c-Fos |
| 32 | GCAGAAAGGACCATCCCTTATA | −32 muIFN-β coding strand |
| 90 | TTTTCCTCTGTCATTTTCTCTT | −90 muIFN-β noncoding strand |
| mut90 | TTTTCCTCTGTaATTTTCTCTT | |
| 122 | CTTCTAATATTCATTTTATTCA | −122 muIFN-β noncoding strand |
| mut122 | CTTCTAATATTgATTTTATTCA | |
| 161 | TTAACCCAGTACATAGCATATA | −161 muIFN-β coding strand |
In the consensus sequence, as defined by Hyde-DeRuyscher et al. (15), uppercase letters are the preferred nucleotides and lowercase letters are nucleotides tolerated to a lesser extent. Boldface type indicates the nucleotides corresponding to the YY1 core motif. Underlining indicates the nucleotides present in the consensus core motif.
Cell line and transfection.
L929 wt330 and wt110 cell lines have been described previously (30). Cell culture, transient transfection by the calcium phosphate method, Newcastle disease virus infection, and CAT assays were as described previously (3). Plasmid pCMV-YY1 was a generous gift of Jianping Ye and has been described previously by Flanagan et al. (10). YY1 overexpressing plasmid pCMV-YY1 or the corresponding empty vector pCMV was transiently transfected into L929 wt330 or L929 wt110 cells in six-well plates (200,000 cells/well in 2 ml of medium). When indicated, TSA (Sigma) was added to the medium 6 h after transfection at a final concentration of 100 ng/ml. Cells were infected with Newcastle disease virus 48 h after transfection and collected 10 h after infection. Stable transfection of plasmids mut122, mut90, and mut122/90 into L929 cells was carried out as previously described (30). Geneticin-resistant clones were isolated, propagated, and tested for virus-induced CAT activity. Eight to 12 positive clones were pooled, and the integrated IFN-β promoter was sequenced. For each pool, its phenotype was compared to the phenotypes of three corresponding individual positive clones. The results shown in Fig. 4, 5, 6, 7, and 8 correspond to those obtained with the pool (which was very similar to that of the corresponding independent clones), they correspond to the average of two or three independent experiments with each point in duplicate.
FIG. 4.
YY1 down-regulates the transcriptional capacity of the muIFN-β promoter via histone deacetylation. (A) Murine L929 wt330 cells, carrying the stably integrated muIFN-β promoter (from position −330 to +20) fused upstream of a CAT reporter gene, were transiently transfected with 1 μg/well, final concentration, of YY1 expressing plasmid (pCMV-YY1) or the corresponding empty vector (pCMV). When indicated, TSA (100 ng/ml final concentration) was added to the medium 4 h after transfection. Cells were virus induced 48 h after transfection and collected 18 h after virus infection. TSA was removed from the medium after virus infection and thereafter until the collection of the cells. (B) Murine L929 wt330 cells were transiently transfected and virus infected as described for panel A with increasing amounts (62, 125, 250, 500, or 1,000 ng/well) of YY1 expressing a plasmid (pCMV-YY1) or the corresponding empty vector (pCMV). Repression fold corresponds to the CAT activity measured from pCMV-transfected cells divided by the CAT activity measured from pCMV-YY1 transfected cells. (C) Murine L929 wt110 cells, carrying the stably integrated muIFN-β promoter (from position −110 to +20) fused upstream of a CAT reporter gene, were transiently transfected and virus infected as described for panel A with 1 μg/well, final concentration, of YY1 expressing plasmid (pCMV-YY1) or the corresponding empty vector (pCMV).
FIG. 5.
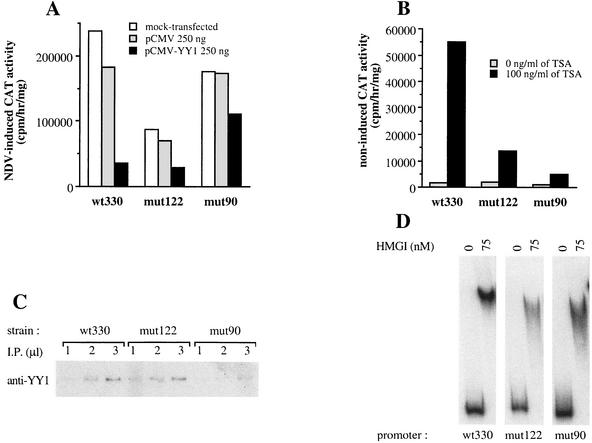
An intact YY1 −90 site is required for YY1-induced repression and TSA-dependent activation of the IFN-β promoter activity. (A) Murine L929 wt330, mut122, and mut90 strains carrying, respectively, integrated wild-type or mutated muIFN-β promoters fused to a CAT reporter gene were transiently transfected with 250 ng/well, final concentration, of pCMV or pCMV-YY1 plasmid. Cells were virus induced 48 h after transfection and collected 18 h after infection. NDV, Newcastle disease virus. (B) Noninfected murine L929 wt330, mut122, and mut90 cells were treated or not with 100 ng of TSA/ml (final concentration) for 48 h before being collected. (C) Equivalent amounts of genomic DNA isolated from L929 wt330, mut122, and mut90 strains were immunoprecipitated (I.P.) with anti-YY1 antibodies. Immunoprecipitated DNA (1, 2, and 3 μl) was amplified with primers specific for the integrated IFN-β region. (D) Gel retardation assay of labeled wt330, mut122, and mut90 muIFN-β promoters (from −330 to +20) incubated with 75 ng of recombinant protein HMGI in the presence of 250 ng of sonicated, unlabeled salmon sperm DNA as a random, nonspecific competitor DNA.
