Abstract
RNA interference (RNAi) is a conserved mechanism in which double-stranded, small interfering RNAs (siRNAs) trigger a sequence-specific gene-silencing process. Here we describe the inhibition of murine herpesvirus 68 replication by siRNAs targeted to sequences encoding Rta, an immediate-early protein known as an initiator of the lytic viral gene expression program, and open reading frame 45 (ORF 45), a conserved viral protein. Our results suggest that RNAi can block gammaherpesvirus replication and ORF 45 is required for efficient viral production.
RNA interference (RNAi) is a mechanism conserved among different species, in which double-stranded RNAs trigger a sequence-specific gene-silencing process. Long double-stranded RNAs are processed into 21- to 23-nucleotide (nt) small interfering RNAs (siRNAs) by the Dicer enzyme (8, 26, 40) and then incorporated into a multicomponent nuclease called RNA-induced silencing complex. This complex, when activated, can specifically down regulate gene expression. RNAi has been used to study gene function in multiple model organisms, including flies (13), trypanosomes (20), zebra fish (36), mice (37), plants (33), and Caenorhabditis elegans (6). However, in most mammalian cells, double-stranded RNAs longer than 30 nt activate an interferon response, leading to nonspecific degradation of RNA transcripts and a general shutdown of host cell protein translation (2, 28). This nonspecific effect can be circumvented by the use of synthetic siRNAs that are 21 nt long with short 3′ overhangs (5). The synthesized siRNAs have been shown to induce homology-dependent degradation of cognate mRNA and used to knock down expression of endogenous and heterologous genes in mammalian cell lines (3, 7, 11, 14, 21). Although evidence suggests that viruses have evolved proteins that suppress RNA silencing, RNAi is believed to have evolved as a host defense mechanism against transposable elements and infectious viruses (15, 16). The effect of RNAi on herpesvirus replication has yet to be reported.
Two human gammaherpesviruses, Kaposi's sarcoma-associated herpesvirus (KSHV), also referred to as human herpesvirus 8, and Epstein-Barr virus (EBV), are associated with several types of malignancies and lymphoproliferative disorders. KSHV is linked to Kaposi's sarcoma (19), multicentric Castleman's disease (27), and primary effusion lymphoma (4). EBV is associated with nasopharyngeal carcinoma, Burkitt's lymphoma, Hodgkin's disease, lymphoproliferative disease, and certain types of T-cell lymphomas (24). The life cycle of herpesviruses is divided into two phases: latency and lytic replication. Rta, an immediate-early viral protein, is known to be a switch between the latent and lytic phases of the gammaherpesvriuses (9, 17, 29, 30, 38, 39). Herpesvirus lytic genes are transcribed in three stages: (i) the immediate-early stage, during which transcription occurs in the absence of de novo protein synthesis; (ii) the early stage, during which transcription is independent of viral DNA synthesis; and (iii) the late stage, during which transcription is dependent on viral DNA synthesis. The KSHV open reading frame 45 (ORF 45) has been shown to be transcribed in the absence of de novo protein synthesis (41). Analysis of KSHV global gene expression by other groups revealed that ORF 45 is transcribed at the early stage of virus reactivation (12, 25). The gene product of KSHV ORF 45 was suggested to inhibit virus-mediated interferon response by interacting with cellular interferon-regulatory factor 7 (42), a transcription activator up-regulated in KSHV-infected endothelial cells (22). The KSHV ORF 45 protein was also reported to interact with a human immunodeficiency virus type 1 transactivator, Tat (10). However, studies on the role of ORF 45 during productive human gammaherpesvirus infection have been very limited due to the lack of cell lines that can support the replication of these viruses.
