Abstract
Retrovirus entry into cells is mediated by the viral envelope glycoproteins which, through a cascade of conformational changes, orchestrate fusion of the viral and cellular membranes. In the absence of membrane fusion, viral entry into the host cell cannot occur. For human T-cell leukemia virus type 1 (HTLV-1), synthetic peptides that mimic a carboxy-terminal region of the transmembrane glycoprotein (TM) ectodomain are potent inhibitors of membrane fusion and virus entry. Here, we demonstrate that this class of inhibitor targets a fusion-active structure of HTLV-1 envelope. In particular, the peptides bind specifically to a core coiled-coil domain of envelope, and peptide variants that fail to bind the coiled-coil lack inhibitory activity. Our data indicate that the inhibitory peptides likely function by disrupting the formation of a trimer-of-hairpins structure that is required for membrane fusion. Importantly, we also show that peptides exhibiting dramatically increased potency can be readily obtained. We suggest that peptides or peptide mimetics targeting the fusion-active structures of envelope may be of therapeutic value in the treatment of HTLV-1 infections.
Retrovirus entry into cells is achieved through fusion of the lipid bilayers that surround both the virus and the target cell, thus releasing the viral core into the host cell cytoplasm. Membrane fusion is mediated by the virally encoded envelope glycoproteins (Env), which are presented on the surface of the virus or infected cell as a trimer of surface glycoprotein (SU) subunits anchored to a trimer of transmembrane glycoproteins (TM). Models for Env function suggest that binding of SU to a cell surface receptor induces global changes in Env conformation that convert Env from a “resting” nonfusogenic state to a fusion-active conformation (reference 13 and references therein; 38). The receptor-stimulated conformational changes within Env facilitate insertion of the N-terminal hydrophobic fusion peptide of TM into the target cell membrane and subsequently promote membrane fusion (10, 18). In agreement with this model, mutations within Env (33), anti-Env antibodies (32, 37), recombinant competitive ligands (19), and Env-derived synthetic peptides (5, 22, 35) all interfere with Env activity and potently block retroviral infection of cells. Thus, inhibition of virus entry by small-molecule antagonists of Env function appears to be a viable objective in the development of clinically relevant antiretroviral therapies.
Crystal structures of fusion proteins from a number of viruses (2, 6, 9, 14, 27, 28, 34, 42, 45), including human T-cell leukemia virus type 1 (HTLV-1) (24), have provided considerable insight into the mechanism of Env-catalyzed membrane fusion. A common feature of fusion protein structure is the formation of a trimer-of-hairpins motif. For HTLV-1 TM (gp21), the N-terminal helices from three gp21 ectodomains form a central triple-stranded coiled-coil. At the base of the coiled-coil, the peptide backbone of each monomer forms a disulfide-bonded 180° loop that reverses the chain direction; finally, the C-terminal sequences run antiparallel to the coiled-coil, fold into an extended structure that includes a short helical region, and pack into the grooves formed by the core coiled-coil to complete the trimer of hairpins (24). It is likely that the trimer of hairpins represents a postfusion TM conformation, suggesting a model for membrane fusion in which insertion of the N-terminal fusion peptide into the target cell membrane results in the formation of a transient prehairpin intermediate. In the prehairpin conformation, one end of TM is anchored in the viral membrane while the other is embedded within the target membrane. Ultimately, the prehairpin intermediate resolves to the trimer-of-hairpins structure, which draws the viral and cellular membranes together, facilitates their destabilization, and induces fusion (13).
Interestingly, synthetic peptides that potently inhibit HTLV-1Env-mediated membrane fusion and virus entry into cells have been identified (5, 22, 35). One of the inhibitory peptides, P-400, models amino acids 400 to 429 of HTLV-1 Env. These residues span the C-terminal region of the TM ectodomain that packs into the grooves formed by the core coiled-coil. Jinno et al. (22) have suggested that P-400 exerts its inhibitory effect by blocking the interaction of HTLV-1 Env with a cellular receptor, or alternatively, by disrupting the interaction of gp21 with a nonprotein membrane component that supports membrane fusion. In support of that interpretation, it was noted that the crystal structure of TM places amino acid residues 400 to 429 at the surface of the trimeric hairpin, thereby allowing amino acid residues within this region to interact with components on the target cell surface (22).
An alternative, and perhaps simpler, view is that P-400 inhibits Env-mediated membrane fusion by directly interacting with the core coiled-coil of TM, thereby disrupting the conformational transitions required for Env-mediated membrane fusion. Importantly, a precedent for this type of inhibitory activity has been extensively documented for peptide inhibitors of HIV-1 (8, 12, 15, 23, 25, 43, 44; S. Jiang, N. Strick, and A. R. Neurath, Letter, Nature 365:113, 1993). Since there is a pressing need for effective antiretroviral therapies to treat HTLV-1-associated disease, we have examined in detail the inhibitory properties of the TM-mimetic P-400. Specifically, we set out to test the hypothesis that the target for the inhibitory peptide is a fusion-active structure of HTLV-1 Env. Here, we demonstrate that P-400-related peptides with increased potency can be readily obtained, that these peptides interact directly with the core coiled-coil domain of HTLV-1 TM, and that the inhibitory properties of the peptides correlate directly with their ability to bind to the coiled-coil region of TM.
MATERIALS AND METHODS
Cells.
HeLa and HOS cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum. Jurkat, MOLT-4, and SupT1 T-cell lines were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum.
Plasmids.
The plasmid pHTE-1 has been previously described (11). The plasmid pMAL-gp21hairpin was constructed by PCR amplification of the HTLV-1 TM region, using pHTE-1 as the template and the primers FPgp21 M338 (5′-CGGAATTCATGTCCCTCGCCTCAGGAAAGAGC-3′) and RPgp21T425 (5′-GCTCTAGAAAGCTTTCAAGTCAGGACTCGATTTTCAAGG-3′). The PCRfragment was cloned into the vector pMALc2 (New England Biolabs) using the EcoRI and HindIII sites introduced by the primers (indicated by underlined letters in the primer sequences). The resulting plasmid encodes amino acids Met338 to Thr425 of HTLV-1 TM fused in frame to maltose binding protein (MBP). Similarly, pMAL-gp21fishhook was made using the primer pair FPgp21 M338 and RPgp21C400 (5′-GCTCTAGAAAGCTTTCAGCACTGTTCTTGTAATGC-3′). In order to express the control MBP protein, a stop codon was introduced into the pMALc2 vector immediately after the EcoRI site by site-directed mutagenesis using the Quikchange method (Stratagene) to generate the plasmid pMAL-stop.
Peptides.
The peptides used in this study were chemically synthesized using standard techniques and are listed in Table 1. P-80, derived from the amino-terminal region of HTLV-1 gp46, has been previously described (5). P-400 was modeled on amino acids 400 to 429 of gp21 (gp68 numbering starting at the initiation codon) based on the published sequence of HTLV-1 strain ATK (36). Pcr-400 and its derivative peptides were synthesized based on our sequence analyses of pHTE-1. All peptides were dissolved in dimethyl sulfoxide for use at the appropriate concentrations.
TABLE 1.
Amino acid sequences of synthetic peptides used in this study
| Peptide | Amino acid positions | Sequence | MW |
|---|---|---|---|
| P-80 | gp46 80-96 | SLYLFPHWTKKPNRNGG | 2,118 |
| P-197 | gp46 197-216 | DHILEPSIPWKSKLLTLVQL | 2,331 |
| P-400 | gp21 400-429 | CRFPNITNSHVPILQERPPLENRVLTGWGL | 3,458 |
| Pcr-400a | gp21 400-429 | CCFLNITNSHVSILQERPPLENRVLTGWGL | 3,411 |
| Pcr-400L/Ab | gp21 400-429 | ---A---------A-----A----A----A | 3,200 |
| Pcr-NS407YFb | gp21 400-429 | -------YF--------------------- | 3,520 |
| Pcr-W427Ab | gp21 400-429 | ---------------------------A-- | 3,296 |
Amino acid substitutions are indicated in boldface.