FIG. 6.
Mutated promoters mut122 and mut90 display reduced virus-induced activities. Cells from the L929 wt330 strain (top), mut122 strain (middle), and mut90 strain (bottom) were virus infected and collected at 0, 2, 4, 6, 8, 10, and 18 h after infection. The corresponding absolute CAT activities (i − mi) were measured.
FIG. 8.
Mutation of both YY1-binding sites (mut122/90) strongly reduces the virus-induced transcriptional capacity of the IFN-β promoter. (A) Cells from the wild-type L929 wt330 strain or the double mutant L929 mut122/90 strain were mock transfected or transiently transfected with 250 ng/well, final concentration, of pCMV or pCMV-YY1 plasmid. Cells were virus induced 48 h after transfection and collected 18 h after infection. NDV, Newcastle disease virus. (B) Noninfected wild-type murine L929 wt330 cells and double mutant L929 mut122/90 cells were treated or not with 100 ng of TSA/ml (final concentration) for 48 h before being collected. TSA was removed from the medium after virus infection and thereafter until collection of the cells. (C) Cells from the double mutant L929 mut122/90 strain were virus infected and collected at 0, 2, 4, 6, 8, 10, and 18 h after infection. The corresponding absolute CAT activities (i − mi) were measured.
Titration of IFN activity.
IFNs present in the supernatants of virus-infected L929 wt330 cells, transfected or mock-transfected with plasmids pCMV-YY1 or pCMV, were titrated by using the antiviral activity assay described by Mogensen and Bandu (24) against an IFN reference, which had itself been standardized against the international reference MRC 69/19.
RESULTS
YY1 binds to the muIFN-β promoter.
Four potential YY1-binding sites containing the conserved 5′-C/a/tCAT-3′ YY1 DNA-binding core motif are present at positions −32, −90, −122, and −161 of the muIFN-β promoter (Fig. 1 and Table 1). In order to analyze the specific binding of YY1 to these sites, gel shift assays were carried out in the presence of the corresponding double-stranded DNA probes and murine L929 nuclear extracts. After migration, the gels were either dried and autoradiographed or transferred to a nitrocellulose membrane and submitted to Western blotting with a polyclonal anti-YY1 antibody raised against the entire protein to selectively identify among the retarded protein-DNA complexes those containing YY1. Figure 2 shows the results obtained during Western blot analysis of gel retardation assays. As shown in Fig. 2A, a protein-DNA complex containing YY1 was formed with oligonucleotides 32, 90, and 122. These complexes migrated at the same position as the one formed with the c-Fos probe containing the sequence of a previously described YY1 DNA-binding site present in the promoter region of the c-Fos gene (31). Protein YY1 displayed a strong affinity for its sites present in oligonucleotides 90 and 122, whereas the complex formed with oligonucleotide 32 was of very weak intensity and no complex at all was observed with probe 161 (Fig. 2A). Mutations introduced in the YY1 DNA-binding core motifs of oligonucleotides 90 and 122 (Table 1, sequences mut90 and mut122) disrupted the complex formed between YY1 and the corresponding oligonucleotides (Fig. 2B). In Fig. 2C we show that a second more-retarded complex of less intensity can also be observed with probe 122. The results shown on this figure also indicate that nuclear extracts loaded in the absence of DNA probes give no specific signal equivalent to those observed after incubation of nuclear extracts with probe c-Fos, 90, or 122 (Fig. 2C).
FIG. 1.
Four potential YY1-binding sites are present in the muIFN-β promoter. The DNA sequence of the muIFN-β promoter region spanning from the TATA box to position −210 is shown (35). Positions of the VRE and NRDs are indicated. Arrows indicate the presence of the YY1 core motifs (5′ to 3′) of the four potential YY1-binding sites.
FIG. 2.
Protein YY1 forms protein-DNA complexes with sequences present in the muIFN-β promoter. (A) Nuclear extracts were incubated with the indicated unlabeled double-stranded DNA probes (whose sequences are listed in Table 1), submitted to a gel retardation assay, and transferred on a nitrocellulose membrane. The presence of protein YY1 in the retarded protein-DNA complexes was revealed with anti-YY1 H-414 raised against the full-length YY1 as the primary antibody. (B) Same experiment as described for panel A but with probe mut90 containing the −90 YY1-binding site mutated in its YY1 core motif and probe mut122 containing the −122 YY1-binding site mutated in its YY1 core motif. (C) Same experiment as described for panels A and B, except that in the last lane nuclear extracts were loaded in the presence of poly(dI-dC) but in the absence (−) of DNA.
In Fig. 3 we show the autoradiograms of gel shift experiments carried out parallel to and under the same conditions as the ones presented in Fig. 2, except that DNA probes were labeled with 32P and gels were dried and autoradiographed instead of being transferred to a nitrocellulose membrane. Contrary to the results observed after Western blotting, several protein-DNA complexes were visible after incubation of L929 nuclear extracts with labeled probes 122 (Fig. 3A) and 90 (Fig. 3B). Introduction of a mutation on the corresponding YY1 core motifs present at position −122 or −90 led to the disruption of the complexes indicated by arrows. The formation of these complexes was also inhibited by a monoclonal anti-YY1 antibody, therefore confirming the presence of YY1. In the case of probe 122, the lower migrating complex corresponds to the YY1-122 complex observed in Fig. 2. The second more-retarded complex observed in Fig. 3A was only weakly detected during Western blot analysis of gel retardation (Fig. 2C). In the case of probe 90, the upper migrating complex corresponds to the YY1-90 complex observed in Fig. 2, whereas the second less-retarded complex observed in Fig. 3B was not detected during Western blot analysis of gel retardation.