Murine herpesvirus 68 (MHV68), also known as gammaherpesvirus 68 (γHV68), is a natural pathogen of wild rodents (18, 23). Complete sequence and genomic analyses indicate that MHV68 is closely related to KSHV and EBV (34). For example, amino acid sequence alignments revealed that the MHV68 ORF 45 has 37.3 and 22.2% identity to the homologue of ORF45 in KSHV and EBV, respectively. Unlike KSHV and EBV, MHV68 establishes productive infections in a variety of fibroblast and epithelial cell lines, facilitating the examination of gammaherpesvirus replication and de novo infection. MHV68 ORF 45 is conserved among all gammaherpesviruses, but it has no extensive similarity to other cellular or viral proteins with known functions, making it relatively difficult to predict its functional roles during virus replication. Analysis of the roles of conserved viral genes, i.e., ORF 45, will allow us to gain a greater understanding of the functions of these genes in the human gammaherpesvirus life cycle. Use of the RNAi approach to examine functional roles of viral genes during gammaherpesvirus replication may provide an efficient way to screen genes that are essential for virus replication.
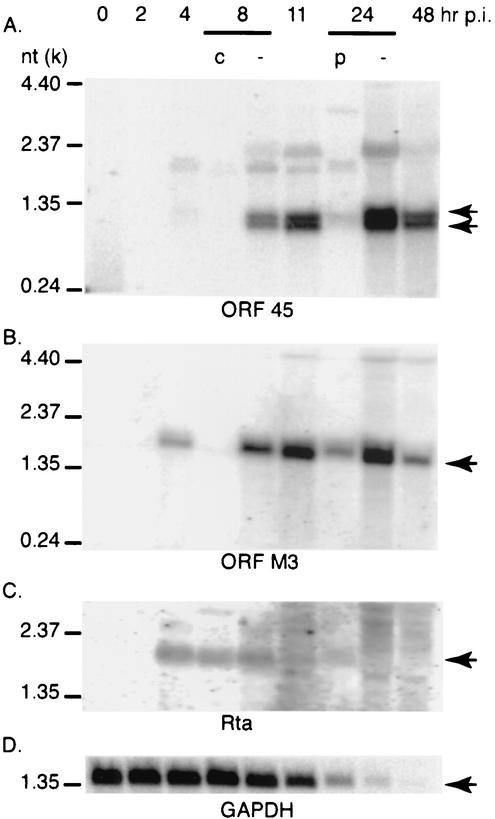
To assess the kinetics of MHV68 ORF 45 transcription, baby hamster kidney (BHK-21) cells were infected with MHV68 at a multiplicity of infection (MOI) of 5 in the presence or absence of inhibitors of protein synthesis (cycloheximide, 100 μg/ml) or viral DNA polymerase activity (phosphonoacetic acid, 200 μg/ml). Total cellular RNA (20 μg) was collected at various time points postinfection and analyzed by Northern blotting. Double-stranded DNA probes were prepared from PCR products of ORF 45, ORF M3, Rta, or GAPDH in the presence of [α32-P]dCTP. The 0.6-kb double-stranded ORF 45 probe (nt 63652 to 64225) detected multiple signals, with two major bands of 0.6 and 0.7 kb. These transcripts were detectable at 4 h, reached maximum expression at 24 h, and decreased at 48 h postinfection. The major transcripts of ORF 45 were eliminated by cycloheximide and significantly reduced by phosphonoacetic acid treatment (Fig. 1A), which is consistent with reported MHV68 DNA array analysis (1). A single-stranded DNA probe complementary to ORF 45 transcripts was also used for Northern blotting and confirmed that the major 0.6- and 0.7-kb transcripts of ORF 45 were in the sense orientation (data not shown). To verify that the concentrations of cycloheximide and phosphonoacetic acid used in this experiment were appropriate, the membrane was sequentially rehybridized with probes to sequences encoding MHV68 ORF M3, cellular GAPDH, and the SacI fragment of MHV68 Rta (38). The 1.2-kb probe to M3 (nt 6051 to 7282) detected one major transcript of 1.4-kb. The expression of this transcript was inhibited by cycloheximide and partially inhibited by phosphonoacetic acid (Fig. 1B), which is consistent with its classification as an early-late gene (32). Probing with the 0.7-kb probe to Rta detected one major transcript of 2.0 kb, which reached maximum expression at 4 h and decreased at 11 h postinfection. This major transcript level was not changed in the presence of cycloheximide, consistent with its expression pattern as an immediate-early gene (Fig. 1C) (38). As expected, the level of GAPDH expression declined during the course of infection (Fig. 1D). These results indicate that the drug concentrations used in this experiment fall within an appropriate range and the inhibition of ORF 45 transcripts by cycloheximide and phosphonoacetic acid was not due to nonspecific toxicity of the inhibitors. Therefore, our data suggest that MHV68 ORF 45 was transcribed at an early-late stage of virus lytic replication.
