Abstract
Since it emerged in Japan in the 1870s, Japanese encephalitis has spread across Asia and has become the most important cause of epidemic encephalitis worldwide. Four genotypes of Japanese encephalitis virus (JEV) are presently recognized (representatives of genotypes I to III have been fully sequenced), but its origin is not known. We have determined the complete nucleotide and amino acid sequence of a genotype IV Indonesian isolate (JKT6468) which represents the oldest lineage, compared it with other fully sequenced genomes, and examined the geographical distribution of all known isolates. JKT6468 was the least similar, with nucleotide divergence ranging from 17.4 to 19.6% and amino acid divergence ranging from 4.7 to 6.5%. It included an unusual series of amino acids at the carboxy terminus of the core protein unlike that seen in other JEV strains. Three signature amino acids in the envelope protein (including E327 Leu→Thr/Ser on the exposed lateral surface of the putative receptor binding domain) distinguished genotype IV strains from more recent genotypes. Analysis of all 290 JEV isolates for which sequence data are available showed that the Indonesia-Malaysia region has all genotypes of JEV circulating, whereas only more recent genotypes circulate in other areas (P < 0.0001). These results suggest that JEV originated from its ancestral virus in the Indonesia-Malaysia region and evolved there into the different genotypes which then spread across Asia. Our data, together with recent evidence on the origins of other emerging viruses, including dengue virus and Nipah virus, imply that tropical southeast Asia may be an important zone for emerging pathogens.
Japanese encephalitis virus (JEV) is the most important cause of epidemic encephalitis worldwide, with an estimated 35,000 to 50,000 cases and 10,000 deaths annually (36). The virus is a member of the JE serogroup of the genus Flavivirus, family Flaviviridae, and is transmitted between vertebrate hosts by mosquitoes, principally by Culex tritaeniorhynchus. Other important members of the same serogroup of flaviviruses include West Nile virus (WNV), which has recently spread to cause outbreaks of encephalitis in North America (2, 18), St. Louis encephalitis virus (SLEV), Kunjin virus (KUNV), and Murray Valley encephalitis virus (MVEV). Like other flaviviruses, JEV consists of a small (50 nm) glycoprotein-containing lipid envelope surrounding a nucleocapsid which encloses one molecule of single-stranded positive-sense RNA. This 11-kb molecule comprises 5′- and 3′-untranslated regions (UTRs), between which lie a single open reading frame carrying genes for three structural proteins (capsid [C], premembrane [PrM], and envelope [E]) and seven nonstructural (NS) proteins (3, 34).
Epidemics of encephalitis were described in Japan from the 1870s onwards. JEV was first isolated in 1935 and has subsequently been found across most of Asia (30). The origins of the virus are uncertain, but phylogenetic comparisons with other flaviviruses suggest it evolved from an African ancestral virus, perhaps as recently as a few centuries ago (9, 10). Clinical features of infection with JEV range from a nonspecific febrile illness to a severe meningoencephalopmyelitis, often associated with seizures (31), or a polio-like flaccid paralysis (32). Two epidemiological patterns of JE are recognized (39). In northern temperate areas JE occurs in summer epidemics, whereas in southern tropical areas the disease is endemic and occurs year-round. Based on limited nucleotide sequencing of C/PrM and E genes, four genotypes of virus have been identified (5, 6), and representatives of three of which have been fully sequenced. Genotype I includes isolates from northern Thailand, Cambodia, and Korea, genotype II includes isolates from southern Thailand, Malaysia, Indonesia, and Northern Australia, genotype III includes isolates from mostly temperate regions of Asia, including Japan, China, Taiwan, the Philippines, and the Asian subcontinent, and genotype IV includes isolates from Indonesia. In addition, a strain of JEV isolated in Singapore in 1952 from a patient who originated in Muar, Malaysia (Muar strain), may represent a fifth genotype, according to cross-neutralization and limited phylogenetic evidence (13, 38). Because genotypes I and III occur in epidemic regions, whereas II and IV are associated with endemic disease, it has been postulated that differences in strain virulence may explain the clinical epidemiology (5, 6, 42). However, strains are increasingly being identified that do not fit this paradigm. For example, in Vietnam epidemic disease occurs in the north and endemic disease occurs in the south, yet genotype III strains have been isolated in both areas (14, 30). More recently, a genotype I strain has been found in northern Australia (26). To understand better the origin and geographical distribution of JEV genotypes we have determined the complete nucleotide and deduced amino acid sequence of a representative of a genotype IV strain (Indonesian isolate JKT6468), compared it with other full-length and partial genomes, and investigated the geographical distribution of all known JEV isolates.
MATERIALS AND METHODS
Virus culture, sequencing, and phylogenetic analysis.
JEV strain JKT6468, isolated in 1981 from C. tritaeniorhynchus mosquitoes in Flores, Indonesia, was inoculated onto confluent monolayers of C6/36 cells in 2% fetal calf serum. Culture supernatant containing the virus was harvested at 4 to 5 days postinfection, when cytopathic effects were observed, and virus RNA was extracted as described previously (23). RNA was reverse transcribed by using RAV2 reverse transcriptase (RT; Amersham Pharmacia Biotech Inc., Piscataway, N.J.) and was PCR amplified by using Taq DNA polymerase (Roche Diagnostics Corp.). Primers were designed to amplify overlapping fragments of the genome on the basis of a consensus alignment of complete JEV nucleotides available or on sequence of JKT6468 already derived. RT-PCR products were purified by using the QIAGEN gel extraction kit. Purified cDNA was directly sequenced in both directions by using the appropriate sense and antisense primers and were ligated into pGEM-T Easy vector according to the manufacturer's protocol (Promega Corp., Madison, Wis.). Plasmids were screened for possession of the insert by diagnostic restriction digests with EcoRI. At least three clones of plasmids containing inserts were sequenced in both directions by using universal M13 forward and reverse primers. The full-length JKT6468 strain genome was compiled by using the Vector NTI suite software package (version 7.0; Informax Inc.). The complete sequences of JKT6468 and the derived polyprotein were aligned with the other complete JEV genomes (Table 1) by using the Vector NTI sequence analysis program and percentage differences were calculated. Attenuated strains were not used if the sequence of their parental wild-type isolates were available (e.g., SA14-14-2 and SA14). Phylogenetic trees were constructed by using the neighbor-joining, the maximum parsimony, and maximum likelihood methods on PAUP* version 4.04a (Sinauer Associates, Sutherland, Mass.). The robustness of phylograms was evaluated by 1,000 bootstrap replicates.
