Abstract
We designed a novel single-chain chimeric protein, designated sCD4-17b, for neutralization of human immunodeficiency virus type 1 (HIV-1). The recombinant protein contains domains 1 and 2 of soluble CD4 (sCD4), connected via a flexible polypeptide linker to a single-chain variable region construct of 17b, a human monoclonal antibody that targets a conserved CD4-induced epitope on gp120 overlapping the coreceptor binding region. We hypothesized that the sCD4 moiety would bind gp120 and expose the 17b epitope; the 17b moiety would then bind, thereby blocking coreceptor interaction and neutralizing infection. The sCD4-17b protein, expressed by a recombinant vaccinia virus, potently neutralized a prototypic R5 clade B primary isolate, with a 50% inhibitory concentration of 3.2 nM (0.16 μg/ml) and >95% neutralization at 32 nM (1.6 μg/ml). The individual components (sCD4 and 17b, singly or in combination) had minimal effects at these concentrations, demonstrating that the activity of sCD4-17b reflected the ability of a single chimeric molecule to bind gp120 simultaneously via two independent moieties. sCD4-17b was highly potent compared to the previously characterized broadly cross-reactive neutralizing monoclonal antibodies IgGb12, 2G12, and 2F5. Multiple primary isolates were neutralized, including two previously described as antibody resistant. Neutralization occurred for both R5 and X4 strains and was not restricted to clade B. However, several primary isolates were insensitive over the concentration range tested, despite the known presence of binding sites for both CD4 and 17b. sCD4-17b has potential utility for passive immunization against HIV-1 in several contexts, including maternal transmission, postexposure prophylaxis, and sexual transmission (topical microbicide).
The primary neutralization target on the human immunodeficiency type 1 (HIV-1) virion is the envelope glycoprotein (Env), which promotes virus entry by catalyzing fusion between the virion and target cell membranes. Env is thus the major focus for humoral vaccine and antibody-based immunotherapeutic strategies against HIV-1 (reviewed in references 29 and 49). Passive immunization studies in murine and nonhuman primate models have suggested the protective potential of Env-targeted neutralizing antibodies against establishment of infection and possibly against subsequent disease progression. However, such efforts have been frustrated by the difficulties in eliciting antibodies with potent neutralizing activities against genetically diverse HIV-1 isolates.
Env has evolved a multilayered strategy to carry out its fusogenic function in the face of a persistent humoral immune response (29, 49). Potential neutralizing epitopes on the gp120 external subunit are occluded by genetically variable loops, by extensive glycosylation, and by subunit interactions within the surface Env trimer. Moreover, conformational features of gp120 protect the conserved determinants involved in sequential binding to specific target cell receptors, i.e., first to CD4 and then to the coreceptor (chemokine receptor CCR5 or CXCR4 [4]). The invariant gp120 residues that form the CD4 binding site are located within a conformationally dependent pocket that is poorly accessible to antibody and is probably highly unstable prior to the CD4 interaction (18, 19, 26). Moreover, the highly conserved “bridging sheet” of the gp120 core that constitutes a critical component of the coreceptor binding site (18, 31) is masked (or unformed) prior to CD4 binding and is exposed (or formed) only after a CD4-induced conformational change(s).
The latter point is supported by several experimental findings with HIV-1 and the related simian immunodeficiency virus, as follows. (i) The CD4 interaction greatly enhances binding of soluble gp120 to coreceptor (2, 14, 16, 21, 22, 36, 43, 47). (ii) Soluble CD4 (sCD4) induces Env to promote fusion/entry with cells bearing coreceptor but lacking surface CD4 (32, 36, 37, 39). (iii) Structural, kinetic, and thermodynamic analyses suggest that CD4 binding induces major structural rearrangements in the gp120 core, which in the absence of CD4 is unlikely to adopt a conformation with the bridging sheet exposed (or formed) (18, 19, 26). (iv) The CD4 interaction enhances binding of monoclonal antibodies (MAbs) directed against highly conserved gp120 epitopes overlapping the conserved bridging sheet (e.g., human MAbs17b and 48d) (38, 40, 41, 48, 50); such epitopes are referred to as CD4 inducible (CD4i). (v) MAbs 17b and 48d block binding of CD4-activated gp120 to coreceptor (15, 47). (vi) MAbs 17b and 48d only weakly neutralize Env function, but the activities are greatly enhanced in the presence of sCD4 (32, 40). (vii) HIV-1 isolates selected in vitro for CD4 independence display stable exposure of the coreceptor binding site and enhanced sensitivity to neutralizing antibody (11, 16). The favored interpretation is that the conserved CD4i epitopes of gp120 are only transiently exposed in standard infectivity or cell fusion assays, after CD4 binding but before the coreceptor interaction; kinetic and/or steric factors limit the accessibility of the corresponding antibodies and hence their efficacy at neutralization. Indeed, recent immunostaining studies demonstrated that the 17b epitope is inaccessible (to immunoglobulin G [IgG] or Fab) at the site of Env-target cell interaction (12). Thus, antibodies against the conserved CD4i determinants of gp120 involved in coreceptor binding have the potential to neutralize infection, if only their epitopes can be accessed.
In this report, we describe a novel neutralizing agent based on the ability of sCD4 to render the CD4i epitopes on the conserved bridging sheet accessible to antibody-mediated blockade. The agent, designated sCD4-17b, is a recombinant chimeric protein containing sCD4 attached via a flexible polypeptide linker to a single-chain variable region construct (SCFv) of MAb 17b. The concept is that binding of the sCD4 moiety to gp120 on virions would induce exposure of the 17b epitope; the 17b SCFv moiety would then bind, thereby blocking the gp120-coreceptor interaction and neutralizing infectivity. The bifunctional binding of the chimeric protein, coupled with the conserved nature of both the CD4 and 17b binding sites on gp120, would be expected to confer high potency and broad cross-reactivity. Moreover, the fact that both the sCD4 and 17b sequences are human would be expected to minimize immunogenicity. The sCD4-17b protein would therefore have critical features for preventing and treating HIV-1 infection.