FIG. 1.
MHV68 ORF 45 is transcribed at an early-late stage of virus lytic replication. BHK-21 cells were infected with MHV68 at an MOI of 5 in the absence (−) or presence of cycloheximide (c) or phosphonoacetic acid (p). Total cellular RNA was harvested at the indicated times postinfection (p.i.), as shown at the top of the panels, and analyzed by Northern blotting. The membrane was sequentially hybridized with probes to ORF 45, M3, GAPDH (providing a loading control), and Rta. Arrows indicate major transcripts of ORF 45, M3, Rta, and GAPDH. The sizes of RNA ladder are indicated on the right.
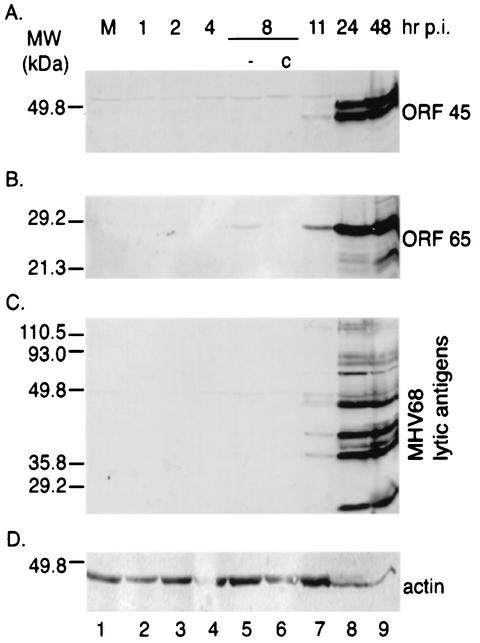
To examine the kinetics of ORF 45 protein expression, a polyclonal antibody against ORF 45 was prepared. A bacterial expression plasmid, pET-45, in which the full-length MHV68 ORF 45 coding sequence without a translation termination codon was fused to the His6 sequence of pET30b(+) (Novagen, Madison, Wis.), was constructed. The pET-45 plasmid was transformed into strain BL21 (Stratagene, La Jolla, Calif.) of Escherichia coli, and protein expression was induced with isopropyl-β-d-thiogalactopyranoside. The His6-tagged ORF 45 protein expressed in E. coli was purified and injected into rabbit to produce polyclonal antibody against ORF 45 (Covance Research Products, Denver, Pa.). To examine the kinetics of ORF 45 protein expression during virus replication, MHV68-infected BHK-21 cell extracts were analyzed by Western blotting with the prepared polyclonal antibody against ORF 45. Based on its putative amino acid sequence, the predicted size of the MHV68 ORF 45 is 22.5 kDa. However, the ORF 45 protein was detected to be approximately 48 kDa at 11 h postinfection. An additional band of 51 kDa was observed at 24 and 48 h postinfection (Fig. 2A). To confirm the specificity of the ORF 45 polyclonal antibody, the membrane was reprobed with polyclonal antibody against M9 (ORF65), as well as against multiple MHV68 lytic proteins raised in rabbits (31, 35, 38). ORF 65, a minor viral capsid protein, was detected at 8 h postinfection and increased during the course of virus replication (Fig. 2B). The expression of multiple lytic viral proteins was detected at 11 h and reached maximum expression at 24 h postinfection (Fig. 2C). To further verify that both the 48- and 51-kDa proteins were expressed from ORF 45, a mammalian expression plasmid, pFlag-45 (containing the full-length ORF 45 coding sequence fused to the Flag sequence of pFlag-CMV2 [Kodak]) was constructed. The sequence of pFlag-45 was determined