TABLE 1.
Details of isolates used in Fig. 1
| Straina | Year | Location | Source | Source or GenBank accession no. (gene)b |
|---|---|---|---|---|
| 691004 | 1969 | Sri Lanka | Human | Z34097 (E) |
| B1065/63 | 1983 | Thailand | Pig | U70388 |
| B-2524 | 1985 | Nepal | Human | U70392 (E) |
| Beijing-1 | 1949 | China | Human | L48961 |
| CH1392 | 1990 | Taiwan | Mosquito | AF254452 |
| CH2195LA | 1994 | Taiwan | Mosquito | AF221499 |
| Chiang Mai | 1964 | Thailand | Human | U70393 (E) |
| CNS138-11 | 1999 | Malaysia | Human | This paper; accession no. AY184213 |
| Den2-New Guinea C | 1944 | New Guinea | Human | M29095 |
| FU | 1995 | Australia | Human | AF217620 |
| GP78 | 1978 | India | Human | AF075723 |
| HVI | 1958 | Taiwan | Human | AF098736 |
| Indonesia | IUc | Indonesia | Mosquito | U03692 (E) |
| Ishikawa | 1998 | Japan | Mosquito | AB051292 |
| JaGAr01 | 1959 | Japan | Mosquito | AF069076 |
| JaOArS982 | 1982 | Japan | Mosquito | NC_001437 |
| JE827 | 1968 | Malaysia | Mosquito | D00994 |
| JKT1724 | 1979 | Indonesia | Mosquito | U70404 (E) |
| JKT5441 | 1981 | Indonesia | Mosquito | U70406 (E) |
| JKT6468 | 1981 | Indonesia | Mosquito | This paper; accession no. AY184212 |
| K87P39 | 1987 | Korea | Mosquito | U34927 (E) |
| K94P05 | 1994 | Korea | Mosquito | AF045551 |
| KPP034-35CT | 1982 | Thailand | Mosquito | U03693 (E) |
| KUN MRM61C | 1960 | Australia | Mosquito | D00246 (ORF) |
| Ling | 1965 | Taiwan | Human | Jan et al. (16) |
| M859 | 1967 | Cambodia | Mosquito | U70410 (E) |
| Muar | 1952 | Malaysia | Human | Hasegawa et al. (13) |
| MVE 1-51 | 1951 | Australia | Human | AF161266 |
| P20778 | 1958 | India | Human | AF080251 (E) |
| P3 | 1950 | China | Mosquito | U47032 |
| PhAn1242 | 1984 | Philippines | Pig | U70417 (E) |
| RP-9 | 1985 | Taiwan | Mosquito | AF014161 |
| SA14 | 1954 | China | Mosquito | U14163 |
| Saigon | 1962 | Vietnam | IUc | U03696 (E) |
| Sarawak | 1965 | Malaysia | Mosquito | This paper; accession no. AY184214 |
| SLE MSI.7 | 1975 | United States | Bird | M16614, AF013414, AF160194, AF242895 |
| T1P1 | 1997 | Taiwan | Mosquito | W.-J. Chen |
| TC | IU | Taiwan | IU | AF098736 |
| ThCMAr4492 | 1992 | Thailand | Mosquito | D45362 (E) |
| Torres Strait 2000 | 2000 | Australia | Pig | AF289818 (E) |
| WN Eg101 | 1950 | Egypt | Human | AF260968 |
| WTP-70-22 | 1970 | Malaysia | Mosquito | U70421 (E) |
All strains are JEV, except for Den2 (Dengue 2), KUN, MVE, SLE, and WN.
GenBank accession numbers refer to the whole genome or the genes indicated in parentheses. (E, envelope gene; ORF, open reading frame).
IU, information unavailable.
Geographic distribution of genotypes.
To understand better the geographic distribution of JEV genotypes, a list of all strains of JEV for which there are any sequence data available was made. This was done by searching GenBank, PubMed, and where necessary, contacting authors directly. In addition we sequenced the structural genes of two strains from Sarawak, Malaysian Borneo, that have not been studied previously: Sarawak strain isolated from mosquitoes in 1965 and CNS138-11 isolated from the brain of a fatal human case in 1999. Based on the available sequence data, phylogenetic trees were constructed for E gene, C/PrM gene, and NS5/3′ UTRs. Each JEV isolate was thus placed into a genotype by using the classification of Chen et al. (5), Ali and Igarashi (1), and Uchil and Satchidanandam (38).
Rates of evolution.
To determine the rate of nucleotide substitutions for JEV, the number of pairwise nonsynonymous substitutions per site, obtained by comparing each member of a clade to its predicted ancestor based on maximum likelihood analysis, was plotted against the time period in years that separates the isolates to determine k, the rate of nonsynonymous substitutions per site. Nucleotide divergence rates were also estimated by comparing sister sequences that were closely related and isolated at least 5 years apart in the same geographical region. The differences in nonsynonymous changes depicted in branch lengths separating each sister sequence from the predicted common ancestor were divided by the number of years between isolates to yield rates expressed as the number of nonsynonymous changes per year.
Mouse neurovirulence studies.