MATERIALS AND METHODS
Cells, viruses, and proteins.
BS-C-1 cells were obtained from the American Type Culture Collection. Indicator cell lines MAGI (HeLa-CD4-LTR-β-gal) (17) and MAGI-CCR5 (9) were obtained from the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program, as were all HIV-1 strains. Two-domain sCD4 was the generous gift of the Upjohn Company. Purified MAbs 2G12 (46) and 2F5 (25) were kindly provided by John Mascola, purified IgGb12 (6) by Dennis Burton, and hybridoma 17b (41) (from which we purified the 17b IgG by affinity chromatography on Hi-Trap Protein G columns [Pharmacia]) by James Robinson.
Genetic constructs.
Standard recombinant DNA methods (33) were used to construct plasmid pCD-3, which contains the sCD4-17b cDNA sequence cloned downstream from a strong synthetic vaccinia virus early/late promoter; the promoter-cDNA is flanked by vaccinia virus thymidine kinase sequences. The sCD4 sequence (including the N-terminal leader) was derived from plasmid pCB-3 (5). For the 17b SCFv (7), we started with a plasmid (generously donated by R. Wyatt, Dana-Farber Cancer Institute) containing the 17b SCFv cDNA followed by a thrombin cleavage site and a 6-His tag, cloned into plasmid vector pmt del0. Synthetic oligonucleotides were used for the linker connecting sCD4 to the 17b SCFv. Plasmid pCD-3 was used to generate vaccinia virus recombinant vCD-3 from the parental WR strain, using standard methods of homologous recombination and thymidine kinase selection (24). Details of the sCD4-17b protein sequence and the corresponding genetic construct are described in Results.
Expression, affinity chromatography, and quantitation of sCD4-17b protein.
BS-C-1 monolayers were infected with vCD-3 at a multiplicity of infection (MOI) of 1 to 2 in MEM plus Earle’s balanced salts supplemented with 2.5% fetal bovine serum; where indicated, control cells were infected in parallel with the parental WR strain. After 2 h at 37°C, the cells were washed, then incubated in low-serum-containing medium (OPTI-MEM; Invitrogen) (106 cells/ml) for 20 to 24 h at 37°C. To remove cell debris and vaccinia virus, the media were centrifuged at 1,000 × g for 10 min at room temperature and filtered through 0.2-mm-pore-size low protein binding filters (Millex-GV; Millipore). The filtrate was concentrated 40-fold at 4°C (Centriprep-10; Millipore).
For analytical comparison of media and cell lysate fractions, infected cell monolayers were dislodged in phosphate-buffered saline (PBS) plus protease inhibitors (1% aprotinin and 1 mM AEBSF) and centrifuged for 10 min at 1,000 × g. Cell pellets were washed once in PBS plus protease inhibitors, resuspended in lysis buffer (10 mM Tris-HCl [pH 8.0], 10 mM MgSO4, 2 mM CaCl2, and 0.1% Triton X-100, supplemented with protease inhibitors and 65 U of micrococcal nuclease/ml), and subjected to three freeze-thaw cycles. Nonreducing sodium dodecyl sulfate (SDS) sample buffer was added, and the samples were incubated 40 min at room temperature with intermittent vortexing.
For affinity chromatography, proteins from concentrated medium supernatants were applied to Ni-nitrilotriacetic acid (NTA) minispin columns (Qiagen) in the presence of 10 mM imidazole. Unbound proteins were removed by four washes with 20 mM imidazole, and bound proteins were eluted with 250 mM imidazole. The eluate was dialyzed against PBS at 4°C (minidialysis units; Pierce).
SDS-polyacrylamide gel electrophoresis (PAGE) was performed on 4 to 20% polyacrylamide gels under reducing conditions (3% β-mercaptoethanol in sample buffer), unless otherwise noted. For immunoblot analysis, proteins were transferred to Protran nitrocellulose membranes (Schleicher and Schuell). CD4 sequences were detected with a 1:7,000 dilution of the T4-4 anti-CD4 rabbit serum (NIH AIDS Research and Reference Reagent Program) followed by 125I-labeled protein A (Amersham Pharmacia); quantitation was performed on a Typhoon 9600 phosphorimager. Alternatively, the second probe was donkey anti-rabbit IgG conjugated to horseradish peroxidase; images were developed with chemiluminescent substrates (Pierce) and exposure to X-ray film. For detection of the His tag, the anti-His4 MAb (Qiagen) followed by peroxidase-conjugated sheep anti-mouse IgG was used.
For determination of sCD4-17b concentrations, comparative immunoblot analyses were performed on serial dilutions of both the sample and a standard preparation of purified two-domain sCD4. CD4 sequences were detected with T4-4 anti-CD4 rabbit serum followed by 125I-labeled protein A, as described above.
HIV-1 infectivity assays.
Infection was quantitated using an indicator cell system (17). MAGI (HeLa-CD4-LTR/β-gal) or MAGI-CCR5 indicator cells were plated in 96-well plates at 104 cells per well in 200 μl of medium (Dulbecco's modified Eagle medium with 10% fetal bovine serum) and incubated overnight at 37°C. HIV-1 dilutions were preincubated with various concentrations of the indicated agents in 50 μl of medium for 30 min at 37°C. The amounts of input virus were chosen to give a readily countable number of blue cells per well in untreated controls. Dextran sulfate was added (final concentration, 20 μg/ml), and the virus-agent mixtures were added to individual wells from which the media had been removed. After 2 h of incubation at 37°C, one of the following treatments was performed (these gave equivalent results). (i) The mixtures were removed, the monolayers were washed twice with medium, and 200 μl of medium was added. (ii) The mixtures were diluted with medium to 200 μl. (iii) The mixtures were diluted to 200 μl with medium containing the original agents at the same concentrations. Following a 2-day incubation at 37°C, the monolayers were fixed and stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal); blue cells were counted under light microscopy.