to confirm that there was no mutation or duplication in the coding region of the Flag-ORF 45 fusion protein. The pFlag-45 was transfected into 293T cells, and Western blotting was performed with a monoclonal antibody against Flag to analyze the protein expression. The anti-Flag monoclonal antibody (Sigma, St. Louis, Mo.) also detected the two bands of 48 and 51 kDa (Fig. 3A). However, the 51-kDa signals were inhibited by treatment with calf intestine alkaline phosphatase (unpublished observation). This result was confirmed when the polyclonal antibody against ORF 45 was used. Our data therefore indicate that ORF 45 is expressed as a phosphoprotein during viral lytic replication. Although the functional significance of the ORF 45 phosphoprotein is not currently known, the kinetics of its phosphorylation is of interest for further analysis.
FIG. 2.
Kinetics of MHV68 viral protein expression. BHK-21 cells were mock infected (M) or infected with MHV68 in the absence (−) or presence of cycloheximide (c). At various times postinfection (p.i.), as shown at the top of the panels, cell lysates were analyzed by Western blotting with rabbit polyclonal antibody against ORF 45 (A), M9 (ORF 65) (B), or multiple MHV68 lytic proteins (C). Actin was probed, providing a loading control (D).
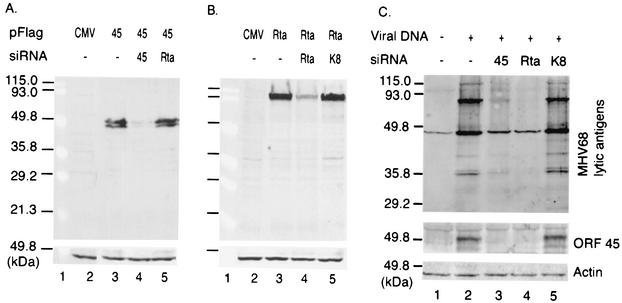
FIG. 3.
siRNAs specifically inhibit the viral protein expression. (A) 293T cells were transfected with pFlag-CMV (lane 2) or pFlag-45 (lane 3). In lane 4 or 5, cells were transfected with pFlag-45 plus RNA duplexes (0.1 μM) targeted to ORF 45 or Rta. Cell extracts were analyzed by Western blotting with anti-Flag monoclonal antibody (upper panel). Actin was probed as a loading control (lower panel). (B) 293T cells were transfected with pFlag-CMV (lane 2) or pFlag-Rta (lane 3). In lane 4 or 5, cells were transfected with pFlag-Rta plus RNA duplexes targeted to Rta or an unrelated transcript (KSHV K8). Cell lysates were analyzed as described for panel A. (C) 293T cells were mock transfected (lane 1) or transfected with MHV68 viral DNA alone (0.1 μg) (lane 2). In lane 3, 4, or 5, cells were transfected with viral DNA plus RNA duplexes (0.1 μM) targeting ORF 45, Rta, or KSHV K8, respectively. Cell extracts were analyzed by Western blotting with rabbit polyclonal antibody against MHV68 (upper panel) or ORF 45 (middle panel). The rabbit polyclonal antibody against MHV68 recognized two cellular proteins from the mock-transfected cell extracts (lane 1). Actin was probed, providing a loading control (lower panel).