To look for differences in pathogenicity, a representative strain of each genotype (KE093-83, genotype I; JKT1724, genotype II; VN118, genotype III; JKT6468, genotype IV) was injected intracerebrally (20 μl at 10-fold serial dilutions) into groups of five female NIH Swiss mice (3 to 4 weeks old). Animal experiments were conducted in accordance with the American Association for Accreditation of Laboratory Animal Care, and an Animal Welfare Assurance is on file with OPRR-NIH.
Statistical analysis.
The relationships between geographical location and genotype were analyzed by using the Fisher's exact test (SAS/STAT version 8; SAS Institute, Cary, N.C.).
Nucleotide sequence accession numbers.
The JEV isolates JKT6468, JKT7003, CNS138-11, and Sarawak were assigned GenBank database accession no. AY184212, AY184215, AY184213, and AY184214, respectively.
RESULTS
Nucleotide and amino acid sequence.
The JKT6468 virus genome was 10,978 nucleotides long (GenBank accession no. AY184212). Comparison with 17 other fully sequenced JEV genomes showed JKT6468 to have the least similarity, with nucleotide divergence ranging from 17.4 to 19.6%, and protein divergence from 4.8 to 6.5%. JKT6468 is compared with a fully sequenced representative of genotype I (K94P05), genotype II (FU) and genotype III (JaOArS982) in Table 2. Analysis by individual genes showed the PrM gene had the greatest divergence (28% compared with that of JaOArS982), while the 3′ noncoding region had the least divergence (9%). Phylogenetic analysis of all 18 full-length genomes for JEV and a representative strain for other viruses in the JE serogroup suggested that SLEV diverged at the deepest node followed by WNV and KUNV, MVEV, and finally JEV (Fig. 1A). (Indeed, recent flavivirus phylogenies based on the NS5 gene suggest SLEV may belong to the Ntaya serogroup rather than the JE serogroup [8, 9]). Similar trees were produced by the neighbor-joining, maximum-parsimony, and maximum likelihood methods, differing only in a few terminal groupings of genotype III. Of the JEV genotypes, genotype IV was the basal group in all trees, diverging at the deepest node with 100% bootstrap support, suggesting that it represents the most ancient lineage, which branched off before genotypes I, II, and III. Sequence data are available only for Muar strain's structural proteins. We therefore derived a phylogenetic tree from structural proteins, which suggested that the Muar strain belonged to a separate genotype (genotype V), which diverged at a deeper node than genotype IV (as is shown in the E gene phylogeny [Fig. 1B]).
TABLE 2.
Sequence divergence among the four genotypes of JEV
| Strain | Genotype | Divergencea (%) with:
|
|||
|---|---|---|---|---|---|
| K94P05 | FU | JaOArS982 | JKT6468 | ||
| K94P05 | I | 11.4 | 11.6 | 19.6 | |
| FU | II | 3.3 | 11.7 | 18.8 | |
| JaOArS982 | III | 2.8 | 2.1 | 17.4 | |
| JKT6468 | IV | 6.5 | 5.6 | 4.8 | |
Nucleotide divergence is in lightface type; amino acid divergence is in boldface type.
FIG. 1.
Phylogenetic trees of JEV. (A) Neighbor-joining phylogeny of complete JEV genomes, with a representative strain from other viruses in the JEV serogroup (KUN, Kunjin; MVE, Murray Valley encephalitis; SLE, St. Louis encephalitis; WN, West Nile), outgrouped by using Dengue-2 strain New Guinea C. (B) Neighbor-joining phylogeny of envelope genes, outgrouped by using MVE strain 1-51. Indonesian isolates are boxed. The tree was initially constructed by using more than 200 isolates from all geographical areas, but for clarity only a representative isolate of each genotype from each geographical area is shown. Isolates sequenced in this paper are shown with arrows. Genotypes are given on the right of each tree (5, 38). Bootstrap support values, given as a percentage of 1,000 replicates, are shown.
Rates of evolution and times of divergence.
To estimate the rate of evolution of JEV we examined full-length genomes with a sister pair comparison method and a regression analysis method (38, 41, 43). The sister pair method estimated the mean (standard deviation) rate of nonsynonymous substitutions per nonsynonymous site per year as 7.4 × 10−5 (4.2 × 10−5), and the regression method estimated it as 8.0 × 10−5. From these rates we determined that genotype IV JEV diverged from the common ancestor approximately 350 (±150) years ago, and genotypes I, II, and III diverged more recently.
Mouse neurovirulence.
Because differences in virulence between the genotypes has been postulated to explain the differing clinical epidemiology of JE, we examined whether the different genotypes have different phenotypes in mice by intracerebral inoculation of representative strains. There was no significant differences between genotypes in mouse neurovirulence. The dose which caused 50% mortality ranged from 0.25 to 3 PFU, with survival times from 5 to 10 days.
Geographical distribution of genotypes.
Most isolates of JEV have been characterized genetically by sequencing the E gene, a region spanning part of C/PrM genes, or a region spanning the NS5/3′ UTR. To examine the geographical distribution of JEV genotypes, we constructed phylogenetic trees for all available sequences using these genes. Figure 1B, the E gene phylogeny, which for clarity includes only a single representative strain for each genotype in each country, shows that whereas some genotypes have been found only in a single country (e.g., genotype IV was found only in Indonesia), others (especially genotype III) are more widely distributed. Similarly, whereas some countries have had several genotypes of JEV isolated (e.g., Indonesia has genotypes II, III, and IV), other countries have only had a single genotype isolated (e.g., India has only genotype III). The figure also shows that Muar strain, representing the putative 5th genotype, diverges at a node more basal than that of genotype IV.