RESULTS
Design of sCD4-17b.
We sought to engineer a recombinant chimeric protein containing the gp120-binding region of sCD4 attached via a flexible polypeptide linker to an SCFv of a MAb whose epitope overlaps the conserved CD4-induced bridging sheet of gp120. We chose the 17b SCFv for several reasons: (i) the epitope is highly conserved on genetically diverse HIV-1 strains (38, 40, 41); (ii) in a cell fusion assay whereby Env is activated by sCD4 to promote fusion with target cells expressing coreceptor but no CD4, 17b potently inhibits fusion for all Envs tested from genetically diverse isolates (32); (iii) the reported X-ray crystallographic structure of the core gp120 in a ternary complex with two-domain sCD4 and the 17b Fab (18) provides a specific guide to the design of the chimeric protein; and (iv) 17b is a human antibody.
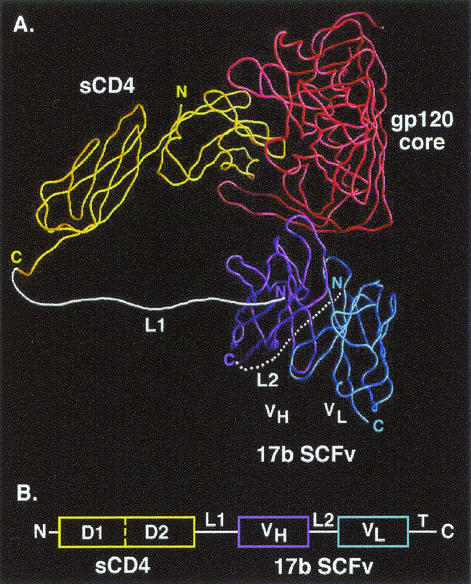
We examined the atomic coordinates of the ternary complex and determined the optimal design (Fig. 1A). The C terminus of two-domain sCD4 (through Ser183) is connected via a flexible polypeptide linker (designated L1) to the N terminus of the 17b VH chain; the C terminus of the VH is attached by a second polypeptide linker (designated L2) to the N terminus of the VL chain, thereby creating the 17b SCFv moiety. There are multiple advantages of this design. (i) The L1 linker spans the shortest distance between available termini on sCD4 and the SCFv (60 Å, compared to 80 Å for the CD4 C terminus to the VL N terminus, 67 Å for the CD4 N terminus to the VH C terminus, and 86 Å for the CD4 N terminus to the VL C terminus). (ii) The L1 linker appears not to interfere sterically with the gp120 core or any other components of the complex (Fig. 1A). (iii) Free 17b SCFv linked in the desired fashion has been reported to have activity comparable to that of the parental antibody (7). We designed the L1 linker conservatively to contain 35 residues (seven repeats of Gly4Ser), which we calculated would comfortably span the required 60 Å. The 17b SCFv (7) contains a 15-residue linker (three repeats of Gly4Ser, commonly used for SCFvs); there is also a thrombin cleavage site and a 6-His tag at the C terminus (not shown in Fig. 1A). The DNA construct of sCD4-17b is depicted schematically in Fig. 1B.
FIG. 1.
sCD4-17b design. (A) Predicted structure of sCD4-17b interacting with gp120. A Cα worm diagram showing the trimeric complex containing the gp120 core (red), two-domain sCD4 (yellow), and the 17b SCFv (VH, purple; VL, blue) was derived from the coordinates of the published X-ray crystallographic complex (18), using the GRASP program (27). The L2 linker is depicted at the back of the complex. The thrombin cleavage site followed by the 6-His tag is not shown. (B) Schematic of the sCD4-17b genetic construct (not drawn to scale), including the CD4 leader at the N terminus and the thrombin cleavage site followed by 6 histidne residues (T) at the C terminus.
Expression and affinity chromatography of sCD4-17b.
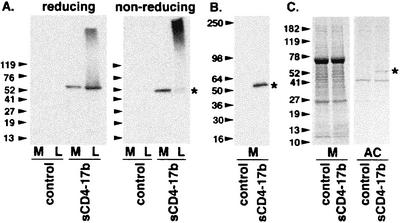
A recombinant vaccinia virus (vCD-3) containing the sCD4-17b cDNA linked to a strong synthetic vaccinia virus promoter was generated and used to infect BS-C-1 cells in low-serum-containing medium; control cells were infected in parallel with the parental vaccinia virus strain WR. Medium and detergent cell lysate samples corresponding to the same number of cells were analyzed by SDS-PAGE and immunoblotting (Fig. 2). On reducing gels (Fig. 2A, left panel), medium from cells infected with vCD-3 (encoding sCD4-17b) displayed a discrete protein species reactive with anti-CD4 antibodies, which migrated near the expected position of the chimeric protein (50.8 kDa; 483 amino acids); the detergent lysate from these cells displayed an even more prominent band at the expected position, plus dense diffuse staining at a higher molecular weight. No bands were observed in the corresponding medium and lysate fractions of cells infected with the WR (control) virus. Under nonreducing conditions (Fig. 2A, right panel), the vCD-3 medium yielded a single anti-CD4-reactive band near the expected monomeric sCD4-17b position, whereas the corresponding lysate gave very dense diffuse staining near the top of the gel. As expected, these reactivities were absent from the control medium and lysate. We conclude that a minor fraction of the total sCD4-17b protein was secreted into the medium; the bulk of the chimeric protein remained cell associated, apparently in aggregated form. As shown in Fig. 2B, a MAb directed against the 6-His tag detected a band at the expected position in medium from cells infected with vCD-3 encoding sCD4-17b, but not with the WR control.
FIG. 2.