Peptides are identical to Pcr-400 sequence except for substitutions shown.
Expression and purification of MBP-TM proteins.
Expression and purification of the MBP-TM proteins (New England Biolabs) were carried out according to the manufacturer's protocols. Briefly, Escherichia coli JM109 cells, transformed with pMAL-stop or the MBP-TM expression plasmids, were grown at 37°C in the presence of ampicillin (100 μg/ml) until the optical density at 600 nm reached 0.6. The cells were induced with isopropylthio-β-d-galactoside (IPTG) at a final concentration of 0.5 mM for 4 h at 37°C. The cells were harvested, resuspended in column buffer (20 mM Tris-HCl [pH 7.5], 200 mM NaCl, 1 mM EDTA) supplemented with phenylmethylsulfonyl fluoride (1 mM) and aprotinin (1 μg/ml), and subsequently lysed by sonication. Cell debris was pelleted by centrifugation at 9,000 × g for 30 min. The crude lysates were diluted 1:5 in column buffer and then loaded onto an amylose column that had been preequilibrated with column buffer. The column was then washed with 12 column volumes of column buffer, and the MBP and MBP-TM fusion proteins were eluted with column buffer containing 10 mM maltose. The concentration of the fusion proteins was estimated by Bradford assay. The recombinant proteins were stored at −80°C in column buffer supplemented with 20% glycerol.
Gel filtration chromatography.
The oligomerization states of our MBP-TM chimeras were assessed by Superdex 200 gel filtration chromatography in PBS. Gel filtration experiments were calibrated with thyroglobulin (669 kDa), ferritin (440 kDa), and catalase (232 kDa) from Pharmacia Biotech and gamma globulin (158 kDa) and ovalbumin (44 kDa) from Bio-Rad.
CD spectra and mass spectrometry analyses.
The circular dichroism (CD) spectrum of each peptide was recorded in H2O and in 50% (vol/vol) trifluoroethanol (TFE) at 20°C on a JASCO J-600 spectropolarimeter. Peptide concentrations of 0.5 mg/ml were used in quartz cells with a path length of 0.02 cm. The results were analyzed by the SELCON procedure (39). Mass spectrometry experiments were conducted using matrix-assisted laser desorption ionization (MALDI) in a Voyager De-Pro MALDI-time of flight mass spectrometer (Applied Biosystems).
Gel shift assays.
The oligomerization states of our MBP-TM chimeras were also investigated by native gel electrophoresis. Equivalent amounts of protein were added to sample buffer that lacked sodium dodecyl sulfate (SDS) and dithiothreitol or 2-mercaptoethanol. The samples were not heat treated prior to separation on an 8% polyacrylamide gel that did not contain SDS. Proteins were subsequently detected by Coomasie blue staining.
The in vitro interaction of the HTLV-1 TM-derived peptides with the MBP-TM chimeras was examined by gel shift on native polyacrylamide gels. Approximately 1.5 to 2.0 μg of MBP-TM fusion protein was incubated with various concentrations of peptide for 45 min at room temperature. The protein complexes formed were analyzed by electrophoresis as described above.
Biotinylation and pull-down assays.
Approximately 1.0 mg of Pcr-400 peptide was biotinylated using EZ-link PEO-Iodoacetyl biotin (Pierce) as directed by the manufacturer. Biotinylated peptide was separated from free biotin using a polyacrylamide gel desalting column (Pierce). Increasing amounts of biotinylated peptide were incubated with ∼20 to 30 μg of MBP-Fishhook or MBP in a reaction volume of 50 μl for 30 min at room temperature. Nine hundred microliters of RIPA buffer (PBS-0.25% Triton X-100) and 50 μl of streptavidin-agarose solution (Sigma) were added, and the reaction mixture was incubated at 4°C for 1 h with gentle rotation. The bound complexes were pelleted by centrifugation and washed five times with RIPA buffer. The proteins were heated in sample buffer and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE).
Syncytium interference assay.
Syncytium interference assays between target T cells (MOLT-4, SupT1, and Jurkat) and HTLV-1-infected MT2 cells has been described previously (5). Syncytium formation in non-T-cell lines was examined as described previously (5). Briefly, 0.5 × 106 to 1.0 × 106 HeLa cells, transfected with the envelope expression vector pHTE-1, were added to an equal number of untransfected target cells (HeLa, HOS, or Cos). Where appropriate, effector and target cells were incubated together in the presence of the P-400-related peptides at the concentrations specified. The cells were incubated for 12 to 15 h at 37°C, washed twice with phosphate-buffered saline (PBS), and then fixed in PBS-2% formaldehyde-0.2% glutaraldehyde. Assays were performed in triplicate, and the number of syncytia from 10 low-power fields per replicate was scored by light microscopy. The effect of preincubating Pcr-400 with MBP-Fishhook on syncytium formation was assayed in a similar manner. Pcr-400 was added to the target cells at a concentration of 1.5 μM. Following a 5-min incubation, up to twofold molar excess of either MBP or MBP-Fishhook was added and allowed to preincubate with the peptide for 40 min prior to the addition of the effector cells.
RESULTS
P-400 inhibits HTLV-1-induced syncytium formation among diverse cellular targets.
A synthetic peptide, P-400, blocks HTLV-1 Env-mediated membrane fusion at a post-receptor-binding step of the entry process (22), but the molecular mechanism by which P-400 functions has not been resolved. Analysis of existing data and examination of the crystal structure of HTLV-1 TM (24) suggested to us that P-400 may function by directly targeting viral Env. We therefore predicted that P-400 should inhibit membrane fusion and syncytium formation for all HTLV-1-permissive cell types and that the potency of the peptide would be independent of the cell type used. Interestingly, it was previously demonstrated that another synthetic peptide, P-197, modeled on a region of HTLV-1 SU, inhibits syncytium formation for all HTLV-1-permissive targets cells tested and is unlikely to block interaction of SU with a cellular receptor (5). We therefore compared the inhibitory activities of P-400 and P-197 in syncytium interference assays.
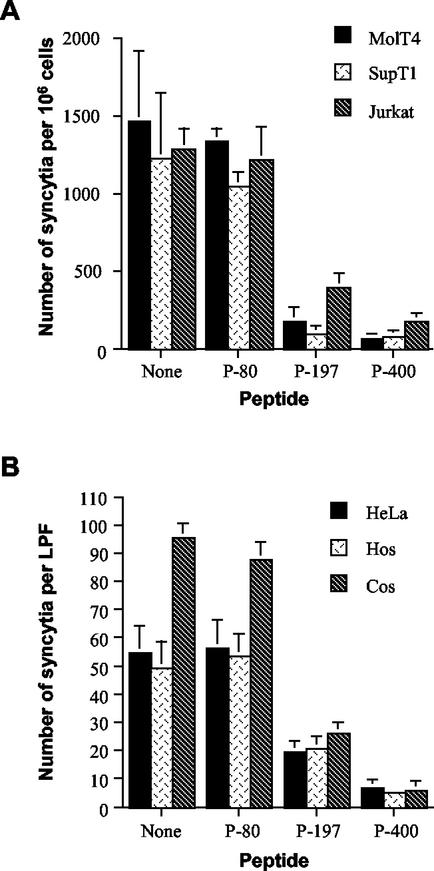
Coculture of HTLV-1-infected MT2 cells with MOLT4, SupT1, or Jurkat T cells resulted in extensive cell-to-cell fusion and syncytium formation. In contrast, when these cells were cocultured in the presence of P-400 or P-197, a dramatic reduction in the number of syncytia was observed (Fig. 1A). These results were specific to P-400 and P-197, as the control peptide, P-80, which was derived from the N-terminal region of HTLV-1 SU, did not inhibit syncytium formation.