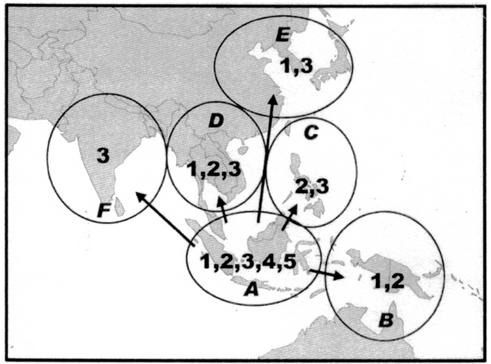
Basing the geographical distribution of isolates on the political boundaries of countries includes many geographical anomalies which are unlikely to have a bearing on the distribution of a virus in nature. For example, the island of Borneo is divided politically into Indonesia (Kalimantan), Malaysia (Sarawak and Sabah), and Brunei; the island of New Guinea is divided into Indonesia (Irian Jaya) and Papua New Guinea. Moreover, some of the political delineations have changed with time. We therefore grouped countries into geographical regions after considering their geographical proximity, temperature, rainfall, and vegetation and examined the distribution of JEV genotypes according to region. This also showed an uneven distribution of genotypes (Fig. 2). Whereas most regions had only one or two genotypes present, the Indonesia-Malaysia region had all five genotypes circulating. The number of different genotypes detected in a region depends to some extent on the number of virus strains that have been isolated in that region (i.e., the more strains looked at, the more likely one is to identify different genotypes). Therefore, to determine whether the observed difference represents a true difference in genotype distribution or is simply a result of sampling bias, we constructed a contingency table to analyze all strains by geographical region and genotype (Table 3). Every strain of JEV for which there is information available was placed into a genotype by using phylogenetic trees constructed from analyzing E, C/PrM, or NS5/3′ UTR genes. For the few strains where there was discordance between the E and C/PrM trees, the E gene was given precedence because it has been shown to be a better predictor of phylogenies determined by complete genomes (38). Comparing the Indonesia-Malaysia region with all other regions combined showed a highly significant difference in the geographical distribution of genotypes (Fisher's exact test, P < 0.0001). An alternative analysis, in which no geographical areas were combined, but genotypes IV and V (which diverged earlier) were compared with I, II, and III (more recently evolved genotypes), confirmed that the observed distribution of genotypes represented a true difference (Fisher's exact test, P < 0.0001). Pairwise analysis showed no significant difference between geographical regions C, E, and F. A further analysis was therefore performed in which these three regions combined were compared with regions A, B, and D for genotypes I, II, III, and IV (the single genotype V isolate was grouped with genotype IV for this analysis). Again this showed a significant difference in the geographical distribution of genotypes (Fisher's exact test, P < 0.001). Alternative geographical groupings, e.g., including Taiwan in region E or D, did not significantly alter these findings. The time intervals during which viruses had been isolated did not differ significantly between regions and thus could not explain the differences in genotypes found. All genotypes, including the most divergent genotypes (IV and V), are present in the Indonesia-Malaysia region, but only the most recently evolved genotypes (I, II, and III) are present in the other geographical regions (Fig. 2).
FIG. 2.
The geographical distribution and spread of JEV genotypes. Countries are grouped into geographical regions: A, Indonesia (excluding New Guinea) and Malaysia; B, Australia and New Guinea; C, Taiwan and the Philippines; D, Thailand, Cambodia, and Vietnam; E, Japan, Korea, and China; F, India, Sri Lanka, and Nepal. Region A contains all genotypes of JEV, including the oldest. The newer genotypes (I, II, and III) have subsequently spread to other geographical areas.
TABLE 3.
Geographical distribution of JEV genotypesa
| Geographical region | Genotype
|
|||||
|---|---|---|---|---|---|---|
| I | II | III | IV | V | Total | |
| Indonesia-Malaysia | 5 | 16 | 17 | 5 | 1 | 44 |
| Australia-New Guinea | 2 | 21 | 23 | |||
| Taiwan-Philippines | 1 | 67 | 68 | |||
| Thailand-Cambodia-Vietnam | 12 | 2 | 26 | 40 | ||
| Japan-Korea-China | 9 | 85 | 94 | |||
| India-Sri Lanka-Nepal | 21 | 21 | ||||
| Total | 28 | 40 | 216 | 5 | 1 | 290 |
Each strain of JEV for which there are sequence data available was categorized according to its genotype and the geographical area from which it came.
Structural differences.
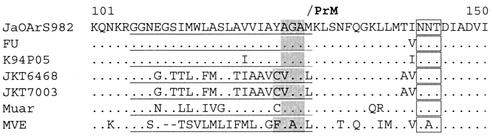
JEV uses a wide range of mosquito species and vertebrate hosts across Asia, and geographical differences in vector and host availability may explain why genotype IV has never spread. A direct comparison of the different mosquito species from which viruses have been isolated was inconclusive because details are known for too few isolates. We therefore examined the virus' complete amino acid sequence to look for molecular determinants that might relate to differences in vector or host preference. Genotype IV had the greatest amino acid divergence compared with other genotypes (Table 2), with 19% of changes occurring in the C protein. These included an unusual series of amino acids near to its carboxy terminus, which are unlike those seen in other JEV strains (Fig. 3). Structural analysis showed that despite these differences, the C protein retained the hydrophobic transmembrane domain at its carboxy terminus (by which it attaches to endoplasmic reticulum during assembly) and its central hydrophobic region (which is attached to membrane in the virion associated form). Because this region of the C protein was so different from the other JEV genotypes, a second JEV isolate from genotype IV (JKT7003 isolated from mosquitoes in Java, Indonesia, in 1981) was also sequenced for this region (GenBank accession no. AY184215). It was found to have an identical sequence to that of JKT6468. Comparison with the Muar strain of JEV and MVEV showed the amino acid sequence of these two viruses was also very different from that of JEV genotypes I to III (Fig. 3).
FIG. 3.