Expression and affinity chromatography of sCD4-17b. BS-C-1 cells were infected with WR (control) or vCD-3 (sCD4-17b). SDS-PAGE was used to resolve samples of media (M) and corresponding detergent lysates (L). In each experiment, arrowheads indicate the migration according to molecular size standards (in kilodaltons), and an asterisk indicates the position of the sCD4-17b band. (A) Immunoblot analysis with anti-CD4 polyclonal antisera, on reducing (left panel) and nonreducing (right panel) gels. (B) Immunoblot analysis of concentrated media with a MAb against the His tag. (C) Coomassie blue staining of concentrated media (M) and affinity chromatography-enriched (AC) fractions. Each lane in panels A and B represents material from equivalent numbers of cells; to facilitate visualization in panel C, the affinity chromatography-enriched samples represent material from three times the number of cells as the concentrated media samples.
Affinity chromatography was performed based on the 6-His tag at the C terminus of sCD4-17b, and the samples were analyzed by Coomassie blue staining (Fig. 2C). The concentrated media from cells infected with vCD-3 (encoding sCD4-17b) or WR (control) displayed similar complex protein profiles. The affinity chromatography-enriched material from vCD-3-infected cells contained a prominent band at the expected position of sCD4-17b, which was absent from the WR-infected cells; in addition, both affinity chromatography-enriched fractions contained a prominent band at a lower molecular weight, plus several minor bands.
Based on immunoblot quantitation, a typical yield from the medium of 20 × 106 BS-C-1 cells after 40-fold concentration was 15 μg in 0.75 ml. Neutralization analyses were performed either with concentrated media or with affinity chromatography-enriched material, as indicated for each experiment.
Neutralization of HIV-1 infection by sCD4-17b.
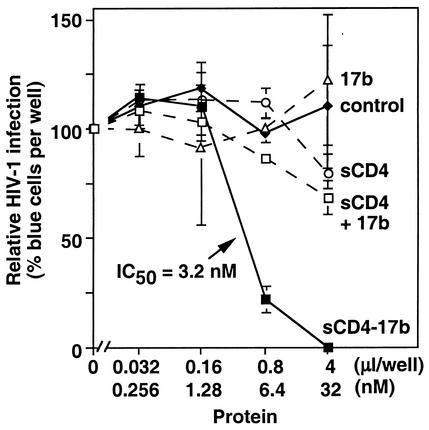
We analyzed the effects of sCD4-17b on infectivity by Ba-L (13), a prototypic primary CCR5-specific (R5) HIV-1 isolate. Infectivity assays were performed with MAGI-CCR5 indicator cells, which contain the Escherichia coli lacZ gene linked to the HIV-1 long terminal repeat (LTR); infection leads to production of β-galactosidase, and blue cells are scored microscopically after X-Gal staining. Figure 3 shows that dose-dependent neutralization occurred when virus was preincubated with concentrated medium from cells infected with vCD-3 (encoding sCD4-17b), but not with that from cells infected with WR (control). The neutralization was highly potent, corresponding to a 50% inhibitory concentration (IC50) of 3.2 nM (∼0.16 μg/ml), and >99% neutralization at 32 nM (∼1.6 μg/ml). We also compared the effects of the individual component proteins comprising the chimeric molecule: neither sCD4 nor 17b IgG neutralized infection when preincubated singly with virus, as expected from the known resistance of primary HIV-1 isolates to these agents. The combination of sCD4 plus 17b IgG had minimal effects over the concentration range examined, consistent with the high sCD4 concentration (200 nM) previously used to render primary Envs susceptible to inhibition by 17b IgG (32).
FIG. 3.
Neutralizing activity of sCD4-17b against a primary HIV-1 isolate. The Ba-L strain was preincubated with increasing volumes of concentrated media from BS-C-1 cells infected with either vCD-3 (sCD4-17b, corresponding to the indicated concentrations) or WR (none); equivalent virus samples were preincubated with increasing amounts of the purified protein sCD4, 17b IgG, or sCD4 plus 17b IgG. The infectivity assay was performed with MAGI-CCR5 cells. Results are expressed as the percentage of blue cells compared to the level (taken as 100%) in samples incubated with no additions (which gave 108 blue cells per well). Error bars indicate standard deviations of duplicate samples. On the x axis, the “μl/well” label refers to the volumes of concentrated media from vaccinia virus-infected cells; the “nM” label indicates the corresponding concentrations of sCD4-17b in the concentrated medium of vCD-3-infected cells, as well as the concentration of each purified protein.
Potency of sCD4-17b compared to those of neutralizing MAbs.
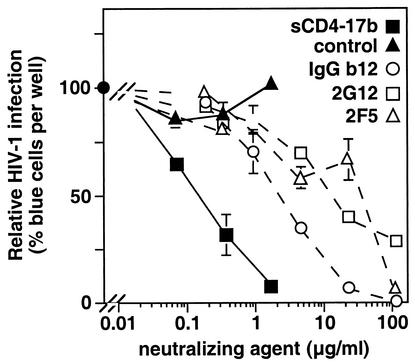
A limited number of MAbs have been described with neutralizing activity against genetically diverse primary HIV-1 isolates; these are considered to be promising candidates for passive immunotherapy (10, 29, 49). IgG1b12 is a recombinant human antibody against the conserved CD4 binding site of gp120 (6, 35); MAb 2G12 (IgG1) recognizes a conserved carbohydrate-dependent conformational epitope on gp120 (34, 46); MAb 2F5 (IgG3) binds to a conserved linear epitope in the ectodomain of gp41, proximal to the membrane (25, 28). As shown in Fig. 4, Ba-L infection of MAGI-CCR5 cells was neutralized by each MAb, with potencies in the ranges previously described in the literature for diverse primary isolates by use of various neutralization assays (6, 10, 30, 45, 46). The sCD4-17b chimeric protein was even more potent (15- to 240-fold on a microgram-per-milliliter basis, corresponding to 5- to 80-fold on a nanomolar basis).
FIG. 4.