FIG. 3.
Protein YY1 binds to the muIFN-β promoter at positions −90 and −122. (A) Nuclear extracts were incubated with labeled probes 122 and mut122 in the presence or absence of 2 μg of anti-YY1 monoclonal antibody (Ab) H-10X raised against the full-length YY1 protein. (B) Nuclear extracts were incubated with labeled probes 90 and mut90 in the presence or absence of 1 or 2 μg of anti-YY1 monoclonal antibody H-10X. (C) Competition experiments were carried out with nuclear extracts incubated with labeled probe 90 in the absence (−) or presence of a 150-fold excess (150X) of unlabeled probes 90, mut90, 122, mut122, 161, and 32. (D) Nuclear extracts were incubated with labeled probe 90 in the absence or presence of 50-, 100-, and 150-fold excesses of unlabeled probes 90 and 122. (E) Chromatin immunoprecipitation assay of genomic DNA from noninfected L929 wt330 cells. Increasing amounts of DNA immunoprecipitated (I.P.) with either anti-YY1 or anti-HMGI antibodies were amplified with primers specific for the integrated IFN-β promoter region.
Gel shift competition experiments were carried out in order to further confirm the different binding affinities of YY1 to the four (90, 122, 161, and 32) YY1-binding sites. In the experiment for which results are presented in Fig. 3C, 32P-labeled probe 90 was incubated with L929 nuclear extracts in the presence of a 150-fold excess of unlabeled probes 90, mut90, 122, mut122, 161, and 32. Unlabeled probes 122 and 90 inhibited the formation of the YY1-90 complexes, whereas neither mutated probes mut122 and mut90 nor wild-type probes 161 and 32 competed for the binding of YY1 to probe 90. This confirms the affinity of YY1 for probes 90 and 122 and the lack of or weak affinity for mutated probes and probes 161 and 32, respectively. As shown in Fig. 3D, higher amounts of unlabeled probe 122 (150-fold excess) than of probe 90 (50-fold excess) were required to completely compete for the formation of the YY1-90 complexes, revealing a higher affinity of YY1 for probe 90 than for probe 122. Similar results were obtained during a reciprocal experiment with labeled probe 122 (data not shown).
Chromatin immunoprecipitation assays were performed in order to investigate the capacity of YY1 to bind to the muIFN-β promoter in vivo. Genomic DNA from noninfected murine L929 wt330 cells was immunoprecipitated with anti-YY1 antibodies and amplified with primers specific for the integrated IFN-β promoter. As an internal negative control, the same fraction of genomic DNA was immunoprecipitated with antibodies directed against HMGI, a protein whose binding to the IFN-β promoter is not observed in vivo in the absence of virus infection (26). As shown in Fig. 3E, the IFN-β promoter region was immunoprecipitated in the presence of anti-YY1 antibodies, indicating that YY1 binds in vivo to the IFN-β promoter in noninfected cells. As expected with genomic DNA isolated from noninfected cells, no signal corresponding to the IFN-β promoter was observed after immunoprecipitation with anti-HMGI antibodies.
YY1 down-regulates the transcriptional capacity of the muIFN-β promoter.
In order to assess the role of YY1 during the regulation of the transcriptional capacity of the IFN-β promoter, YY1 was overexpressed in cells from the murine fibroblastic L929 wt330 cell line carrying a stably integrated wild-type muIFN-β promoter (from position −330 to +20) CAT reporter construct. These cells were transiently transfected with either a YY1 expression vector (pCMV-YY1) or the corresponding empty vector (pCMV) and virus infected 48 h after transfection. As shown in Fig. 4A, overexpression of YY1 strongly repressed the transcriptional capacity of the integrated wt330 muIFN-β promoter. The repression induced by YY1 increased proportionally with the amount of pCMV-YY1 plasmid used during transfection experiments (Fig. 4B), a phenomenon previously described when analyzing YY1-induced transcriptional repression (4, 10). No effect (either activator or repressor) was observed at plasmid concentrations of less than 62 ng/well (data not shown).
The interferon activity present in the medium of either pCMV- or pCMV-YY1-transfected or mock-transfected L929 cells was titrated. The results present in Table 2 clearly indicate that YY1 down-regulates the endogenous interferon activity in a manner very similar to that observed with the integrated CAT reporter construct.
TABLE 2.
Titration of IFN activity before and after YY1 overexpression
| Cell treatment | IFN activity (U/ml/mg)a
|
|
|---|---|---|
| −TSA | +TSA | |
| Mock transfected | 3,753 ± 766 | 4,218 ± 1,500 |
| pCMV-YY1 transfected | 635 ± 62 | 4,483 ± 1,217 |
| pCMV transfected | 5,097 ± 2,334 | 3,782 ± 1,307 |
IFN activity was titrated in the supernatants collected 10 h after infection. +, with; −, without.
In order to analyze whether the repressive effect of YY1 upon the IFN-β promoter was linked to an HDAC activity, we used TSA, which is a specific inhibitor of HDACs. As shown in Fig. 4A, the repression induced by YY1 was fully relieved by TSA, suggesting that histone deacetylation is a key determinant in YY1-induced repression of the IFN-β promoter. As in the case of the integrated construct, YY1-induced repression of the endogenous promoter was also completely neutralized in the presence of TSA (Table 2).