The variable region of JEV genotype IV capsid (C) protein. Genotype IV strain JKT6468 was aligned with JEV strains K94P05 (genotype I), FU (II), JaOArS982 (III), Muar (V), and MVE 1-51. The variable region of 20 amino acids just proximal to the carboxy terminus of genotype IV viruses is shown here. Amino acids are compared with those of JaOArS982; dots indicate conserved residues, dashes indicate gaps. A second genotype IV strain (Indonesian strain JKT7003) was also sequenced for this region and had identical sequence to JKT6468. Although this region of the C protein is markedly different for genotype IV, it still includes a hydrophobic transmembrane domain (underlined). Shaded regions indicate the predicted signalase/protease cleavage motifs. Boxes indicate the predicted N-linked glycosylation sites.
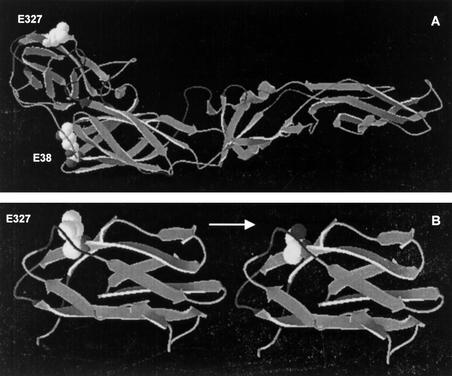
Examination of the E protein of JKT6468 showed it was similar to other flaviviruses, with 12 cysteine residues thought to form six disulfide bridges. The E protein is thought to play a critical role in viral attachment and entry into cells and is highly conserved among flaviviruses (21). X-ray crystallography of the E protein of the flavivirus tick-borne encephalitis virus (TBEV) has shown the three-dimensional structure comprises three domains: domain III, which is the receptor binding domain, domain II, the dimerization domain which also contains the fusion peptide, and domain I, which acts as a hinge between the other two domains (27). A comprehensive alignment of E genes from 120 JEV strains identified three signature amino acid residues that distinguished genotype IV from other genotypes (E38 Arg→Lys, E327 Leu→Thr/Ser, E399 Pro→Ala/Thr) (Fig. 4). The predicted three-dimensional structure of the E protein of JKT6468, modeled on the structure for TBEV, showed E38 mapped to domain I, E399 was in the stem anchor region, and E327 mapped to the exposed lateral surface of domain III in a region thought to be involved in receptor binding (Fig. 3) (22).
FIG. 4.
JEV envelope (E) protein. (A) Predicted three-dimensional model of the E protein of JEV genotype IV strain JKT6468, derived from the crystal structure of TBEV (27), showing E38 in domain I (the hinge domain), and E327 on the exposed surface of domain III. (E399 is in the stem region from domain III into the membrane anchor; not shown). (B) The critical change at residue 327 (in the putative receptor binding region of domain III) from the aliphatic amino acid leucine in genotype IV strains to the hydroxyl-containing serine found in newer genotypes.
DISCUSSION
Arthropod-borne viruses that damage the nervous system are being recognized as increasingly important emerging and reemerging human pathogens, but their origins and evolution remain unclear (29). JE was described in Japan from the 1870s onwards, and the prototype Nakayama strain was isolated from a fatal case in 1935. Since then the disease apparently spread across Asia to affect most of China and the Asian subcontinent, all of southeast Asia, and the Pacific Rim, reaching northern Australia in 1998. Although it is numerically less important than JE, the related WNV has recently reached southern Europe and North America (18, 37), drawing attention to the devastating potential of these viruses to spread. There is a clear need for a better understanding of the origins and spread of these major causes of morbidity and mortality.
Previous attempts to understand the distribution of JEV have tended to consider genotypes according to the countries most frequently represented, which may simply be a function of how many strains have been isolated from a particular place. Hence, genotypes I and III were considered northern epidemic genotypes and genotypes II and IV were considered southern endemic genotypes. And even when a genotype I isolate was identified in the south (26) it was considered that the genotype may have moved from epidemic to endemic areas. In our study, by concentrating on the presence or absence of a particular genotype in each geographical area and by using a robust statistical analysis to determine whether the observed differences are likely to represent the true distribution in nature, we have shown that not only is the Indonesia-Malaysia region the only area where all genotypes are represented, it is also the only region where the most divergent genotypes (IV and V) that are thought to represent the oldest lineages, have so far been found. The Indonesia-Malaysia region is geographically close to Australia, where JEV's closest fully sequenced relative, MVEV, is found. Alfuy virus, another member of the JEV-serogroup, which is thought to occur in Indonesia, New Guinea, and Australia, may actually be genetically closer to JEV (20), but it has not been fully sequenced. (Usutu virus, an African flavivirus, also lies close to JEV in phylogenies based on limited sequence, but it too has not been fully sequenced [17]). Taken together these observations suggest that JEV originated in the Indonesia-Malaysia region from an ancestral virus common to JEV and MVEV. From this ancestral virus JEV genotypes IV and V diverged, followed by the more recent genotypes I, II, and III. Whereas more recent genotypes have spread to other areas, the more divergent genotypes (IV and V) appear geographically confined to the Indonesia-Malaysia region. Indeed, only a single genotype V isolate has ever been found, and that was isolated in 1952 (13). This and its marked serological differences from other JE strains suggests it may represent a transitional lineage between MVEV and JEV, which has subsequently become extinct.
We also considered other explanations for our observations. For example, the most divergent genotypes (IV and V) may yet be found in different geographical areas, though the intensive surveillance in Japan, Taiwan, and Australia suggests that they are unlikely to have been missed in these countries. An alternative possibility is that these genotypes of JEV existed in more Northern latitudes and have subsequently disappeared. However, epidemiological evidence argues against this. Studies have shown that when JEV arrives in new areas, adults are affected as well as children because they have no preexisting immunity (35). The fact that when JE was first seen in Japan all ages were affected thus provides a strong indication that this was the first time the virus (a genotype III virus) arrived here (33). Interestingly, such large epidemics never seem to have occurred in the Indonesia-Malaysia region, which is also consistent with the virus having evolved here. Nevertheless, it is possible that our conclusions will need revising as the complete nucleotide sequences of other JEV strains and other flaviviruses become available.