Comparison of the HIV-1 neutralization potency of sCD4-17b with those of anti-Env MAbs. Virus samples were preincubated with increasing volumes of concentrated media from cells infected with vCD-3 (containing sCD4-17b at the indicated concentrations) or WR (control), or with increasing amounts of purified MAb IgGb12, 2G12, or 2F5. Infectivity was assayed with MAGI-CCR5 cells. Results are expressed as the percentage of blue cells compared to the level (taken as 100%) in a parallel control in which virus was preincubated with no additional proteins (which gave 770 blue cells per well). Error bars indicate standard deviations of duplicate samples.
Activity of sCD4-17b against both R5 and X4 HIV-1 strains.
The coreceptor binding determinants on gp120 include both the conserved bridging sheet and several variable loops (V3, as well as V1/V2); the latter regions (particularly V3) determine whether an individual viral strain can enter via CCR5 only (R5), CXCR4 only (X4), or either coreceptor (R5X4). In various assay systems, R5 isolates typically are non-syncytium inducing (NSI) whereas X4 and R5X4 isolates are syncytium inducing (SI) (4). Studies have demonstrated that coreceptor specificity is not a significant determinant of susceptibility to neutralizing anti-Env antibodies (20, 44). Because of the conservation of the 17b epitope on viruses with different coreceptor usage phenotypes, we anticipated that sCD4-17b would be active against both R5 and X4 strains. This prediction is confirmed in Table 1 (experiment 1); the chimeric protein neutralized the prototypic X4 T-cell line-adapted LAV strain (3) as well as the R5 primary Ba-L isolate; the potency was somewhat lower for LAV than for Ba-L.
TABLE 1.
Effects of sCD4-17b against genetically diverse HIV-1 isolates
| Expta | Isolate | Cladeb | Phenotype (coreceptor usage)b,c | % Neutralization by sCD4-17b at:
|
|
|---|---|---|---|---|---|
| 6.4 nM | 32 nM | ||||
| 1 | Ba-L | B | NSI (R5) | 94 | 98 |
| LAV | B | SI (X4) | 50 | 75 | |
| 2 | 92RW023 | A | NSI (R5) | 0 | 0 |
| Ba-L | B | NSI (R5) | 26 | 99 | |
| 91US054 | B | ns | 0 | 0 | |
| 92US714 | B | NSI (R5) | 0 | 80 | |
| 92US076 | B | SI (R5X4) | 11 | 84 | |
| 93IN905 | C | NSI (R5) | 7 | 6 | |
| 92UG024 | D | SI (X4) | 0 | 65 | |
| 93TH073 | E | NSI (R5) | 0 | 0 | |
| 93BR029 | F | NSI (R5) | 14 | 0 | |
In experiment 1, viruses were preincubated with the indicated concentrations of sCD4-17b in medium and assayed for infectivity, by using MAGI-CCR5 cells for Ba-L and MAGI cells for LAV. The numbers of blue cells per well in untreated controls were 350 for Ba-L and 300 for LAV. In experiment 2, viruses were preincubated with the indicated concentrations of affinity chromatography-enriched sCD4-17b and assayed for infectivity by using MAGI-CCR5 cells. For the different viruses, the numbers of blue cells per well in the untreated controls ranged from 50 to 475.
Clade and coreceptor usage phenotypes are as specified in the AIDS Research and Reference Reagent Program catalogue.
ns, not specified.
Breadth of sCD4-17b neutralization against genetically diverse HIV-1 isolates.
The 17b epitope is conserved geographically on diverse HIV-1 isolates of different genetic subtypes (clades). We previously demonstrated that in an assay whereby Env-expressing cells are activated by sCD4 to fuse with cells expressing coreceptor but no CD4, Envs from genetically diverse isolates (clades A through F) were all potently inhibited by MAb 17b (32). We therefore anticipated that the sCD4-17b chimeric protein would display neutralizing activity against a broad array of genotypes. Table 1 (experiment 2) shows the results with a panel of HIV-1 primary isolates from clades A through F. Surprisingly, we observed marked variation in the sCD4-17b sensitivity of different strains; including both experiments 1 and 2, the extent of neutralization at the highest concentration tested (32 nM) ranged from strong (75 to 99% for Ba-L, LAV, 92US714, and 92US076) to moderate (65% for 92UG024) to negligible (<10% for 92RW023, 91US054, 93IN905, 93TH073, and 93BR029). Experiment 2 expands the previous conclusion that sCD4-17b sensitivity was not associated with coreceptor specificity. Nor was there a clear correlation with genetic subtype: four clade B isolates were strongly neutralized but one was unaffected, the single clade D isolate was moderately neutralized, and the single isolates from each of clades A, C, E, and F were minimally affected at the concentrations tested. Interestingly, two strains (92US714 and 92US076) previously reported to be highly MAb resistant (30) were strongly neutralized by sCD4-17b. Especially puzzling was the finding that sCD4-17b resistance was observed for one isolate (93BR029) whose corresponding Env was previously found to be highly sensitive to MAb 17b in an sCD4-activated fusion assay (32).
DISCUSSION
The results presented herein demonstrate the powerful neutralizing activity of sCD4-17b against multiple HIV-1 primary isolates. Based on the minimal activity observed with the monofunctional sCD4 and 17b proteins (individually or in combination), we conclude that the potency of the chimeric protein derives from the ability of two distinct moieties on the same protein to associate simultaneously with their corresponding binding sites on a single gp120 subunit. It is likely that this bifunctional interaction greatly reduces the dissociation rate of the chimeric protein compared to those of the unlinked sCD4 or 17b constituent proteins; indeed, preliminary surface plasmon resonance analyses support this model (D. Hamer, personal communication).
When tested against the Ba-L strain, sCD4-17b proved to be highly potent compared to the well-characterized broadly cross-reactive neutralizing MAbs IgGb12, 2G12, and 2F5. This activity is particularly striking in view of the monomeric nature of the chimeric protein compared to the divalent structure of the MAbs (IgGs). In previous studies of the b12 antibody, the monovalent Fab b12 was converted to the divalent IgGb12 to improve neutralizing acitivity (6); similarly, sCD4 has been rendered dramatically more potent for neutralization by engineering fusion proteins with immunoglobulin constant regions, resulting in divalent (8, 42), tetravalent (45), or dodecavalent (1) derivatives. Thus, the potency of sCD4-17b might be enhanced even further by producing analogous multivalent constructs with immunoglobulin constant regions, most logically by attachment at the C terminus of the SCFv moiety (see Fig. 1). Such modifications probably would also increase the half-life of the protein in the circulation.