We also analyzed the effect of YY1 overexpression on cells from the L929 wt110 cell line which carry a stably integrated short muIFN-β promoter (from position −110 to +20) only containing the virus responsive element (VRE) of the promoter fused to a CAT reporter gene. The VRE region contains the four positive regulatory domains of the IFN-β promoter and corresponds to the minimal region necessary for the virus-induced activation of the promoter (22). The overexpression of YY1 had no effect on the short wt110 IFN-β promoter containing only the VRE region (Fig. 4C), demonstrating that the repressive effect of YY1 upon the IFN-β promoter is not an indirect consequence of YY1 affecting the expression of a regulatory VRE binding factor. It also indicates that YY1-induced repression of the IFN-β promoter requires the presence of the promoter region 5′ of the VRE. This region has been described as a negative regulatory domain (NRDII) which intervenes during the establishment of the promoter constitutive silent state (41). It constitutes a region of nucleosome positioning (1) and of preferential interaction with linker histone H1 (3). Promoters lacking this region are constitutively derepressed (41) and are only weakly affected by TSA (30).
YY1-binding site present at position −90 mediates YY1 repressive effect.
In order to investigate the role of the main YY1-binding sites present in the muIFN-β promoter during the YY1-mediated repression of the promoter, we constructed pBLCAT3-derived plasmids containing the muIFN-β promoters (from position −330 to +20) mutated at either one of the two main YY1-binding sites (−90 and −122) fused to the CAT reporter gene. In the case of the −90 as well as that of the −122 site, a single-base substitution was introduced in the YY1 core motif corresponding to the mut90 and mut122 mutations used during the gel retardation experiments listed in Table 1. The corresponding mutated promoter CAT reporter constructs were stably transfected into L929 cells. Positive independent clones were pooled and analyzed for their capacity to be repressed by YY1 (Fig. 5A). During these experiments only 250 ng of plasmid pCMV or pCMV-YY1 was used for transfection in order to reduce a nonspecific repression induced by the vector alone in strain mut122. The introduction of a mutation on the site present at position −90 almost completely abolished the capacity of YY1 to repress the transcriptional capacity of the IFN-β promoter. On the contrary, YY1 continued to repress, at least partly, promoter mut122.
Treatment of wt330 cells with TSA has been previously described to activate the noninduced, constitutive activity of the promoter (30). In order to analyze the eventual role of YY1 during the TSA-induced activation of the IFN-β promoter, mut122 and mut90 cells were treated with TSA at the same time as the wt330 strain. As shown in Fig. 5B, disruption of YY1 binding to the −90 site completely abolished the capacity of TSA to activate the noninduced constitutive activity of the IFN-β promoter. Disruption of the −122 site also inhibited the capacity of TSA to activate the IFN-β promoter but did not completely abolish it. Nevertheless, even though TSA-induced activation was abolished in mut90 cells, the constitutive noninduced activity of promoter mut90 remained very low.
Chromatin immunoprecipitation assays were carried out in order to compare the in vivo binding capacity of the YY1 protein to the mut122 and mut90 promoters. Equivalent amounts of genomic DNA isolated from noninduced wt330, mut122, and mut90 strains were immunoprecipitated with anti-YY1 antibodies and amplified with primers specific for the integrated IFN-β promoter. The amount of IFN-β promoter immunoprecipitated with anti-YY1 antibodies in the case of the mut90 strain was very low compared to that immunoprecipitated from the wild-type or mut122 strain (Fig. 5C), indicating that introduction of a mutation at site −90 strongly reduced the amount of YY1 bound in vivo to the IFN-β promoter in noninfected cells. Whereas the mutation in the mut122 strain apparently did not affect much the in vivo capacity of YY1 to bind to the promoter in noninfected cells, the almost complete lack of YY1 binding to the mut90 promoter compared to the mut122 promoter in noninfected cells suggests that at this time YY1 is bound to the IFN-β promoter predominantly at the −90 site.
A strong HMGI binding site is present in the muIFN-β promoter next to the YY1-binding site present at position −122, and disruption of this site has been determined to lead to a reduced promoter activity (3). In order to test the eventual effect of the mutation in strain mut122 on the DNA-binding capacity of protein HMGI to the IFN-β promoter, gel retardation experiments were carried out with wt330, mut90, and mut122 promoters (from position −330 to +20) and recombinant HMGI protein. As seen in Fig. 5D, the overall binding capacity of HMGI protein to the IFN-β promoter was not significantly affected by the introduction of the mutations in strains mut122 and mut90. Gel retardation experiments carried out with recombinant HMGI protein and short oligonucleotides 122 and mut122 confirmed that the mutation in strain mut122 did not affect the binding of HMGI to this particular region (data not shown).
YY1 regulates virus-induced activation and transcriptional turn off of the IFN-β promoter.