For most of the major human pathogens, we have little idea about where they originated. Because JEV has recently evolved and has so rapidly spread to new areas, tracing its geographical origin has been possible. JEV is naturally transmitted between vertebrate hosts by Culex and other mosquitoes. The reasons for its spread are uncertain but may include changing agricultural practices, such as increasing irrigation (which provides mosquito breeding sites), and animal husbandry (which provides host animals) (35). Migrating birds, particularly the black-crowned night heron (Nycticorax nycticorax) and the Asiatic cattle egret (Bubulcus ibis coromandus), are thought to be important in the virus' dispersal to new geographic areas (15), but wind-blown mosquitoes may also play a role (12). Interestingly, the Asiatic cattle egret's range dramatically expanded across Asia in the 19th century following changing agricultural practices (11), which coincides with the evolution and spread of the more recent JEV genotypes. JEV continues to spread with recent outbreaks in India, Nepal, and Australia. Genotype III is the most widely distributed genotype and is the only genotype that has so far been found in India, despite the fact that fifteen strains from diverse geographical locations over 26 years have been genotyped. Ultimately we might expect genotypes I and II to be found there also. Similarly, we might predict genotype III will eventually reach Australia, and genotype II will be found further north in China or Japan.
Why the more ancient lineages of JEV have not spread is uncertain. JEV uses a wide range of mosquito species and vertebrate hosts across Asia, and geographical differences in vector and host availability may explain why genotype IV has never spread. We examined the virus' complete amino acid sequence to look for molecular determinants that might relate to differences in host preference. We found an unusual series of amino acids near to the carboxy terminus of the C protein, which was different from that seen in other JEV strains and was consistent with divergent evolution of this genotype. However, structural analysis suggested the C protein retained its hydrophobic domains and its excess of basic amino acids, which are thought to be important for its function in forming the nucleocapsid. There were fewer changes in the E protein, but a comprehensive alignment of E proteins identified three signature amino acids, one of which (E327) mapped to the exposed lateral surface of domain III in a region thought to be involved in receptor binding. These findings are consistent with the hypothesis that the change from an aliphatic amino acid (Leu) to a hydroxyl-containing amino acid (Ser/Thr) in this critical region of the E protein, by altering vector and/or host preference, could have contributed to the wider dispersal of the newer JEV genotypes compared with those of genotype IV. Previously a small number of changes in E protein sequence have been shown to affect mosquito oral infectivity (4, 24). Clearly, further studies are needed to substantiate this hypothesis.
The suggestion that JEV originated in the Indonesia-Malaysia region raises intriguing questions about the emergence of viruses in Southeast Asia. Nearly 150 years ago the evolutionary biologist Wallace recognized that this region, the Malay Archipelago as it was then known, has a unique environment that allows divergent evolution of plants and animals (40). Its tropical climate and great diversity of insect and vertebrate life may also facilitate the emergence and rapid evolution of viruses. This is supported by one hypothesis that in this same region WNV may have evolved into its Australian subtype, KUNV (25, 28), and dengue may have evolved from a sylvatic to a human virus (41). The greater diversity of MVEV strains found in Papua New Guinea than in Australia (19) is consistent with the idea that this flavivirus also spread from here to Australia. Traditionally, Africa is thought to be the zone of emerging pathogens. Our data, together with recent evidence on the origins of dengue virus (41) and Nipah virus (7), indicate that tropical Southeast Asia may also be an important region for emerging pathogens.
Acknowledgments
We thank R. Tesh for providing strain JKT6468 from the World Arbovirus Reference Collection our colleagues in Sarawak for providing the clinical material from which CNS138-11 was isolated; S. Weaver and J. Bryant for assistance with the rates of evolution and times of divergence; S. White and D. Freeman for statistical help; R. Telfair for helpful discussions on egret migration; M. Cutchin for geographical advice; and C. A. Hart, R. Tesh, and R. Shope for useful critiques.
This work was funded in part by the World Health Organization Vaccine Development Initiative, NIH grant AI10986, and the Wellcome Trust of Great Britain. T.S. holds a Wellcome Trust Career Development Award.