FIG. 1.
Peptide P-400 inhibits HTLV-1 envelope-mediated syncytium formation among diverse target cells. (A) MOLT-4, SupT1, and Jurkat T cells were cocultured with HTLV-1-infected MT2 cells in the presence of the indicated peptides (20 μg/ml) or the solvent (dimethyl sulfoxide) alone (None). The data are means and standard deviations from six independent assays. (B) HeLa, HOS, or Cos target cells were cocultured with HTLV-1 Env-expressing HeLa cells in the presence or absence of the indicated peptides. The data are means and standard deviations from three fields from three independent assays. LPF, low-power field.
HTLV-1 entry into cells is highly promiscuous in vitro (31, 40), and the cell surface receptors used by the virus are expressed on a diverse array of tissues and nonhuman cell types (20, 41). Therefore, we examined the ability of P-400 to inhibit membrane fusion among HTLV-permissive non-T-cell targets. The Env expression vector pHTE-1 was transfected into HeLa cells, and the resulting transfectants were used as effector cells in cell fusion assays. Coculture of Cos, HeLa, or HOS cells with Env-expressing HeLa cells induced rampant syncytium formation. Syncytium formation was strongly inhibited in the presence of P-400, whereas the control peptide had no effect on cell fusion (Fig. 1B). Our data indicate that P-400 is a potent antagonist of membrane fusion for all fusion-competent cell lines tested, that there is little cell-specific variation in the inhibitory activity of P-400, and that P-400 inhibited syncytium formation more effectively than the SU-derived peptide P-197. The inhibitory effects of P-400 are specific to HTLV-1, since the peptide did not block membrane fusion induced by feline immunodeficiency virus (data not shown) or human immunodeficiency virus (HIV) (35). Furthermore, P-400 did not block the binding of recombinant gp46 to target cells (data not shown), supporting the results of Jinno et al. (22), which suggest that P-400 acts at a post-receptor-binding step of the entry process.
Substitutions within P-400 improve inhibitory activity.
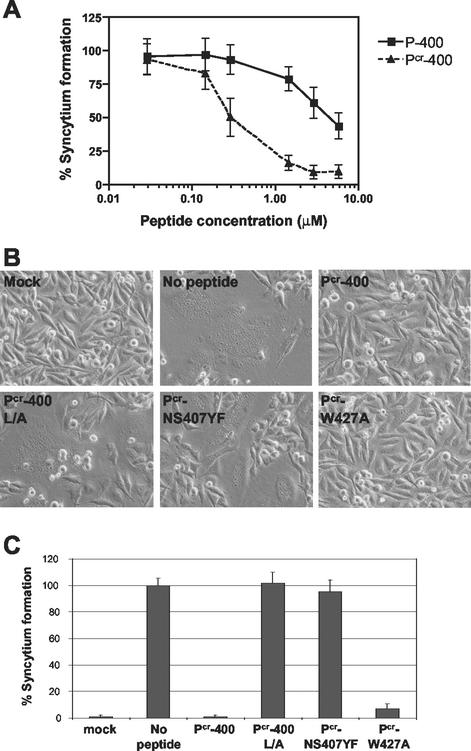
P-400 was originally modeled on the sequence of the prototypic HTLV-1 strain ATK (36). However, sequence analyses of the fusion-competent Env clone, pHTE-1 (HTLV-1 strain CR) (11), used in our studies revealed three strain-specific amino acid substitutions between residues 400 and 429 that differ from the ATK strain. Moreover, analysis of the crystal structure of HTLV-1 TM and molecular modeling of the synthetic peptide suggested to us that the stability of any putative interaction between P-400 and the core coiled-coil region of Env could be improved by optimizing the amino acid sequence of P-400 (see Discussion). To test these ideas, we synthesized an additional peptide based on P-400 but included the following substitutions: Arg401Cys, Pro403Leu, and Pro411Ser. These amino acid substitutions generate a peptide that is identical in sequence to amino acids 400 to 429 of Env from HTLV-1 strain CR (29) and is closer to the consensus amino acid sequence for this region of HTLV-1 Env. The new peptide was therefore designated Pcr-400.
The inhibitory properties of Pcr-400 and P-400 were directly compared in syncytium interference assays using HeLa cells as targets. Coculture of HeLa target cells with Env-expressing cells in the presence of P-400 or Pcr-400 resulted in a dramatic inhibition of syncytium formation (Fig. 2A). The 50% inhibitory concentration (IC50) calculated for P-400 (3.9 μM) was similar to that previously reported by Jinno et al. (22) and Sagara et al. (35) (2.5 and 6.0 μM, respectively). Although both peptides were inhibitory, the optimized Pcr-400 was dramatically and consistently more inhibitory than P-400 in these assays. Comparison of the IC50s for the peptides indicated that Pcr-400 (IC50, 0.28 μM) was 14-fold more active than P-400 (IC50, 3.9 μM). These data also suggest that there may be some strain-specific variability in the sensitivity of HTLV-1 to peptide inhibitors of virus entry.
FIG. 2.
Inhibitory activities of Pcr-400 and derivative peptides. (A) HeLa target cells were cocultured with Env-expressing HeLa cells in the presence of increasing amounts of peptide P-400 or Pcr-400. (B) HeLa target cells were cocultured with mock-transfected HeLa cells or Env-expressing HeLa cells in the absence or presence of Pcr-400 or its mutant derivatives, L/A, NS407YF, and W427A, as indicated. (C) The number of syncytia per low-power field was scored by light microscopy. The error bars indicate standard deviations.
To examine further the biological activity and mechanism of action of Pcr-400, two additional synthetic peptides were generated that contained nonconservative amino acid substitutions within the Pcr-400 sequence. Such substitutions are known to severely impair the inhibitory activity of the parental peptide P-400 (22). In the first mutant peptide, all Leu residues were changed to Ala (Pcr-400L/A). These Leu residues are highly conserved among HTLV-1 isolates and are also conserved between HTLV-1 and HTLV-2 strains. Additionally, the crystal structure of the core of HTLV-1 Env (24) shows that these Leu residues may be important for the correct folding of this region of TM. In the second mutant peptide, Pcr-NS407YF, Asn407 and Ser408 were replaced with Tyr and Phe, respectively. These mutations have been shown to affect HTLV-1 Env maturation, syncytium formation, and infectivity (22, 33). Furthermore, the crystal structure highlights Asn407 as one of the key residues in the interface between the coiled-coil and the C-terminal segment of gp21, forming polar interactions with Gln377 on the coiled-coil (24). Finally, since previous studies have highlighted the importance of Trp residues in the inhibitory activity of anti-HIV peptides (8), a third mutant peptide incorporating a single-amino-acid change converting Trp427 to Ala was synthesized.
The abilities of the peptide derivatives to inhibit Env-dependent membrane fusion were examined in syncytium interference assays. Both the L/A and NS407YF mutations completely abolished the inhibitory activity of the Pcr-400 peptide (Fig. 2B and C). In contrast, the single-amino-acid substitution replacing Trp427 with Ala had little effect on the inhibitory properties of the peptide, as Pcr-W427A antagonized cell-to-cell fusion almost as efficiently as the parental peptide, Pcr-400 (Fig. 2B and C). These substitutions begin to define the amino acid residues that are important for the inhibitory properties of P-400-related peptides.
Pcr-400 peptides have the potential to form ordered secondary structures.