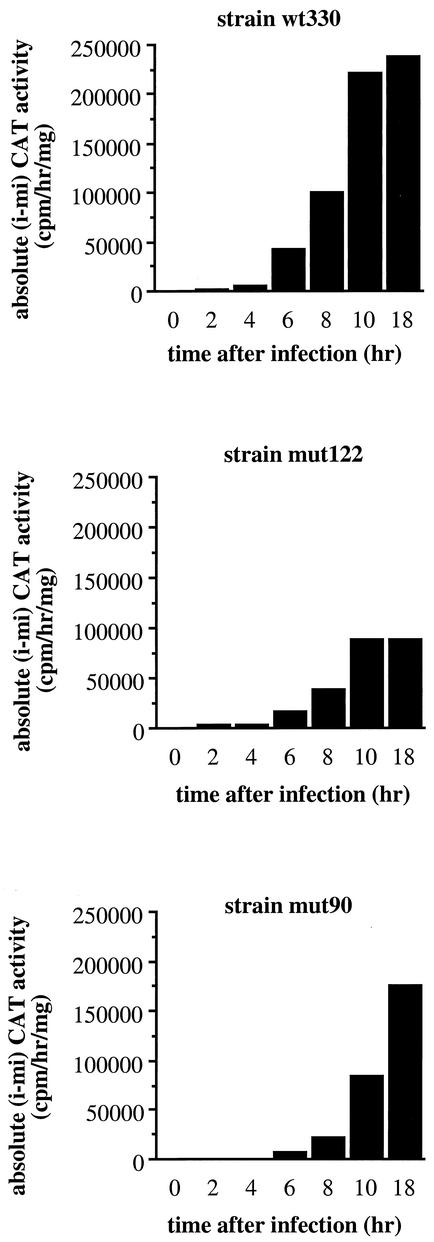
Protein YY1 has been shown to have bifunctional activator/repressor transcriptional properties (19). As shown in Fig. 5A, the mut122 strain promoter displayed reduced virus-induced activity 18 h after infection. In order to investigate the effect of protein YY1 on the virus-induced transcriptional capacity of the IFN-β promoter, cells from the wild-type wt330 and mutated mut122 and mut90 strains were virus infected and collected 0, 2, 4, 6, 8, 10, and 18 h after infection. The corresponding CAT activities are shown in Fig. 6. In this figure, the virus-induced activities of the wt330, mut122, and mut90 IFN-β promoters were analyzed independently of the corresponding constitutive noninduced activities. For this purpose, we subtracted the corresponding mock-induced CAT activities (mi) from the final CAT activities obtained after virus infection (i). In Fig. 6, we have called this value the absolute (i − mi) CAT activity. In agreement with previously described results (3, 14), activation of the IFN-β promoter started 6 h after virus infection and this was observed with the wild-type wt330 strain as well as with the mutated mut122 and mut90 strains (Fig. 6). Nevertheless, strains mutated at either one of the two YY1-binding sites displayed reduced virus-induced activities compared to the wild-type strain, and this started as early as 6 h after infection. Ten hours after infection, strains mut122 and mut90 displayed virus-induced activities corresponding to a maximum of 40% of the activity reached by the wild-type strain at this time.
The maximum transcriptional capacity of the IFN-β promoter is reached between 10 and 12 h after infection (14). After this time, transcription stops as a consequence of the postinduction transcriptional turn off of the IFN-β promoter (37). Since the half-life of CAT mRNA is quite long, the value reached 10 h after infection remained constant until 18 h after infection (Fig. 6). This was observed in the case of the wild-type wt330 strain as well as with the strain carrying a promoter mutated at the YY1-binding site present at position −122. On the contrary, the activity of the strain carrying a promoter mutated at the YY1-binding site present at position −90 continued to progress beyond 10 h after infection. In order to study the kinetics of induction of each promoter, the percentage of activity of the promoters of the wt330, mut122, and mut90 strains were measured at 2, 4, 6, 8, 10, 24, 32, and 54 h after infection. We have considered the absolute CAT activity reached by each promoter 10 h after infection to be 100%. As it is shown in Fig. 7, between 2 and 10 h after infection, the kinetics of induction of the three promoters were identical. Both the wt330 and the mut122 promoters reached their maximum activity near 10 h after infection. Because of the stability of the CAT mRNA, the corresponding activities slowly decreased thereafter and reached levels around 70% of their 10-h-postinfection value at 54 h after infection. In the case of the mut90 promoter, its activity continued to strongly increase between 10 and 24 h after infection and the mut90 promoter displayed a value still superior to 100% of its 10-h-postinfection value at 54 h after infection.
FIG. 7.
Promoter mut90 displays a retarded postinfection transcriptional turn off. Cells from the L929 wt330 (○), mut122 (▪), and mut90 (▴) strains were virus infected and collected at 0, 2, 4, 6, 8, 10, 24, 32, and 54 h after infection. Percentages of the corresponding absolute CAT activities (i − mi) were measured. We have considered the activity reached by each strain 10 h after infection to be 100%.
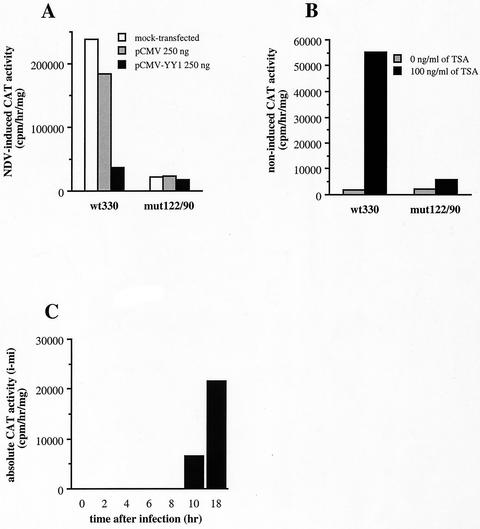
In Fig. 8 are shown the results obtained with cells from the mut122/90 strain which contains a muIFN-β promoter (from position −330 to +20) mutated in both −122 and −90 YY1-binding sites fused to the CAT reporter gene and stably integrated into the genome of L929 cells. The mut122/90 promoter mutated in both sites displayed an extremely weak virus-induced activity, corresponding 18 h after infection to no more than 10% of the activity of the wild-type promoter at this time. Also, similar to the results obtained with the mut90 promoter, the mut122/90 promoter was not affected by YY1 overexpression (Fig. 8A) or by TSA treatment (Fig. 8B) and displayed a retarded postinduction transcriptional turn off (Fig. 8C). The results obtained with the doubly mutated promoter mut122/90 confirmed those obtained with the singly mutated promoters mut122 and mut90. They also very clearly indicated that in the context of a promoter spanning from position −330 to +20, containing the VRE and the NRDII region, YY1 binding is essential for the virus-induced promoter activation.