To examine the functional role of ORF 45 during virus replication, we analyzed the loss of function of ORF 45 via RNAi methods. We designed siRNAs to target against ORF 45 (siRNA-45, 5′-AACUCCAGACUCAGUGUUUGA-3′, 120 to 140 nt downstream of the start codon of the ORF 45 coding sequence) and Rta (siRNA-Rta, 5′-AACCUCUGGCCUGCAGUCUGU-3′, nt 161 to 181). Since Rta is known to play a critical role during gammaherpesvirus replication, it can serve as a positive control. An siRNA targeted to KSHV ORF K8 (K-bZIP) (siRNA-K8, 5′-AAGCCUCAACGGGCAACCAUU-3′, nt 87 to 107) was used as a negative control, since it has no homologous gene in MHV68. All these siRNA sequences start with AA and have similar GC contents in their targeting sequences. The selected siRNA targeting sequences were then subjected to a BLAST search against nonredundant nucleotide sequences to ensure that only the intended viral gene was targeted. The 21-nt RNA oligonucleotides were purchased from Dharmacon Research, Inc. (Lafayette, Colo.), and RNA duplexes were prepared according to the manufacturer's recommendations. 293T cells were used for all transfections involved in RNAi for the following reasons: (i) they are permissive for MHV68 replication and produce titers of viral progenies similar to those produced in BHK-21 cells (unpublished data), (ii) they show higher transfection efficiency with the reagents used in this study, and (iii) they have been successfully used to knock down endogenous and heterologous genes by the RNAi method. We transfected 293T cells with pFlag-45 or pFlag-Rta in the presence or absence of various annealed RNA duplexes (0.1 μM) using Lipofectamine 2000 (Invitrogen, Carlsbad, Calif.). The protein expression was examined by Western blot analysis using anti-Flag monoclonal antibody. As shown in Fig. 3A, the expression of both forms of the ORF 45 protein (48 and 51 kDa) was dramatically blocked by siRNA-45 but not by siRNA-Rta. Similarly, when 293T cells were transfected with RNA duplexes plus pFlag-Rta (38), the expression of Rta protein (approximately 100 kDa) was markedly down regulated by siRNA-Rta but not by siRNA-K8 (Fig. 3B) or siRNA-45 (data not shown). These results demonstrate that siRNAs can specifically and efficiently inhibit ORF 45 or Rta protein expression in mammalian cells.
To examine the effects of siRNAs targeting against ORF 45 or Rta on lytic viral protein expression, MHV68 viral genomic DNA was transfected into 293T cells with or without various RNA duplexes. Western blotting was used to analyze the cell extracts. By use of a polyclonal antibody against MHV68, two cellular and multiple viral proteins were detected (Fig. 3C, upper panel, lanes 1 and 2). Cotransfection of 293T cells with MHV68 viral DNA plus siRNA-Rta significantly blocked the expression of other viral lytic proteins (lane 4), which is consistent with the known function of Rta as a transactivator of the downstream genes required for virus replication (17, 29, 38). Interestingly, siRNA targeted to ORF 45 also inhibited the expression of other lytic viral proteins (lane 3), indicating the critical role of ORF 45 in the process for those viral protein expressions. The siRNA targeted to KSHV gene K8 (siRNA-K8) did not show inhibition of viral lytic protein expression (lane 5). Another nonspecific siRNA targeting against the KSHV ORF 57 also failed to inhibit the expression of MHV68 viral proteins (data not shown). To confirm the specificity of RNAi-induced inhibition of viral lytic protein expression, the membrane was reprobed with the polyclonal antibody against ORF 45. The results showed that ORF 45 expression was greatly depleted by siRNA-45, but not by siRNA-K8 (Fig. 3C, middle panel, lanes 3 and 5). As we expected, siRNA-Rta also inhibited the expression of ORF 45 from the viral genome (lane 4) but did not affect that from the protein expression plasmid (Fig. 3A, lane 5). This result further confirmed that ORF 45 is expressed as a downstream gene of Rta during virus replication. We therefore conclude that siRNAs are capable of specifically inhibiting ORF 45 expression from the viral genome and this inhibition leads to reduction of virus replication.