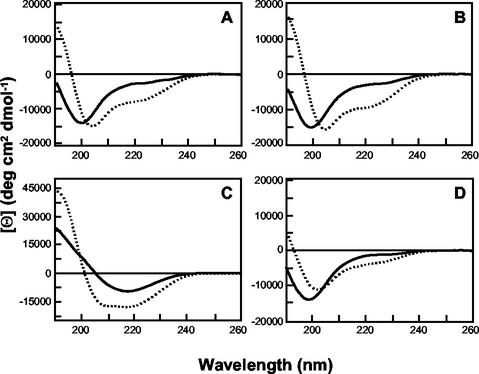
The crystal structure of the HTLV-1 TM core reveals that amino acids Cys400 to Thr425 adopt an extended structure that includes a short α-helical coil between amino acids 408 and 415 (24). To determine if the synthetic peptides have the ability to adopt helical structures, we examined the structure of Pcr-400 and its derivatives using far-UV CD spectroscopy. The inhibitory peptides Pcr-400 (Fig. 3A) and Pcr-W427A (Fig. 3B) have very similar CD properties. Both peptides show little tendency to form helices in aqueous solution but a significant tendency to form helical structures in TFE, which generally acts as a helix-promoting solvent (21). In the presence of TFE, the proportion of helical structure for both peptides increases by ∼20 to 25% (as determined by SELCON analysis [data not shown]). In contrast, the peptides that do not inhibit fusion, Pcr-NS407YF (Fig. 3C) and Pcr-400L/A (Fig. 3D), exhibit strikingly different properties. Notably, Pcr-400L/A has little propensity to adopt a helical structure in either aqueous solution or TFE. These observations suggest that the inhibitory properties of Pcr-400-related peptides can be correlated with their structures. Thus, the synthetic peptides appear to adopt a conformation that accurately models an important structural feature of HTLV-1 TM.
FIG. 3.
CD spectra are shown for 0.5-mg/ml concentrations of Pcr-400 (A), Pcr-W427A (B), Pcr-NS407YF (C), and Pcr-400L/A (D) in H2O (dotted lines) and 50% (vol/vol) TFE (solid lines). deg, degrees.
Modeling the core coiled-coil structure of HTLV-1 TM.
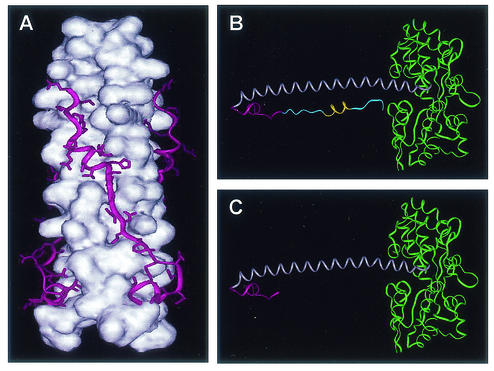
Elegant studies of HIV Env show that synthetic peptides which model a C-terminal region of the gp41 ectodomain inhibit virus entry by binding to a core coiled-coil domain of HIV Env (8, 15, 25, 38, 44). To test the hypothesis that synthetic peptides modeling amino acids 400 to 429 of HTLV-1 Env interact directly with the central coiled-coil of HTLV-1 TM, we first generated recombinant TM-derived Env chimeras by the method of Center et al. (7). Briefly, MBP was fused in frame to amino acids Met338 to Thr425 of TM, which include all of the amino acid sequences and structural features required for formation of the trimer-of-hairpins motif (Fig. 4A); hence, we refer to this fusion as MBP-Hairpin (Fig. 4B). We also generated an additional chimera fusing MBP to amino acids Met338 to Cys400 of TM, encompassing the N-terminal α-helical coiled-coil domain and including the region of chain reversal. We predicted that this recombinant gp21-derived structure would be trimeric and that the core α-helical coiled-coil structure would be exposed to solvent. The predicted monomeric structure for this chimera resembles a fishhook (Fig. 4C) and, for simplicity, is referred to as MBP-Fishhook. These MBP-TM fusion proteins were expressed in E. coli and purified using standard techniques.
FIG. 4.
Structures of the trimeric hairpin of HTLV-1 envelope and the MBP-TM chimeras. (A) The core coiled-coil (white) is shown as a space-filling model, while the C-terminal ectodomain (magenta) is illustrated as a ribbon showing the amino acid side chains. (B) Ribbon diagram of the MBP-Hairpin monomer with MBP (green), the N-terminal coiled-coil-forming helix (grey), the region of chain reversal (magenta), and the C-terminal sequences (blue). The short α-helical segment within the C-terminal ectodomain is also shown (yellow). (C) Ribbon diagram of MBP-Fishhook monomer. The colors are as in panel B. All structures were modelled from coordinates provided by Kobe et al. (24) in the Protein Data Bank, Brookhaven National Laboratory, PDB ID c ode 1MG1.
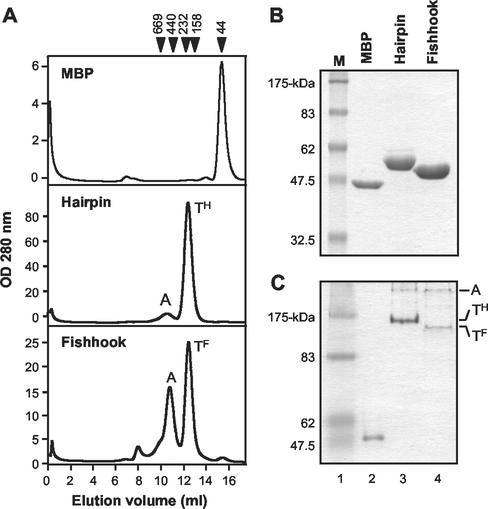
The C-terminal sequences of the TM ectodomain pack into grooves along the exterior of the central coiled-coil (24). These grooves exist only in the context of a trimer. Therefore, the oligomerization status of the MBP-TM fusion proteins was verified using gel filtration chromatography. While MBP eluted as a monomer (43 kDa), MBP-Hairpin and MBP-Fishhook eluted as a discrete peak between the 232- and 158-kDa molecular mass markers, indicating that these chimeras adopt higher-order structures that are likely trimeric (Fig. 5A). In addition, a considerable fraction of MBP-Fishhook eluted as a high-molecular-weight complex, or aggregate. Presumably, the large amount of aggregated MBP-Fishhook is due to exposure of the hydrophobic surfaces of the central coiled-coil domain to the solvent. In keeping with published results for other MBP-gp21 chimeras (26), analysis of MBP-Hairpin and MBP-Fishhook by MALDI-time of flight mass spectroscopy confirmed the presence of trimers (data not shown).
FIG. 5.
Oligomerization states of MBP-TM-derived chimeras. (A) Gel filtration chromatography profiles of MBP, MBP-Hairpin, and MBP-Fishhook. OD, optical density; A, aggregate; TH, trimeric hairpin; TF, trimeric fishhook. Callibration markers are indicated in kilodaltons at the top of the panel. (B) MBP-TM chimeras analyzed by SDS-PAGE under denaturing conditions followed by staining with Coomasie blue. The predicted molecular mass of each protein is as follows: MBP, 43-kDa (lane 2); MBP-Hairpin, 53-kDa (lane 3); MBP-Fishhook, 50-kDa (lane 4). Lane M, molecular-mass markers. (C) Native-PAGE analysis of MBP (lane 2), MBP-Hairpin (lane 3), and MBP-Fishhook (lane 4). The mobilities of the aggregate, trimeric hairpin, and trimeric fishhook are indicated.
Finally, denaturing SDS-PAGE demonstrated that MBP-Hairpin and MBP-Fishhook ran as discrete monomers of 53 and 50 kDa, respectively (Fig. 5B). In contrast, under nondenaturing PAGE conditions, MBP ran as a monomer while MBP-Hairpin and MBP-Fishhook ran as higher-order structures (Fig. 5C). Importantly, both MBP-Hairpin and MBP-Fishhook were resolved as two discrete bands, which, based on the predicted molecular weights of the monomers, are likely to represent a trimer and a higher-order aggregate. Compared with MBP-Hairpin, it was apparent that more of MBP-Fishhook ran as higher-order aggregates.