Given the broad conservation of the sCD4 binding site and the 17b epitope, we anticipated that sCD4-17b would be active against diverse HIV-1 isolates. As expected, we found that the chimeric protein functioned against viruses using either CCR5 or CXCR4. When primary isolates from different geographic regions were examined, no clear correlation was evident between sCD4-17b sensitivity and genetic clade; however, only a small number of non-B isolates were tested, and more extensive studies are required to establish this point. Particularly encouraging was the strong activity observed against two primary strains previously shown to be highly resistant to the broadly cross-reactive MAbs.
In contrast with these favorable findings, we were surprised that many of the primary isolates appeared resistant to the chimeric protein, at least at the relatively low maximum concentration tested (32 nM, ∼1.6 μg/ml). Studies are under way to improve the expression, secretion, and purification protocols so that higher concentrations can be tested. Several explanations can be envisioned to explain the marked variations in sensitivity of different strains, each involving inherent properties of the corresponding Envs. (i) The affinity of the 17b SCFv may be relatively weak for some isolates. While possible, this hypothesis would not explain the enigmatic observation that for one of the sCD4-17b-resistant strains (93BR029), the corresponding Env was previously found to be highly sensitive to MAb 17b in an sCD4-activated cell fusion assay (32); this finding rules out the interpretation that the 17b epitope is simply absent from the gp120 subunit. (ii) Kinetic and thermodynamic barriers may limit the sensitivity of some Envs to sCD4-17b. In studies of neutralization of different HIV-1 isolates by MAbs and sCD4, there is a good correlation between neutralization activity and binding to the oligomeric Env expressed on the virion surface (but not to the corresponding soluble gp120 monomers); the association rate appears to be of particular significance (reviewed in reference 23). Recent thermodynamic studies have indicated that the structural rearrangements associated with CD4 binding to gp120 are accompanied by uncharacteristically high entropic changes compared to those for typical protein-protein interactions; this entropic barrier may provide a major resistance mechanism against HIV-1 neutralization by antibodies or sCD4 derivatives (26; P. Kwong, personal communication). Perhaps the varying sCD4-17b sensitivities of different HIV-1 strains may reflect variation in these kinetic and thermodynamic parameters. However, this argument is difficult to reconcile with our consistent observation that sCD4-17b was somewhat less potent against the prototypic T-cell line-adapted LAV strain than against the primary Ba-L strain, despite the fact that T-cell line-adapted viruses have long been known to be much more susceptible than primary viruses to neutralization by sCD4, and their oligomeric Envs bind sCD4 with considerably higher affinities and faster association rates (23). Nevertheless, if kinetic and thermodynamic barriers are responsible for the sCD4-17b resistance of some isolates, then multimeric fusion proteins with sCD4-17b linked to immunoglobulin constant regions (see above) may help to overcome this problem. (iii) Perhaps for some strains, the L1 linker is of insufficient length to enable simultaneous binding of the sCD4 and 17b moieties to the same gp120 subunit. Our design of the chimeric protein was guided by the X-ray crystallographic structure of the strain HXBc-2 gp120 core in complex with sCD4 and the 17b Fab (18). From this we proposed that the L1 linker would not interfere sterically with any components of the complex (Fig. 1A). However, additional considerations may complicate this interpretation. For one, determination of the crystallographic structure necessitated deliberate removal of multiple determinants on gp120, notably the V1/V2 and V3 variable loops as well as the complex oligosaccharide chains (18). Perhaps in gp120 from some HIV-1 isolates, these determinants impart steric hindrance with the L1 linker, thereby preventing bifunctional binding of the sCD4-17b construct. Examination of the gp120 primary sequences of some of the resistant strains (i.e., numbers of residues in the variable loops) failed to provide any insight (data not shown), but such analyses cannot reveal the critical three-dimensional structure information. A related issue is the relative orientation of sCD4 and 17b when bound to gp120; perhaps there is variation such that in the resistant isolates, the distances between the sCD4 C terminus and the 17b VH N terminus are greater than reported in the crystallographic structure for the complex with HXBc-2 gp120. Thus, the 35-residue L1 linker might not be sufficiently long to span the required distance. We are addressing these possibilities by designing constructs of sCD4-17b with different L1 linker lengths.
The potent neutralizing activity of sCD4-17b suggests its potential use for passive immunization against HIV-1. The fact that both the sCD4 and 17b sequences are human should minimize immunogenicity problems, although the linker may be of concern. Therapeutic applications can be envisioned, perhaps using the chimeric protein in combination with other anti-HIV agents. Particularly intriguing is the notion of using sCD4-17b to prevent acute HIV-1 infection in various circumstances, including blockade of maternal transmission, postexposure prophylaxis, and use as a topical microbicide against sexual transmission. Future efforts to broaden the activity of sCD4-17b against diverse HIV-1 variants will greatly influence the viability of these prospects.
Acknowledgments
This work was funded in part by the NIH Intramural AIDS Targeted Antiviral Program.
Paul Kennedy provided invaluable technical contributions. We thank Marius Clore, Chih-Chin Huang, and Peter Kwong for assistance with analysis of the gp120 structural coordinates; Shirley Leow (Upjohn), John Mascola, Dennis Burton, Richard Wyatt, and James Robinson for donation of reagents; and Peter Kwong, Richard Wyatt, and Laurel Lagenaur for critical comments.