DISCUSSION
YY1 binds to the muIFN-β promoter at positions −90 and −122.
Four potential YY1-binding sites are present in the muIFN-β promoter region at positions −22, −90, −122, and −161. Using gel retardation assays and YY1 antibodies we demonstrate here that the sites present at positions −122 and −90 are specific YY1-binding sites. The site present at position −32 appeared as a very weak site, and no binding at all was observed with the −161 site. The presence of two thymidines 3′ of the core motif (C/t/aCATNTT) has been shown to be important for binding affinity and specificity during YY1-DNA complex formation (15). Sequence −90 (GTCATTTT) and −122 (TTCATTTT) carry two T's at this position, whereas they are absent from sequences −32 (ACCATCCC) and −161 (TACATAGC). Sequence −161 is the only sequence with an A as the first nucleotide of the core motif. The absence of a T in the 3′ position and the presence of an A as the first nucleotide of the core motif, could be responsible for the complete lack of affinity of YY1 for sequence −161. YY1 displayed a stronger affinity towards the −90 site than to the −122 site. Such a difference could be related to the presence of a G immediately 5′ of the core motif in the case of site −90 (GTCATTTT). In fact, a G at this position has also been described as intervening during the establishment of strong interactions in YY1-DNA complex formation (15).
Chromatin immunoprecipitation experiments carried out with noninfected cells indicated that, before virus infection occurs, YY1 binds in vivo to the wild-type IFN-β promoter predominantly at its −90 site, which appeared to be the strongest YY1-binding site during in vitro experiments.
It is interesting that potential YY1-binding sites are also present in the human; bovine 1, 2, and 3; and horse IFN-β promoters (35). Binding of YY1 to IFN-β promoters is therefore expected to be a feature highly conserved among different species.
YY1 represses the muIFN-β promoter.
During YY1 overexpression experiments, the A/T-rich region of the promoter positioned 5′ of the VRE corresponding to NRDII and the YY1 −90 site appeared to be necessary to mediate YY1-induced repression. The NRDII sequence is a region of nucleosome positioning (1) that regulates the establishment of the promoter constitutive silent state (41) and partly mediates the TSA-induced constitutive derepression of the IFN-β promoter (30). The role played by this region during YY1-induced repression of the IFN-β promoter was a dominant one since promoter wt110 lacking NRDII was not affected by YY1 overexpression despite the presence of an intact −90 site on this promoter. The NRDII region could be the targeted region undergoing YY1-dependent modifications such as histone deacetylation during YY1-induced repression of the transcriptional activity of the IFN-β promoter (Fig. 9).
FIG. 9.
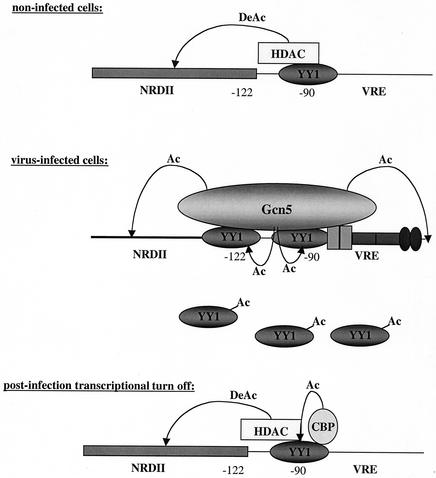
The dual activator/repressor role of YY1 could be related to its capacity to interact with HATs and HDACs. We propose here a model that attempts to explain the bifunctional role of YY1 during the regulation of the transcriptional capacity of the IFN-β promoter. Several points remain to be demonstrated, setting up directions for future work. Besides the data obtained during this work, the model we propose here relies on (i) previous results indicating that before virus infection HDAC participates in the establishment of the promoter constitutive repression state (30); (ii) the work of Agalioti et al. (1) which indicates that Gcn5 is recruited by the promoter 3 h after infection, peaks 6 h after infection, and is released from the promoter 9 h after infection, whereas CBP peaks between 9 and 12 h after infection, does not participate during promoter histone acetylation, and remains bound to the promoter 24 h after infection; and (iii) the work of Munshi et al. (25) that describes CBP as essential for the transcriptional turn off of the IFN-β promoter. We suppose that in noninfected cells YY1 is predominantly bound to its −90 site and participates in promoter repression through an HDAC activity that deacetylates (DeAc) histones positioned in the NRDII region (top panel). Shortly after infection, YY1, alongside virus-activated factors bound to the VRE, participates in the recruitment of Gcn5. Gcn5 induces histone acetylation (Ac) necessary for nucleosome sliding and promoter transcriptional activation (1, 21) as well as YY1 acetylation (Ac) that disrupts YY1-DNA interactions (39) and could therefore induce YY1 promoter unbinding (middle panel). After release of Gcn5 from the promoter, nonacetylated forms of YY1 bind the promoter at its strongest −90 site and participate in the promoter transcriptional turn off in association with CBP and HDAC. Acetylation (Ac) of YY1 by CBP stabilizes YY1-HDAC interactions (39) (bottom panel).