Bioactive peptides bind directly to a trimeric coiled-coil domain of gp21.
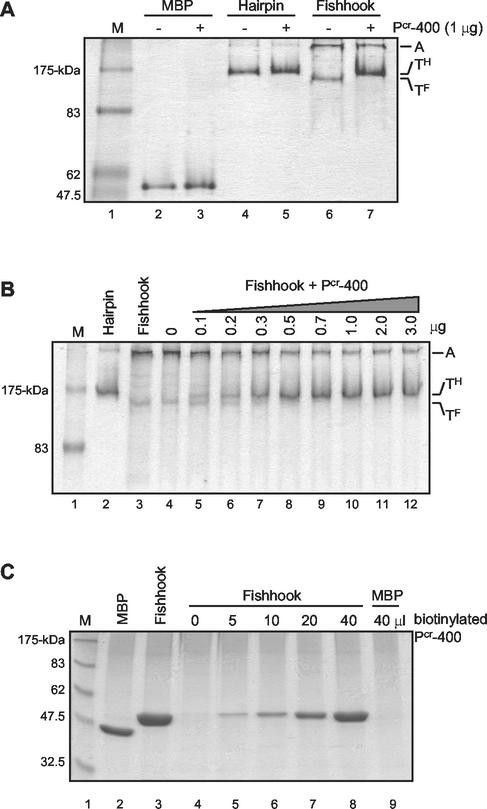
To examine the abilities of Pcr-400 and its derivative peptides to bind to the central coiled-coil of TM, we employed a novel assay that relies upon the ability of the synthetic peptide to bind the target protein and alter its electrophoretic mobility under nondenaturing PAGE conditions. The MBP and MBP-TM derivatives were incubated in the presence or absence of Pcr-400, and subsequently, the mobility of the target protein was examined by native PAGE. Pcr-400 had no effect on the mobility of the control MBP or MBP-Hairpin, indicating that Pcr-400 did not interact with these target proteins under the conditions used (Fig. 6A, lanes 2 to 5). In contrast, compared to controls lacking peptide (Fig. 6A, lane 6), a shift in the mobility of MBP-Fishhook was observed in the presence of Pcr-400 (Fig. 6A, lane 7). Moreover, Pcr-400 appears to drive a substantial fraction of the aggregated MBP-Fishhook into a complex that is likely a trimer; the mobility of this complex is equivalent to that of the trimeric MBP-Hairpin. Further analysis indicated that the shift in mobility of MBP-Fishhook induced by Pcr-400 is dependent on the amount of Pcr-400 used (Fig. 6B). These data indicate to us that Pcr-400 binds directly to the central coiled-coil structure of HTLV-1 TM and that binding of Pcr-400 alters the mobility of the trimeric MBP-Fishhook under native-PAGE conditions.
FIG. 6.
Pcr-400 interacts with the coiled-coil of MBP-Fishhook. (A) MBP (lanes 2 and 3), MBP-Hairpin (lanes 4 and 5), or MBP-Fishhook (lanes 6 and 7) was incubated without (−) or with (+) Pcr-400, and the complexes were analyzed by native PAGE and stained with Coomasie blue. Pcr-400 induces a shift in the electrophoretic mobility of the trimeric MBP-Fishhook (TF), causing it to migrate similarly to trimeric MBP-Hairpin (TH). A, aggregate. Lane M, molecular-mass markers. (B) Increasing amounts of Pcr-400 were incubated with MBP-Fishhook, and the complexes were analyzed by native PAGE. Lanes 2 and 3, MBP-Hairpin and MBP-Fishhook controls. The amount of MBP-Fishhook (TF) shifting to a complex that mimics MBP-Hairpin (TH) increases with the amount of Pcr-400 added (lanes 4 to 12). (C) Increasing amounts of biotinylated Pcr-400 were incubated with MBP-Fishhook (lanes 4 to 8). As a control, MBP was also incubated with biotinylated Pcr-400 (lane 9). Streptavidin-agarose was then added to the reaction mixture, and the bound complexes were precipitated, heat denatured, and analyzed by SDS-PAGE.
We used pull-down assays to further demonstrate that Pcr-400-related peptides bind directly to the core coiled-coil domain of TM. Biotinylated Pcr-400 was incubated with the control MBP or with MBP-Fishhook. Subsequently, the biotinylated peptide and any bound proteins were captured on streptavidin-agarose beads, washed extensively, and examined by SDS-PAGE. In the absence of Pcr-400, little MBP-Fishhook bound to the streptavidin-agarose beads (Fig. 6C, lane 4). However, in the presence of the biotinylated peptide, the amount of MBP-Fishhook bound increased dramatically, and the amount of MBP-Fishhook recovered was directly proportional to the concentration of Pcr-400 in the reaction mixture (Fig. 6C, lanes 5 to 8). Significantly, MBP did not bind to Pcr-400 but remained in the supernatant (Fig. 6C, lane 9). Thus, Pcr-400 interacts directly with MBP-Fishhook, representing the recombinant core coiled-coil of HTLV-1 TM, but does not bind to the MBP control.
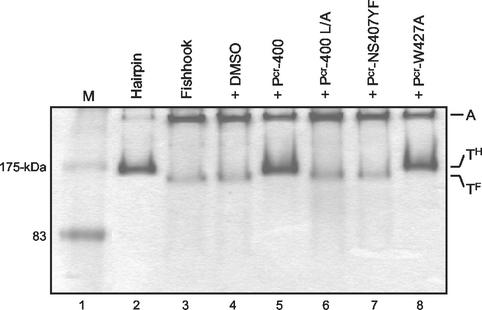
To determine if binding of the P-400-related peptides to the coiled-coil domain correlates with the biological activity of the peptide, we examined the binding of Pcr-400 and its derivatives to MBP-Fishhook using the band shift assay. While the inhibitory peptides Pcr-400 and Pcr-W427A induced a change in mobility of MBP-Fishhook (Fig. 7, lanes 5 and 8), the biologically inactive peptides Pcr-400L/A and Pcr-NS427YF had no effect on the migration of the chimera (lanes 6 and 7). Taken together, our collected data are consistent with the notion that P-400-related peptides bind directly to the central coiled-coil domain of HTLV-1 TM and that binding to the central coiled-coil correlates directly with the antiviral properties of the synthetic peptides.
FIG. 7.
Inhibitory activity of P-400-related peptides correlates with binding to MBP-Fishhook. MBP-Fishhook was incubated with Pcr-400 (lane 5) and its derivative peptides L/A (lane 6), NS407YF (lane 7), and W427A (lane 8) or with solvent alone (lane 4). The complexes were analyzed by native PAGE and stained with Coomassie blue. The controls MBP-Hairpin (lane 2) and MBP-Fishhook (lane 3) are shown. The mobilities of the aggregate (A), trimeric hairpin (TH), and trimeric fishhook (TF) are indicated. Lane M, molecular-mass markers.
Recombinant gp21-derived coiled-coil reduces the inhibitory properties of Pcr-400.
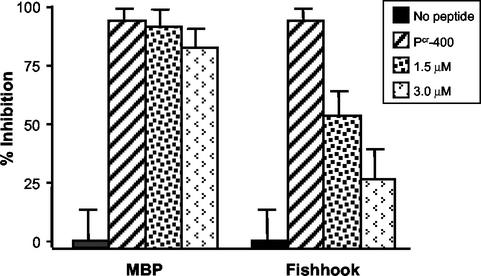
Studies of peptide antagonists of HIV Env-mediated membrane fusion have demonstrated that peptides modeling a C-terminal region of the HIV TM ectodomain bind to the core coiled-coil of TM and prevent formation of the fusion-active trimer-of-hairpins structure (8, 15, 25, 38, 44). Our results suggest that HTLV-1 P-400-related peptides function through a similar trans-dominant-negative mechanism of action. Our interpretation of the collected data predicts that preincubation of Pcr-400 with the core coiled-coil should disrupt the inhibitory activity of this peptide. We find that this is indeed the case. Incubation of target cells with Pcr-400 and MBP-Fishhook, prior to the addition of Env-expressing effector cells, resulted in a marked decrease in the inhibitory activity of Pcr-400 in syncytium interference assays (Fig. 8). In contrast, preincubation of Pcr-400 with MBP had little or no effect on the inhibitory properties of the antiviral peptide. Thus, the recombinant coiled-coil competes effectively with the natural target of Pcr-400, providing further support for the view that peptides modeling amino acids 400 to 429 of HTLV-1 Env block Env-mediated membrane fusion by targeting the core coiled-coil of fusion-active TM.