REFERENCES
- 1.Arthos, J., C. Cicala, T. D. Steenbeke, T.-W. Chun, D. Dela Cruz, D. B. Hanback, P. Khazanie, D. Nam, P. Schuck, S. M. Selig, D. Van Ryk, M. A. Chaikin, and A. S. Fauci. 2002. Biochemical and biological characterization of a dodecameric CD4-Ig fusion protein. J. Biol. Chem. 277:11456-11464. [DOI] [PubMed] [Google Scholar]
- 2.Bandres, J. C., Q. F. Wang, J. O'Leary, F. Baleaux, A. Amara, J. A. Hoxie, S. Zolla-Pazner, and M. K. Gorny. 1998. Human immunodeficiency virus (HIV) envelope binds to CXCR4 independently of CD4, and binding can be enhanced by interaction with soluble CD4 or by HIV envelope deglycosylation. J. Virol. 72:2500-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barre-Sinoussi, F., J.-C. Chermann, F. Rey, M. T. Nugeyre, S. Chamaret, J. Gruest, C. Dauguet, C. Axler-Blin, F. Vezinet-Brun, C. Rouzioux, W. Rozenbaum, and L. Montagnier. 1983. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220:868-871. [DOI] [PubMed] [Google Scholar]
- 4.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 5.Broder, C. C., and E. A. Berger. 1993. CD4 molecules with a diversity of mutations encompassing the CDR3 region efficiently support human immunodeficiency virus type 1 envelope glycoprotein-mediated cell fusion. J. Virol. 67:913-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. H. I. Parren, L. S. W. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, M. Lamacchia, E. Garratty, E. R. Stiehm, Y. J. Bryson, Y. Z. Cao, J. P. Moore, D. D. Ho, and C. F. Barbas. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 7.Cao, J., N. Sullivan, E. Desjardin, C. Parolin, J. Robinson, R. Wyatt, and J. Sodroski. 1997. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J. Virol. 71:9808-9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capon, D. J., S. M. Chamow, J. Mordenti, S. A. Marsters, T. Gregory, H. Mitsuya, R. A. Byrn, C. Lucas, F. M. Wurm, and J. E. Groopman. 1989. Designing CD4 immunoadhesins for AIDS therapy. Nature 337:525-531. [DOI] [PubMed] [Google Scholar]
- 9.Chackerian, B., E. M. Long, P. A. Luciw, and J. Overbaugh. 1997. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J. Virol. 71:3932-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Souza, M. P., D. Livnat, J. A. Bradac, S. H. Bridges, Y. Bryson, C. Hanson, T. Matthews, J. Moore, A. Trkola, S. Zolla-Pazner, M. Gorny, D. Burton, T. Merigan, J. Mcnamara, M. Norcross, M. Posner, J. Robinson, N. Virani-Ketter, C. Barbas, P. Parren, and H. Katinger. 1997. Evaluation of monoclonal antibodies to human immunodeficiency virus type 1 primary isolates by neutralization assays: performance criteria for selecting candidate antibodies for clinical trials. J. Infect. Dis. 175:1056-1062. [DOI] [PubMed] [Google Scholar]
- 11.Edwards, T. G., T. L. Hoffman, F. Baribaud, S. Wyss, C. C. Labranche, J. Romano, J. Adkinson, M. Sharron, J. A. Hoxie, and R. W. Doms. 2001. Relationships between CD4 independence, neutralization sensitivity, and exposure of a CD4-induced epitope in a human immunodeficiency virus type 1 envelope protein. J. Virol. 75:5230-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finnegan, C. M., W. Berg, G. K. Lewis, and A. L. DeVico. 2001. Antigenic properties of the human immunodeficiency virus envelope during cell-cell fusion. J. Virol. 75:11096-11105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gartner, S., P. Markovits, D. M. Markovitz, M. H. Kaplan, R. C. Gallo, and M. Popovic. 1986. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science 233:215-219. [DOI] [PubMed] [Google Scholar]
- 14.Hill, C. M., H. Deng, D. Unutmaz, V. N. Kewalramani, L. Bastiani, M. K. Gorny, S. Zolla-Pazner, and D. R. Littman. 1997. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J. Virol. 71:6296-6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffman, T. L., G. Canziani, L. Jia, J. Rucker, and R. W. Doms. 2000. A biosensor assay for studying ligand-membrane receptor interactions: binding of antibodies and HIV-1 env to chemokine receptors. Proc. Natl. Acad. Sci. USA 97:11215-11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffman, T. L., C. C. Labranche, W. T. Zhang, G. Canziani, J. Robinson, I. Chaiken, J. A. Hoxie, and R. W. Doms. 1999. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc. Natl. Acad. Sci. USA 96:6359-6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwong, P. D., R. Wyatt, Q. J. Sattentau, J. Sodroski, and W. A. Hendrickson. 2000. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J. Virol. 74:1961-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LaCasse, R. A., K. E. Follis, T. Moudgil, M. Trahey, J. M. Binley, V. Planelles, S. Zolla-Pazner, and J. H. Nunberg. 1998. Coreceptor utilization by human immunodeficiency virus type 1 is not a primary determinant of neutralization sensitivity. J. Virol. 72:2491-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lapham, C. K., J. Ouyang, B. Chandrasekhar, N. Y. Nguyen, D. S. Dimitrov, and H. Golding. 1996. Evidence for cell-surface association between fusin and the CD4-gp120 complex in human cell lines. Science 274:602-605. [DOI] [PubMed] [Google Scholar]
- 22.Martin, K. A., R. Wyatt, M. Farzan, H. Choe, L. Marcon, E. Desjardin, J. Robinson, J. Sodroski, C. Gerard, and N. P. Gerard. 1997. CD4-independent binding of SIV gp120 to rhesus CCR5. Science 278:1470-1473. [DOI] [PubMed] [Google Scholar]
- 23.Moore, J. P., and D. D. Ho. 1995. HIV-1 neutralization: the consequences of viral adaptation to growth on transformed T cells. AIDS 9(Suppl. A):S117-S136. [PubMed] [Google Scholar]
- 24.Moss, B., P. L. Earl, N. Cooper, L. S. Wyatt, M. W. Carroll, and O. Elroy-Stein. 1998. Expression of protein in mammalian cells using vaccinia viral vectors, p. 16.15.1-16.19.11. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 2, chapter 16, suppl. 43. John Wiley & Sons, Inc., New York, N.Y.