REFERENCES
- 1.Ali, A., and A. Igarashi. 1997. Antigenic and genetic variations among Japanese encephalitis virus strains belonging to genotype 1. Microbiol. Immunol. 41:241-252. [DOI] [PubMed] [Google Scholar]
- 2.Briese, T., X.-Y. Jia, C. Huang, L. J. Grady, and W. I. Lipkin. 1999. Identification of a Kunjin/West Nile-like flavivirus in brains of patients with New York encephalitis. Lancet 354:1261-1262. [DOI] [PubMed] [Google Scholar]
- 3.Chambers, T. J., C. S. Hahn, R. Galler, and C. M. Rice. 1990. Flavivirus genome organisation, expression and replication. Annu. Rev. Microbiol. 44:649-688. [DOI] [PubMed] [Google Scholar]
- 4.Chen, B. Q., and B. J. Beaty. 1982. Japanese encephalitis vaccine (2-8 strain) and parent (SA 14 strain) viruses in Culex tritaeniorhynchus mosquitoes. Am. J. Trop. Med. Hyg. 31:403-407. [DOI] [PubMed] [Google Scholar]
- 5.Chen, W. R., R. Ricco-Hesse, and R. B. Tesh. 1992. A new genotype of Japanese virus from Indonesia. Am. J. Trop. Med. Hyg. 47:61-69. [DOI] [PubMed] [Google Scholar]
- 6.Chen, W. R., R. B. Tesh, and R. Ricco-Hesse. 1990. Genetic variation of Japanese encephalitis virus in nature. J. Gen. Virol. 71:2915-2922. [DOI] [PubMed] [Google Scholar]
- 7.Chua, K. B., W. J. Bellini, P. A. Rota, B. H. Harcourt, A. Tamin, S. K. Lam, T. G. Ksiazek, P. E. Rollin, S. R. Zaki, W.-J. Shieh, C. S. Goldsmith, D. J. Gubler, J. T. Roehrig, B. Eaton, A. R. Gould, J. Olson, H. Field, P. Daniels, A. E. Ling, C. J. Peters, L. J. Anderson, and B. W. J. Mahy. 2000. Nipah virus: a recently emergent deadly paramyxovirus. Science 288:1432-1435. [DOI] [PubMed] [Google Scholar]
- 8.Gaunt, M. W., A. A. Sall, X. Lamballerie, A. K. Falconar, T. I. Dzhivanian, and E. A. Gould. 2001. Phylogenetic relationships of flaviviruses correlate with their epidemiology, disease association and biogeography. J. Gen. Virol. 82:1867-1876. [DOI] [PubMed] [Google Scholar]
- 9.Gould, E. A. 2002. Evolution of the Japanese serological group viruses, p. 391-404. In J. S. Mackenzie, A. D. Barrett, and V. Deubel (ed.), Current topics in microbiology and immunology: Japanese encephalitis and West Nile virus infections, vol. 267. Springer-Verlag, Berlin, Germany.
- 10.Gould, E. A., P. M. Zanotto, and E. C. Holmes. 1997. The genetic evolution of flaviviruses, p. 51-63. In J. F. Saluzzo and B. Dodet (ed.), Factors in the emergence of arbovirus diseases. Elselvier, Paris, France.
- 11.Hancock, J., and J. Kushlan. 1984. The herons handbook. Harper and Row, New York, N.Y.
- 12.Hanna, J., S. Ritchie, D. A. Phillips, J. Shield, M. C. Bailey, J. S. Mackenzie, M. Poidinger, B. J. McCall, and P. J. Mills. 1996. An outbreak of Japanese encephalitis in the Torres Trait, Australia, 1995. Med. J. Aust. 165:256-260. [DOI] [PubMed] [Google Scholar]
- 13.Hasegawa, H., M. Yoshida, S. Fujita, and Y. Kobayashi. 1994. Comparison of structural proteins among antigenically different Japanese encephalitis virus strains. Vaccine 12:841-844. [DOI] [PubMed] [Google Scholar]
- 14.Huong, V. T. Q., D. Q. Ha, and V. Deubel. 1993. Genetic study of Japanese encephalitis viruses from Vietnam. Am. J. Trop. Med. Hyg. 49:538-544. [DOI] [PubMed] [Google Scholar]
- 15.Innis, B. L. 1995. Japanese encephalitis, p. 147-174. In J. S. Porterfield (ed.), Exotic viral infections. Chapman & Hall, London, United Kingdom.
- 16.Jan, L. R., K. L. Chen, C. F. Lu, Y. C. Wu, and C. B. Horng. 1996. Complete nucleotide sequence of the genome of Japanese encephalitis virus ling strain: the presence of a 25-nucleotide deletion in the 3′-nontranslated region. Am. J. Trop. Med. Hyg. 55:603-609. [DOI] [PubMed] [Google Scholar]
- 17.Kuno, G., G. J. Chang, K. R. Tsuchiya, N. Karabatsos, and C. B. Cropp. 1998. Phylogeny of the genus Flavivirus. J. Virol. 72:73-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lanciotti, R. S., J. T. Roehrig, V. Deubel, J. Smith, M. Parker, K. Steele, B. Crise, K. E. Volpe, M. B. Crabtree, J. H. Scherret, R. A. Hall, J. S. MacKenzie, C. B. Cropp, B. Panigrahy, E. Ostlund, B. Schmitt, M. Malkinson, C. Banet, J. Weissman, N. Komar, H. M. Savage, W. Stone, T. McNamara, and D. J. Gubler. 1999. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science 286:2333-2337. [DOI] [PubMed] [Google Scholar]
- 19.Lobigs, M., I. D. Marshall, R. C. Weir, and L. Dalgarno. 1988. Murray Valley encephalitis virus field strains from Australia and Papua New Guinea: studies on the sequence of the major envelope protein gene and virulence for mice. Virology 165:245-255. [DOI] [PubMed] [Google Scholar]
- 20.Mackenzie, J. S., C. A. Johansen, S. A. Ritchie, A. F. van den Hurk, and R. A. Hall. 2002. Japanese encephalitis virus as an emerging virus: the emergence and spread of Japanese encephalitis virus in Australasia, p. 49-73. In J. S. Mackenzie, A. D. Barrett, and V. Deubel (ed.), Current topics in microbiology and immunology: Japanese encephalitis and West Nile virus infections, vol. 267. Springer-Verlag, Berlin, Germany. [DOI] [PubMed]
- 21.Monath, T. P., and F. X. Heinz. 1996. Flaviviruses, p. 961-1034. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 22.Ni, H., and A. D. Barrett. 1998. Attenuation of Japanese encephalitis virus by selection of its mouse brain membrane receptor preparation escape variants. Virology 241:30-36. [DOI] [PubMed] [Google Scholar]