Promoters mutated at the −90 site displayed a retarded postinfection transcriptional turn off, indicating that binding of YY1 to its −90 site is required for the postinfection repression of the muIFN-β promoter. YY1 is able to interact with cofactor CBP (39) that intervenes during the transcriptional turn off of the IFN-β promoter (25). As illustrated in Fig. 9, the postinfection capacity of YY1 to repress the IFN-β promoter activity could depend on the establishment of a YY1-CBP interaction starting around 9 h after infection and occurring predominantly through the YY1 −90 site. CBP-induced acetylation of YY1 residues 170 to 200 activates the interaction of YY1 with HDACs without affecting its capacity to bind to DNA (39). This could enhance YY1 repressor function during postinfection turn off.
YY1 is an activator of the muIFN-β promoter.
Mutated promoters, mut122 and mut90, displayed reduced virus-induced activities, and this started shortly after infection. The HAT Gcn5, which belongs to the same family as PCAF, itself a YY1-interacting protein, is essential for the virus-induced transcriptional activation of the IFN-β promoter. It is recruited by the IFN-β promoter starting 3 h after infection, it peaks 6 h after infection, and it is released from the promoter 9 h after infection (1). The factor(s) responsible(s) for the recruitment of Gcn5 on the IFN-β promoter have not been identified. We suggest that the protein YY1 could intervene during the recruitment of Gcn5 and by doing so act as a transcriptional activator shortly after infection (Fig. 9).
Dual activator/repressor role of YY1.
The protein YY1 has the dual capacity to interact with corepressors such as HDACs as well as coactivators such as HATs. Besides acetylating or deacetylating histones, these cofactors can acetylate or deacetylate YY1 itself. Acetylation of YY1 residues 170 to 200 stabilizes YY1-HDAC interactions and therefore activates YY1 transcriptional repression activity. Acetylation of the YY1 C terminus by PCAF leads to the disruption of the interaction of YY1 with DNA (39). Therefore, changes concerning the local concentrations of these cofactors and of YY1 itself as well as variations on the DNA-binding affinities of YY1 for different promoter sites could influence whether YY1 acts as a repressor or an activator.
Investigation of the precise patterns of acetylation of histones H3 and H4 in mut90 and mut122 promoters before and after virus infection as well as comparative analysis of the effects linked to overexpression of Gcn5 in wt330 versus mut90 and mut122 promoters should help us to clarify the bifunctional role of YY1 during IFN-β gene expression in relation to histone acetylation/deacetylation.
Acknowledgments
We are grateful to Jianping Ye for the gift of plasmid pCMV-YY1, Fédéric Rathouis for participation in the construction of mutated promoters, Marie-Thérèse Bandu for the IFN activity test, and Eugenio Prieto for photographic work.
This work was supported by the Centre National de la Recherche Scientifique and by a grant from the Association pour la Recherche sur le Cancer (ARC 5828). E.S. was a recipient of a fellowship from the Fondation pour la Recherche Medicale (FRM) until April 2002 and is a recipient of a fellowship from the Association Française contre les Myopathies (AFM).
REFERENCES
- 1.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 2.Austen, M., B. Luscher, and J. M. Luscher-Firzlaff. 1997. Characterization of the transcriptional regulator YY1. J. Biol. Chem. 272:1709-1717. [DOI] [PubMed] [Google Scholar]
- 3.Bonnefoy, E., M.-T. Bandu, and J. Doly. 1999. Specific binding of high-mobility-group I (HMGI) protein and histone H1 to the upstream AT-rich region of the murine beta interferon promoter: HMGI protein acts as a potential antirepressor of the promoter. Mol. Cell. Biol. 19:2803-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bushmeyer, S., K. Park, and M. L. Atchison. 1995. Characterization of functional domains within the multifunctional transcription factor YY1. J. Biol. Chem. 270:30213-30220. [DOI] [PubMed] [Google Scholar]
- 5.Coull, J. J., F. Romerio, J.-M. Sun, J. L. Volker, K. M. Galvin, J. R. Davie, Y. Shi, U. Hansen, and D. M. Margolis. 2000. The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J. Virol. 74:6790-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davie, J. R., and D. N. Chadee. 1998. Regulation and regulatory parameters of histone modifications. J. Cell. Biochem. Suppl. 30-31:203-213. [PubMed] [Google Scholar]
- 7.Davie, J. R., and V. A. Spencer. 1999. Control of histone modifications. J. Cell. Biochem. Suppl. 32-33:141-148. [DOI] [PubMed] [Google Scholar]
- 8.DeMayer, E., and J. DeMayer-Guignard. 1988. Interferons and other regulatory cytokines. John Wiley and Sons, New York, N.Y.