FIG. 8.
MBP-Fishhook reverses the effects of Pcr-400 in syncytium formation assays. Target HeLa cells were incubated in the absence or presence of Pcr-400 at a concentration of 1.5 μM. As indicated, MBP or MBP-Fishhook, at 1.5 and 3.0 μM concentrations, was preequilibrated with the peptide prior to the addition of the Env-bearing HeLa effector cells. The error bars indicate standard deviations.
DISCUSSION
Inhibitory peptides bind directly to the core coiled-coil of HTLV-1 TM.
In this study, we provide compelling evidence that synthetic peptides, which mimic a C-terminal region of the TM ectodomain, inhibit HTLV-1 Env-mediated membrane fusion by targeting and disrupting the fusion-active structures of Env. Several lines of evidence are consistent with this view. First, the peptides target a component that is common to the fusion reactions of all HTLV-1-permissive cell lines tested, and there appears to be little cell-specific variation in the inhibitory potency of the peptide. These observations suggest that the component recognized by the synthetic peptide is present at similar densities in all of the fusion reactions, and they point to a factor that is likely present on the effector (Env-expressing) cell population. Second, the secondary structures of the inhibitory peptides appear to be important for biological activity. Our CD analysis indicates that the biologically active peptides have the capacity to adopt a helical structure, suggesting that the synthetic peptides accurately model important structural features of the C-terminal region of the TM ectodomain as revealed by the crystal structure. Third, and most significant, the inhibitory peptides directly bind to a recombinant coiled-coil derived from HTLV-1 TM (MBP-Fishhook) in vitro. Moreover, peptides that fail to bind the recombinant coiled-coil do not inhibit Env-catalyzed membrane fusion. Fourth, amino acid substitutions that are predicted to improve the stability of the interaction between the peptide and coiled-coil (see below) increase the potency of the inhibitory peptides. Fifth, and finally, preincubation of Pcr-400 with the recombinant coiled-coil greatly reduces the inhibitory activity of the TM-mimetic peptide, indicating that the recombinant coiled-coil competes effectively with the natural target for binding of the peptide ligand.
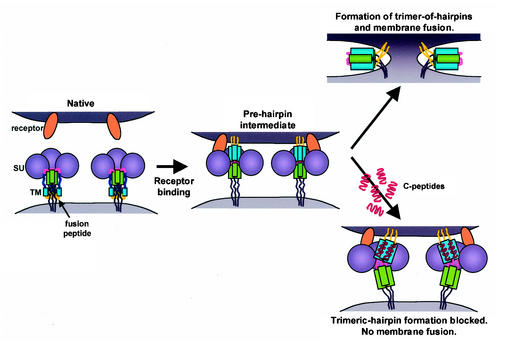
Taken together, our results are consistent with the view that P-400-related peptides target the core coiled-coil of HTLV-1 TM and most likely inhibit membrane fusion in a trans-dominant-negative manner by blocking the formation of the fusion-active trimer-of-hairpins structure. The trimeric-hairpin structure represents a highly stable conformation of TM (25), and once formed, it is unlikely to be disrupted by insertion of the inhibitory peptide. Indeed, Pcr-400 is unable to alter the mobility of the recombinant trimeric hairpin (MBP-Hairpin) in native gels, suggesting that it does not bind the hairpin. Instead, P-400-related inhibitory peptides likely act prior to the formation of the trimeric hairpin, binding to the transiently exposed prehairpin intermediate and blocking its subsequent resolution to the fusion-active state (Fig. 9). Thus, P-400-related peptides that antagonize HTLV-1 entry and membrane fusion appear to be functionally analogous to the C-peptide inhibitors of HIV infection.
FIG. 9.
Model of retroviral envelope-mediated membrane fusion. In its native conformation, HTLV-1 Env consists of a trimer of SU subunits noncovalently linked to a trimer of TM subunits. Binding of SU to its cellular receptor triggers conformational changes in Env that result in the insertion of the fusion peptide into the target cell membrane and the concomitant formation of the prehairpin intermediate. TM then resolves into the trimer-of-hairpins structure, drawing the cellular and viral membranes into apposition and inducing membrane fusion. C-peptides, such as P-400 and Pcr-400, are able to bind the grooves of the central coiled-coil, thereby blocking formation of the trimeric-hairpin structure and disrupting membrane fusion.
Peptides with improved potency.
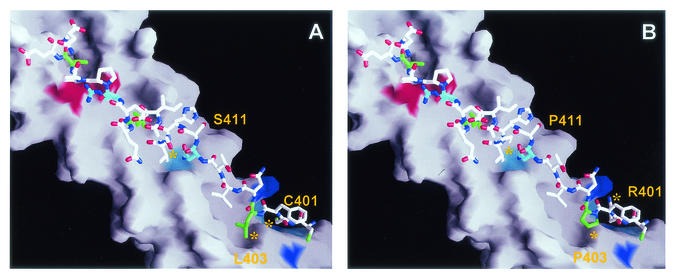
A surprising finding of our study is that replacement of relatively few amino acids in the inhibitory peptide, P-400, resulted in a dramatic gain in potency. In fact, replacement of only three amino acids generated a peptide (Pcr-400) displaying a 14-fold decrease in IC50. Examination of the TM crystal structure suggests that the gain in potency is due to the stabilizing effects these substitutions have on the interaction of the peptide with the coiled-coil (Fig. 10). Consistent with this view, we have observed that P-400 is less efficient at binding the coiled-coil than Pcr-400 (unpublished results). In Pcr-400, substitution of Arg401 for Cys removes the larger, positively charged guanidinium group from an environment defined by coiled-coil residues Lys370, Gln373, Tyr374, Gln377, and Arg380 of one N-helical subunit and Arg379 and Glu398 of an adjacent subunit. The small polar Cys residue of Pcr-400 is likely more suited to the surrounding environment, reducing possible instability due to unfavorable charge interactions associated with the positively charged Arg side chain. Moreover, for Pcr-400 the Leu403 residue is expected to make favorable contacts with the central coil by extending down into a pocket defined by the side chains Ile371 and Tyr374 of one helix and Ala372 and Ala375 of an adjacent coil. By contrast, the Pro403 residue of P-400 is unlikely to extend into this pocket, possibly leaving a cavity and removing any stabilizing contacts. The third amino acid substitution defined by Pcr-400 replaces Pro377 with Ser. Within Env, residue 377 occurs within the first helical turn of a short α-helical region in the C-terminal half of the TM ectodomain. In a surface-exposed position, it is difficult to rationalize the effect(s) of this substitution with respect to the loss or gain of interactions that may influence the stability of the peptide. While it is not yet possible to identify which of the three substitutions makes the greatest contribution to the increased activity of Pcr-400, we suspect that the Pro403-to-Leu and Arg401-to-Cys substitutions are largely responsible for the gain in potency.
FIG. 10.