- 23.Ni, H., and A. D. Barrett. 1995. Nucleotide and deduced amino acid sequence of the structural protein genes of Japanese encephalitis viruses from different geographical locations. J. Gen. Virol. 76:401-407. [DOI] [PubMed] [Google Scholar]
- 24.Ni, H., N. J. Burns, G. J. Chang, M. J. Zhang, M. R. Wills, D. W. Trent, P. G. Sanders, and A. D. Barrett. 1994. Comparison of nucleotide and deduced amino acid sequence of the 5′ noncoding region and structural protein genes of the wild-type Japanese encephalitis virus strain SA14 and its attenuated vaccine derivatives. J. Gen. Virol. 75:1505-1510. [DOI] [PubMed] [Google Scholar]
- 25.Poidinger, M., R. A. Hall, and J. S. Mackenzie. 1996. Molecular characterization of the Japanese encephalitis serocomplex of the flavivirus genus. Virology 218:417-421. [DOI] [PubMed] [Google Scholar]
- 26.Pyke, A. T., D. T. Williams, D. J. Nisbet, A. F. van den Hurk, C. T. Taylor, C. A. Johansen, J. Macdonald, R. A. Hall, R. J. Simmons, R. J. Mason, J. M. Lee, S. A. Ritchie, G. A. Smith, and J. S. Mackenzie. 2001. The appearance of a second genotype of Japanese encephalitis virus in the Australasian region. Am. J. Trop. Med. Hyg. 65:747-753. [DOI] [PubMed] [Google Scholar]
- 27.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 28.Scherret, J. H., J. S. Mackenzie, R. A. Hall, V. Deubel, and E. A. Gould. 2002. Phylogeny and molecular epidemiology of West Nile and Kunjin viruses. Curr. Top. Microbiol. Immunol. 267:373-390. [DOI] [PubMed] [Google Scholar]
- 29.Solomon, T., and M. J. Cardosa. 2000. Emerging arboviral encephalitis. Br. Med. J. 321:1484-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Solomon, T., N. M. Dung, R. Kneen, M. Gainsborough, D. W. Vaughn, and V. T. Khanh. 2000. Japanese encephalitis. J. Neurol. Neurosurg. Psychiatry 68:405-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solomon, T., N. M. Dung, R. Kneen, L. T. Thao, M. Gainsborough, A. Nisalak, N. P. Day, F. J. Kirkham, D. W. Vaughn, S. Smith, and N. J. White. 2002. Seizures and raised intracranial pressure in Vietnamese patients with Japanese encephalitis. Brain 125:1084-1093. [DOI] [PubMed] [Google Scholar]
- 32.Solomon, T., R. Kneen, N. M. Dung, V. C. Khanh, T. T. N. Thuy, D. Q. Ha, N. P. J. Day, A. Nisalak, D. W. Vaughn, and N. J. White. 1998. Poliomyelitis-like illness due to Japanese encephalitis virus. Lancet 351:1094-1097. [DOI] [PubMed] [Google Scholar]
- 33.Solomon, T., and D. W. Vaughn. 2002. Clinical features and pathophysiology of Japanese encephalitis and West Nile virus infections, p. 171-194. In J. S. Mackenzie, A. D. Barrett, and V. Deubel (ed.), Current topics in microbiology and immunology: Japanese encephalitis and West Nile virus infections, vol. 267. Springer-Verlag, Berlin, Germany. [DOI] [PubMed]
- 34.Sumiyoshi, H., C. Mori, I. Fuke, K. Morita, S. Kuhara, J. Kondou, Y. Kikuchi, H. Nagamtu, and A. Igarashi. 1987. Complete nucleotide sequence of the Japanese encephalitis virus genome RNA. Virology 161:497-510. [DOI] [PubMed] [Google Scholar]
- 35.Tsai, T. F. 1997. Factors in the changing epidemiology of Japanese encephalitis and West Nile fever, p. 179-189. In J. F. Saluzzo and B. Dodet (ed.), Factors in the emergence of arbovirus diseases. Elselvier, Paris, France.
- 36.Tsai, T. F. 2000. New initiatives for the control of Japanese encephalitis by vaccination: minutes of a W. H. O./CVI meeting, Bangkok, Thailand, 13-15 October 1998. Vaccine 18(Suppl. 2):1-25. [DOI] [PubMed] [Google Scholar]
- 37.Tsai, T. F., F. Popovici, C. Carnescu, G. L. Campbell, and N. I. Nedlelcu. 1998. West Nile encephalitis epidemic in Southeastern Romania. Lancet 352:767-771. [DOI] [PubMed] [Google Scholar]
- 38.Uchil, P. D., and V. Satchidanandam. 2001. Phylogenetic analysis of Japanese encephalitis virus: envelope gene based analysis reveals a fifth genotype, geographic clustering, and multiple introductions of the virus into the Indian subcontinent. Am. J. Trop. Med. Hyg. 65:242-251. [DOI] [PubMed] [Google Scholar]
- 39.Vaughn, D. W., and C. H. Hoke. 1992. The epidemiology of Japanese encephalitis: prospects for prevention. Epidemiol. Rev. 14:197-221. [DOI] [PubMed] [Google Scholar]
- 40.Wallace, A. R. 1869. The Malay Archipelago. The land of the Orang-utan and the bird of paradise. A narrative of travel, with studies of man and nature, vol. I. MacMillan and Co., London, United Kingdom.
- 41.Wang, E., H. Ni, R. Xu, A. D. Barrett, S. J. Watowich, D. J. Gubler, and S. C. Weaver. 2000. Evolutionary relationships of endemic/epidemic and sylvatic dengue viruses. J. Virol. 74:3227-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams, D. T., L. F. Wang, P. W. Daniels, and J. S. Mackenzie. 2000. Molecular characterization of the first Australian isolate of Japanese encephalitis virus, the FU strain. J. Gen. Virol. 81:2471-2480. [DOI] [PubMed] [Google Scholar]
- 43.Zanotto, P. M., E. A. Gould, G. F. Gao, P. H. Harvey, and E. C. Holmes. 1996. Population dynamics of flaviviruses revealed by molecular phylogenies. Proc. Natl. Acad. Sci. USA 93:548-553. [DOI] [PMC free article] [PubMed] [Google Scholar]