- 9.Donohoe, M. E., X. Zhang, L. McGinnis, J. Biggers, E. Li, and Y. Shi. 1999. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol. Cell. Biol. 19:7237-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flanagan, J. R., K. G. Becker, D. L. Ennist, S. L. Gleason, P. H. Driggers, B. Z. Levi, E. Appella, and K. Ozato. 1992. Cloning of a negative transcription factor that binds to the upstream conserved region of Moloney murine leukemia virus. Mol. Cell. Biol. 12:38-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galvin, K. M., and Y. Shi. 1997. Multiple mechanisms of transcriptional repression by YY1. Mol. Cell. Biol. 17:3723-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grozinger, C. M., C. A. Hassig, and S. L. Schreiber. 1999. Three proteins define a class of human histone deacetylases related to yeast Hdap1. Proc. Natl. Acad. Sci. USA 96:4868-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo, B., P. R. Odgren, A. J. van Wijnen, T. J. Last, J. Nickerson, S. Penman, J. B. Lian, J. L. Stein, and G. S. Stein. 1995. The nuclear matrix protein NMP-1 is the transcription factor YY1. Proc. Natl. Acad. Sci. USA 92:10526-10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higashi, Y. 1985. Changes of chromatin conformation around mouse interferon-β gene associated with induction of interferon synthesis. Nucleic Acids Res. 13:5157-5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hyde-DeRuyscher, R. P., E. Jennings, and T. Shenk. 1995. DNA binding sites for the transcriptional activator/repressor YY1. Nucleic Acids Res. 23:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imai, S., C. M. Armstrong, M. Kaeberlein, and M. Guarante. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795-800. [DOI] [PubMed] [Google Scholar]
- 17.Jones, P. L., G. J. C. Veenstra, P. A. Wade, D. Vermaak, S. U. Kass, N. Landsberger, J. Strouboulis, and A. P. Wolffe. 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19:187-191. [DOI] [PubMed] [Google Scholar]
- 18.Khochbin, S., A. Verdel, C. Lemercier, and D. Seigneurin-Berny. 2001. Functional significance of histone deacetylation diversity. Curr. Opin. Genet. Dev. 11:162-166. [DOI] [PubMed] [Google Scholar]
- 19.Lee, T.-C., Y. Zhang, and R. J. Schwartz. 1994. Bifunctional transcriptional properties of YY1 in regulating muscle actin and c-myc gene expression during myogenesis. Oncogene 9:1047-1052. [PubMed] [Google Scholar]
- 20.Lewis, B. L., G. Tullis, E. Seto, N. Horikoshi, R. Weinmann, and T. Shenk. 1995. Adenovirus E1A proteins interact with cellular YY1 transcription factor. J. Virol. 69:1628-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lomvardas, S., and D. Thanos. 2001. Nucleosome sliding via TBP DNA binding in vivo. Cell 106:685-696. [DOI] [PubMed] [Google Scholar]
- 22.Maniatis, T., J. V. Falvo, T. H. Kim, T. K. Kim, C. H. Lin, B. S. Parekh, and M. G. Wathelet. 1998. Structure and function of the interferon-beta enhanceosome. Cold Spring Harbor Symp. Quant. Biol. 63:609-620. [DOI] [PubMed] [Google Scholar]
- 23.Miska, E. A., C. Karlsson, E. Langley, S. J. Nielsen, J. Pines, and T. Kouzarides. 1999. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 18:5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mogensen, K. E., and M.-T. Bandu. 1983. Kinetic evidence for an activation step following binding of human interferon α2 to the membrane receptors of Daudi cells. Eur. J. Biochem. 134:355-364. [DOI] [PubMed] [Google Scholar]
- 25.Munshi, N., M. Merika, J. Yie, K. Senger, G. Chen, and D. Thanos. 1998. Acetylation of HMGI(Y) by CBP turns off IFNβ expression by disrupting the enhanceosome. Mol. Cell 2:457-467. [DOI] [PubMed] [Google Scholar]
- 26.Munshi, N., T. Agalioti, S. Lomvardas, M. Merika, G. Chen, and D. Thanos. 2001. Coordination of a transcriptional switch by HMGI(Y) acetylation. Science 293:1133-1136. [DOI] [PubMed] [Google Scholar]
- 27.Natesan, S., and M. Z. Gilman. 1993. DNA bending and orientation-dependent function of YY1 in the c-fos promoter. Genes Dev. 7:2497-2509. [DOI] [PubMed] [Google Scholar]
- 28.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, M. Nakaya, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-α/β gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 29.Seto, E., B. Lewis, and T. Shenk. 1993. Interaction between transcription factors Sp1 and YY1. Nature 365:462-464. [DOI] [PubMed] [Google Scholar]
- 30.Shestakova, E., M.-T. Bandu, J. Doly, and E. Bonnefoy. 2001. Inhibition of histone deacetylation induces constitutive derepression of the beta interferon promoter and confers antiviral activity. J. Virol. 75:3444-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shrivastava, A., and L. Calame. 1994. An analysis of genes regulated by multi-functional transcriptional regulator Yin Yang-1. Nucleic Acids Res. 22:5151-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stark, G. R., I. M. Kerr, B. R. G. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 33.Therrien, M., and J. Drouin. 1993. Cell-specific helix-loop-helix factor required for pituitary expression of the pro-opiomelanocortin gene. Mol. Cell. Biol. 13:2342-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas, M. J., and E. Seto. 1999. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene 236:197-208. [DOI] [PubMed] [Google Scholar]
- 35.Vodjdani, G., C. Coulombel, and J. Doly. 1988. Structure and characterization of a murine chromosomal fragment containing the interferon β gene. J. Mol. Biol. 204:221-231. [DOI] [PubMed] [Google Scholar]
- 36.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol. Cell 1:507-518. [DOI] [PubMed] [Google Scholar]
- 37.Whittemore, L. A., and T. Maniatis. 1990. Postinduction repression of the β-interferon gene is mediated through two positive regulatory domains. Proc. Natl. Acad. Sci. USA 87:7799-7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Workman, J. L., and R. E. Kingston. 1998. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 67:545-579. [DOI] [PubMed] [Google Scholar]
- 39.Yao, Y.-L., W.-M. Yang, and E. Seto. 2001. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol. Cell. Biol. 21:5979-5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ye, J., M. Cippitelli, L. Dorman, J. R. Ortaldo, and H. A. Young. 1996. The nuclear factor YY1 suppresses the human gamma interferon promoter through two mechanisms: inhibition of AP1 binding and activation of a silencer element. Mol. Cell. Biol. 16:4744-4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zinn, K., D. DiMaio, and T. Maniatis. 1983. Identification of two distinct regulatory regions adjacent to the human β-interferon gene. Cell 34:865-879. [DOI] [PubMed] [Google Scholar]