Molecular interactions between P-400-related peptides and the core coiled-coil. (A) Pcr-400 is shown against the surface of the coiled-coil. The Leu residues are colored green. The buried residues Asn407 and Arg416 are shown in cyan. The colored regions of the coiled-coil represent polar and charged residues interacting with the peptide. The binding groove for Cys401 is shown in blue; residues contributing to the base and wall of the groove are colored cyan. Asn364, which interacts with the buried side chain of Asn407, is represented by the cyan region beneath Asn407, while the coiled-coil side chains involved in coordinating Arg416 are shown in red. (B) P-400 is illustrated using the same color coding as for Pcr-400. The three amino acid differences between P-400 and Pcr-400 are indicated. Most notable are the cavities generated by Pro403 and Arg401. Structures were modelled from coordinates provided by Kobe et al. (24) in the Protein Data Bank, Brookhaven National Laboratory, PDB ID code 1MG1.
Mutant peptides.
It is now possible to account in molecular terms for the inability of some peptides carrying substitutions in critical amino acids to inhibit membrane fusion. CD analysis indicates that Pcr-400L/A, carrying Ala residues in place of all Leu residues, is unlikely to adopt the stable helical structures typically formed by the inhibitory peptide Pcr-400 or by amino acids 400 to 425 of TM. Moreover, examination of the TM crystal structure reveals that the Leu residues after the region of chain reversal occupy orientations in which the side chain is predominantly buried between the interface of the C-terminal extended ribbon and the core coiled-coil. Of the five Leu residues present in this region after Cys400, only Leu403, Leu413, and Leu419 are observed in the TM crystal structure. However, it would appear that removal of all but the Cβ from the Leu side chains, using Ala substitutions, generates a series of cavities between the C-terminal extended ribbon and the central coiled-coil. Such substitutions would be expected to greatly reduce the ability of the peptide to bind in an optimum conformation, an expectation that is corroborated by the experimental data.
Notably, for TM, the interaction between the C-terminal extended ribbon and the core coiled-coil uses residues Asn407 and Arg416 to make polar and ionic contacts across the buried interface. The residues defining the environment into which the Asn407 side chain binds include His365, His409, and Val410 of one subunit and Ala360, Ile361, Lys363, Asn367, and Asn364 of an adjacent subunit. Interactions between the amino group of the amide side chain of Asn407 and the carbonyl group of the amide side chain of Asn364, located at the base of the cavity, as well as the His409 side chain located on the cavity wall, may aid the interaction of Asn407 with its environment. Modeling the Tyr substitution of Pcr-NS407YF suggests that the bulky aromatic side chain of Tyr cannot be accommodated within the cavity of the coiled-coil. A substantial conformational rearrangement of the peptide against the coiled-coil would be necessary in order to remove unfavorable steric interactions and to provide a hydrogen bond acceptor for the OH group of Tyr.
Implications for HTLV-1 therapy.
Our results suggest that there is scope for the development of P-400-related peptides as therapeutic agents targeting HTLV-1. Worldwide, HTLV-1 infections have considerable clinical impact. HTLV-1 is the etiological agent of an aggressive adult T-cell leukemia and a chronic neurodegenerative disease, HTLV-1-associated myelopathy, or tropical spastic paraparesis. Despite considerable clinical effort, these virus-associated diseases remain difficult to treat, and therapeutic strategies specifically tailored to target HTLV-1 replication are unavailable. Nevertheless, the observations that viral replication persists in HTLV-associated disease (1, 17, 30, 46) and that a combined antiretroviral drug therapy shows promise in the treatment of HTLV-associated disease (3, 4, 16) indicate that novel antiretroviral compounds targeting HTLV entry may be of therapeutic value. Importantly, our study reveals that it is possible to identify synthetic peptides that specifically target the fusion-active structures of HTLV-1 Env and that more potent inhibitors of HTLV-1 entry can be readily obtained. Based upon these observations, we suggest that synthetic peptides that target the fusion-active structures of HTLV-1 Env represent promising leads in the search for pharmacologically relevant therapies to combat HTLV-1 infections. Further analysis of HTLV-1-induced membrane fusion will improve our understanding of Env function during virus entry into cells and will facilitate the rational design of low-molecular-weight antagonists of HTLV infection.
Acknowledgments
We thank members of the laboratory for helpful discussions and Patrick Whitty for assistance with size exclusion chromatography.
This work was generously supported by grants to D.W.B. from the Leukemia Research Fund (LRF 9827) and from the University of Glasgow.
REFERENCES
- 1.Asquith, B., E. Hanon, G. P. Taylor, and C. R. Bangham. 2000. Is human T-cell lymphotropic virus type I really silent? Philos. Trans. R. Soc. Lond. B 355:1013-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. [DOI] [PubMed] [Google Scholar]
- 3.Bazarbachi, A., and O. Hermine. 2001. Treatment of adult T-cell leukaemia/lymphoma: current strategy and future perspectives. Virus Res. 78:79-92. [DOI] [PubMed] [Google Scholar]
- 4.Bazarbachi, A., and O. Hermine. 1996. Treatment with a combination of zidovudine and alpha-interferon in naive and pretreated adult T-cell leukemia/lymphoma patients. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 13:S186-S190. [DOI] [PubMed] [Google Scholar]
- 5.Brighty, D. W., and S. R. Jassal. 2001. The synthetic peptide P-197 inhibits human T-cell leukemia virus type 1 envelope-mediated syncytium formation by a mechanism that is independent of Hsc70. J. Virol. 75:10472-10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37-43. [DOI] [PubMed] [Google Scholar]
- 7.Center, R. J., B. Kobe, K. A. Wilson, T. Teh, G. J. Howlett, B. E. Kemp, and P. Poumbourios. 1998. Crystallization of a trimeric human-Tcell leukaemia virus type 1 gp21 ectodomain fragment as a chimera with maltose binding protein. Protein Sci. 7:1612-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan, D. C., C. T. Chutkowski, and P. S. Kim. 1998. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc. Natl. Acad. Sci. USA 95:15613-15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 10.Damico, R. L., J. Crane, and P. Bates. 1998. Receptor-triggered membrane association of a model retroviral glycoprotein. Proc. Natl. Acad. Sci. USA 95:2580-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dokhelar, M. C., H. Pickford, J. Sodroski, and W. A. Haseltine. 1989. HTLV-1 p27rex regulates gag and env protein expression. J. Acquir. Immune Defic. Syndr. 2:431-440. [PubMed] [Google Scholar]
- 12.Eckert, D. M., V. N. Malashkevich, L. H. Hong, P. A. Carr, and P. S. Kim. 1999. Inhibiting HIV-1 entry: discovery of D-peptide inhibitors that target the gp41 coiled coil pocket. Cell 99:103-115. [DOI] [PubMed] [Google Scholar]
- 13.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 14.Fass, D., S. C. Harrison, and P. S. Kim. 1996. Retrovirus envelope domain at 1.7 Å resolution. Nat. Struct. Biol. 3:465-469. [DOI] [PubMed] [Google Scholar]
- 15.Furuta, R. A., C. T. Wild, Y. Wend, and C. D. Weiss. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat. Struct. Biol. 5:276-279. [DOI] [PubMed] [Google Scholar]