- 25.Muster, T., F. Steindl, M. Purtscher, A. Trkola, A. Klima, G. Himmler, F. Ruker, and H. Katinger. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myszka, D. G., R. W. Sweet, P. Hensley, M. Brigham-Burke, P. D. Kwong, W. A. Hendrickson, R. Wyatt, J. Sodroski, and M. L. Doyle. 2000. Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. USA 97:9026-9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicholls, A., K. A. Sharp, and B. Honig. 1991. Protein folding and association—insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct. Funct. Genet. 11:281-296. [DOI] [PubMed] [Google Scholar]
- 28.Parker, C. E., L. J. Deterding, C. Hager-Braun, J. M. Binley, N. Schulke, H. Katinger, J. P. Moore, and K. B. Tomer. 2001. Fine definition of the epitope on the gp41 glycoprotein of human immunodeficiency virus type 1 for the neutralizing monoclonal antibody 2F5. J. Virol. 75:10906-10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parren, P. W. H. I., J. P. Moore, D. R. Burton, and Q. J. Sattentau. 1999. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS 13:S137-S162. [PubMed] [Google Scholar]
- 30.Parren, P. W. H. I., M. Wang, A. Trkola, J. M. Binley, M. Purtscher, H. Katinger, J. P. Moore, and D. R. Burton. 1998. Antibody neutralization-resistant primary isolates of human immunodeficiency virus type 1. J. Virol. 72:10270-10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 32.Salzwedel, K., E. D. Smith, B. Dey, and E. A. Berger. 2000. Sequential CD4-coreceptor interactions in human immunodeficiency virus type 1 Env function: soluble CD4 activates Env for coreceptor-dependent fusion and reveals blocking activities of antibodies against cryptic conserved epitopes on gp120. J. Virol. 74:326-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Sanders, R. W., M. Venturi, L. Schiffner, R. Kalyanaraman, H. Katinger, K. O. Lloyd, P. D. Kwong, and J. P. Moore. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76:7293-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saphire, E. O., P. W. H. I. Parren, R. Pantophlet, M. B. Zwick, G. M. Morris, P. M. Rudd, R. A. Dwek, R. L. Stanfield, D. R. Burton, and I. A. Wilson. 2001. Crystal structure of a neutralizing human IgG against HIV-1: a template for vaccine design. Science 293:1155-1159. [DOI] [PubMed] [Google Scholar]
- 36.Schenten, D., L. Marcon, G. B. Karlsson, C. Parolin, T. Kodama, N. Gerard, and J. Sodroski. 1999. Effects of soluble CD4 on simian immunodeficiency virus infection of CD4-positive and CD4-negative cells. J. Virol. 73:5373-5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Speck, R. F., U. Esser, M. L. Penn, D. A. Eckstein, L. Pulliam, S. Y. Chan, and M. A. Goldsmith. 1999. A trans-receptor mechanism for infection of CD4-negative cells by human immunodeficiency virus type 1. Curr. Biol. 9:547-550. [DOI] [PubMed] [Google Scholar]
- 38.Stamatatos, L., M. Lim, and C. Cheng-Mayer. 2000. Generation and structural analysis of soluble oligomeric gp140 envelope proteins derived from neutralization-resistant and neutralization-susceptible primary HIV type 1 isolates. AIDS Res. Hum. Retrovir. 16:981-994. [DOI] [PubMed] [Google Scholar]
- 39.Sullivan, N., Y. Sun, J. Binley, J. Lee, C. F. Barbas, P. W. H. I. Parren, D. R. Burton, and J. Sodroski. 1998. Determinants of human immunodeficiency virus type 1 envelope glycoprotein activation by soluble CD4 and monoclonal antibodies. J. Virol. 72:6332-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sullivan, N., Y. Sun, Q. Sattentau, M. Thali, D. Wu, G. Denisova, J. Gershoni, J. Robinson, J. Moore, and J. Sodroski. 1998. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J. Virol. 72:4694-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thali, M., J. P. Moore, C. Furman, M. Charles, D. D. Ho, J. Robinson, and J. Sodroski. 1993. Characterization of conserved human immunodeficiency virus type-1 -gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Traunecker, A., J. Schneider, H. Kiefer, and K. Karjalainen. 1989. Highly efficient neutralization of HIV with recombinant CD4-immunoglobulin molecules. Nature 339:68-70. [DOI] [PubMed] [Google Scholar]
- 43.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184-187. [DOI] [PubMed] [Google Scholar]
- 44.Trkola, A., T. Ketas, V. N. Kewalramani, F. Endorf, J. M. Binley, H. Katinger, J. Robinson, D. R. Littman, and J. P. Moore. 1998. Neutralization sensitivity of human immunodeficiency virus type 1 primary isolates to antibodies and CD4-based reagents is independent of coreceptor usage. J. Virol. 72:1876-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trkola, A., A. B. Pomales, H. Yuan, B. Korber, P. J. Maddon, G. P. Allaway, H. Katinger, C. F. Barbas, D. R. Burton, D. D. Ho, and J. P. Moore. 1995. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J. Virol. 69:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu, L., N. P. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardoso, E. Desjardin, W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179-183. [DOI] [PubMed] [Google Scholar]
- 48.Wyatt, R., J. Moore, M. Accola, E. Desjardin, J. Robinson, and J. Sodroski. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, W. T., G. Canziani, C. Plugariu, R. Wyatt, J. Sodroski, R. Sweet, P. Kwong, W. Hendrickson, and L. Chaiken. 1999. Conformational changes of gp120 in epitopes near the CCR5 binding site are induced by CD4 and a CD4 miniprotein mimetic. Biochemistry 38:9405-9416. [DOI] [PubMed] [Google Scholar]