- 16.Gill, P., W. Harrington, J. Kaplan, R. C. Ribeiro, J. M. Bennett, H. A. Liebman, M. Bernstein-Singer, B. M. Esoina, L. Cabral, S. Allen, S. Kornblau, M. C. Pike, and A. M. Levine. 1995. Treatment of adult T-cell leukaemia-lymphoma with a combination of interferon-α and zidovudine. N. Engl. J. Med. 332:1744-1748. [DOI] [PubMed] [Google Scholar]
- 17.Hanon, E., R. E. Asquith, G. P. Taylor, Y. Tanaka, J. N. Weber, and C. R. Bangham. 2000. High frequency of viral protein expression in human T cell lymphotropic virus type 1-infected peripheral blood mononuclear cells. AIDS Res. Hum. Retrovir. 6:1711-1715. [DOI] [PubMed] [Google Scholar]
- 18.Hernandez, L. D., R. J. Peters, S. E. Delos, J. A. T. Young, D. A. Agard, and J. M. White. 1997. Activation of a retroviral membrane fusion protein: soluble receptor-induced liposome binding of the ALSV envelope glycoprotein. J. Cell Biol. 39:1455-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jassal, S. R., M. D. Lairmore, A. J. Leigh-Brown, and D. W. Brighty. 2001. Soluble recombinant HTLV-1 surface glycoprotein competitively inhibits syncytia formation and viral infection of cells. Virus Res. 78:17-34. [DOI] [PubMed] [Google Scholar]
- 20.Jassal, S. R., J. R. G. Pöhler, and D. W. Brighty. 2001. Human T-cell leukemia virus type 1 receptor expression among syncytium-resistant cell lines revealed by a novel surface glycoprotein-immunoadhesin. J. Virol. 75:8317-8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jassanoff, A., and A. R. Fersht. 1994. Quantitative determination of helical propensities from trifluoroethanol titration curves. Biochemistry 33:2129-2135. [DOI] [PubMed] [Google Scholar]
- 22.Jinno, A., H. H. Shiraki, and H. Hoshino. 1999. Inhibition of cell-free human T-cell leukaemia virus type 1 infection at a postbinding step by the synthetic peptide derived from an ectodomain of the gp21 transmembrane glycoprotein. J. Virol. 73:9683-9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kilby, J. M., S. Hopkins, T. M. Venetta, B. DiMassino, G. A. Cloud, J. Y. Lee, L. Alldredge, E. Hunter, D. Lambert, D. Bolognesi, T. Matthews, M. R. Johnson, M. A. Nowak, G. M. Shaw, and M. S. Saag. 1998. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 4:1302-1307. [DOI] [PubMed] [Google Scholar]
- 24.Kobe, B., R. J. Center, B. E. Kemp, and P. Poumbourios. 1999. Crystal structure of human T-cell leukemia virus type 1 gp21 ectodomain crystallized as a maltose-binding protein chimera reveals structural evolution of retroviral transmembrane proteins. Proc. Natl. Acad. Sci. USA 96:4319-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu, M., S. C. Blacklow, and P. S. Kim. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2:1075-1082. [DOI] [PubMed] [Google Scholar]
- 26.Maerz, A. L., R. J. C. Center, B. E. Kemp, B. Kobe, and P. Poumbourios. 2000. Functional implications of the human T-lymphotropic virus type 1 transmembrane glycoprotein helical hairpin structure. J. Virol. 74:6614-6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malashkevich, V. N., D. C. Chan, C. T. Chutkowski, and P. S. Kim. 1998. Crystal structure of the simian immunodeficiency virus (SIV) gp41 core: conserved helical interactions underlie the broad activity of gp41 peptides. Proc. Natl. Acad. Sci. USA 95:9134-9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malashkevich, V. N., B. J. Scheider, M. L. McNally, M. A. Milhollen, J. X. Pang, and P. S. Kim. 1999. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-Å resolution. Proc. Natl. Acad. Sci. USA 96:2662-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manzari, V., F. Wong-Staal, G. Franchini, S. Colombini, E. P. Gelmann, S. Oroszlan, S. Staal, and R. C. Gallo. 1983. Human T-cell leukemia-lymphoma virus (HTLV): cloning of an integrated defective provirus and flanking cellular sequences. Proc. Natl. Acad. Sci. USA 80:1574-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagai, M., Y. Yamano, M. B. Brennan, C. A. Mora, and S. Jacobson. 2001. Increased HTLV-I proviral load and preferential expansion of HTLV-I tax-specific CD8+ T cells in cerebrospinal fluid from patients with HAM/TSP. Ann. Neurol. 50:807-812. [DOI] [PubMed] [Google Scholar]
- 31.Okuma, K., M. Nakamura, S. Nakano, Y. Niho, and Y. Matsuura. 1999. Host range of human T-cell leukaemia virus type I analysed by a cell fusion-dependent reporter gene activation assay. Virology 254:235-244. [DOI] [PubMed] [Google Scholar]
- 32.Palker, T. J., E. T. Riggs, D. E. Spragion, A. Muir, R. M. Scearce, R. R. Randall, M. W. McAdams, A. McKnight, P. R. Clapham, R. A. Weiss, and B. F. Haynes. 1992. Mapping of homologous, amino-terminal neutralizing regions of human T-cell lymphotropic virus type I and II gp46 envelope glycoproteins. J. Virol. 66:5879-5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenberg, A. R., L. Delmarre, C. Pique, D. Pham, and M.-C. Dokhelar. 1997. The ectodomain of the human T-cell leukemia virus type 1 TM glycoprotein is involved in postfusion events. J. Virol. 71:7180-7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenthal, P. B., X. Zhang, F. Formanowski, W. Fitz, C. H. Wong, H. Meier-Ewert, J. J. Skehel, and D. C. Wiley. 1998. Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature 396:92-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sagara, Y., Y. Inoue, H. Shiraki, A. Jinno, H. Hoshino, and Y. Maeda. 1996. Identification and mapping of functional domains on human T-cell lymphotropic virus type 1 envelope proteins using synthetic peptides. J. Virol. 70:1564-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seiki, M., S. Hattori, Y. Hirayama, and M. Yoshida. 1983. Human adult T-cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc. Natl. Acad. Sci. USA 80:3618-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shida, H., T. Tochikura, T. Sato, T. Konno, K. Hirayoshi, M. Seki, Y. Ito, M. Hatanaka, Y. Hinuma, and M. Sugimoto. 1987. Effect of the recombinant vaccinia viruses that express HTLV-I envelope gene on HTLV-I infection. EMBO J. 6:3379-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sodroski, J. G. 1999. HIV-1 entry inhibitors in the side pocket. Cell 99:243-246. [DOI] [PubMed] [Google Scholar]
- 39.Sreerama, N., and R. W. Woody. 1993. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal. Biochem. 209:32-44. [DOI] [PubMed] [Google Scholar]
- 40.Sutton, R. E., and D. R. Littman. 1996. Broad host range of human T-cell leukemia virus type 1 demonstrated with an improved pseudotyping system. J. Virol. 70:7322-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trejo, S. R., and L. Ratner. 2000. The HTLV receptor is a widely expressed protein. Virology 268:41-48. [DOI] [PubMed] [Google Scholar]
- 42.Weissenhorn, W., A. Carfi, K. H. Lee, J. J. Skehel, and D. C. Wiley. 1998. Crystal structure of the Ebola virus membrane fusion subunit GP2, from the envelope glycoprotein ectodomain. Mol. Cell 2:605-616. [DOI] [PubMed] [Google Scholar]
- 43.Wild, C., T. Oas, C. McDanal, D. Bolognesi, and T. Matthews. 1992. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc. Natl. Acad. Sci. USA 89:10537-10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wild, C. T., T. K. Greenwell, D. Shugars, L. Rimsky-Clarke, and T. J. Matthews. 1995. The inhibitory activity of an HIV type 1 peptide correlates with its ability to interact with a leucine zipper structure. AIDS Res. Hum. Retrovir. 11:323-325. [DOI] [PubMed] [Google Scholar]
- 45.Wilson, I. A., J. J. Skehel, and D. C. Wiley. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature 289:366-373. [DOI] [PubMed] [Google Scholar]
- 46.Yamano, Y., M. Nagai, M. Brennan, C. A. Mora, S. S. Soldan, U. Tomaru, N. Takenouchi, S. Izumo, M. Osame, and S. Jacobson. 2002. Correlation of human T-cell lymphotropic virus type 1 (HTLV-1) mRNA with proviral DNA load, virus-specific CD8+ T cells, and disease severity in HTLV-1-associated myelopathy (HAM/TSP). Blood 99:88-94. [DOI] [PubMed] [Google Scholar]