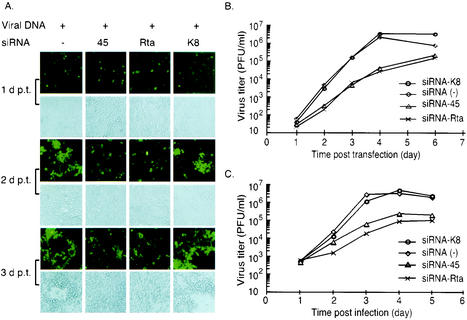
To determine whether inhibiting viral lytic protein expression by RNAi decreases virus replication, MHV68.GFP recombinant viral DNA containing an insertion of an enhanced green fluorescence protein (GFP) expression cassette (38) was transfected into 293T cells with or without RNA duplexes (0.1 μM). At various time points posttransfection, GFP expression from the recombinant viral genome and the morphology of cells were analyzed with fluorescence and light microscopy, respectively. Supernatants from transfected cultures were collected to determine virus titers. As shown in Fig. 4A, at day 1 posttransfection there was no significant difference in GFP expression among cells cotransfected with different siRNAs, indicating that cells were transfected with equal amounts of viral DNA. Accordingly, titers of virus progeny were similar among cells treated with different siRNAs at this time point (Fig. 4B). However, at day 2 posttransfection, viruses from cells transfected with viral DNA alone or cotransfected with siRNA-K8 showed dissemination to neighboring cells, indicated by GFP expression from the recombinant viral genome. This dissemination was greatly impaired by siRNAs targeting ORF 45 or Rta. At day 3 posttransfection, typical signs of cytopathic effect (i.e., rounding up, death, and detachment of the infected cells) were observed in cultures that were transfected with viral DNA alone or cotransfected with siRNA-K8. However, cells cotransfected with either siRNA-Rta or siRNA-45 showed increasing growth (Fig. 4A, columns 2 and 3) and produced only 1 to 2% virus progeny at 4 days posttransfection, compared to those produced in control groups. This inhibition effect was maintained until 6 days posttransfection, when culturing was terminated (Fig. 4B).
FIG. 4.
Inhibiting ORF 45 or Rta expression by RNAi decreases production of MHV68 viral progenies. (A) 293T cells were transfected with MHV68.GFP viral DNA alone (0.1 μg) (column 1), or viral DNA plus RNA duplexes (0.1 μM) targeting ORF 45, Rta, or KSHV K8 (column 2, 3, or 4). At days 1, 2, and 3 posttransfection (p.t.), expression of the GFP from the recombinant viral genome and the morphology of cells were analyzed by fluorescence and light microscopy, respectively. The siRNA targeted to Rta or ORF 45 was shown to inhibit virus dissemination to neighboring cells (as indicated by GFP expression) and block the cytopathic effect caused by virus replication. (B) Supernatants from transfected cell cultures as described for panel A were harvested at the indicated times posttransfection. Virus titers were determined twice in duplicate plaque assays. (C) 293T cells were mock transfected (−) or transfected with RNA duplexes (0.1 μM) targeted to ORF 45, Rta, or KSHV K8. At 1 day posttransfection, cells were infected with MHV68 (MOI of 1). Supernatants were harvested at the indicated times postinfection (p.i.). Virus titers were determined twice in duplicate plaque assays.
To further confirm that inhibiting ORF 45 and Rta expression by specific siRNAs diminishes gammaherpesvirus replication during de novo virus infection, 293T cells were transfected with RNA duplexes and then infected with MHV68 (MOI of 1) at day 1 posttransfection. The kinetics of virus replication showed that pretreatment with siRNA-Rta or siRNA-45 inhibited virus replication from de novo infection. Consistent with the results shown in Fig. 4B, the greatest inhibition was detected at 4 days posttransfection, resulting in 144- and 43-fold reductions of virus titers caused by siRNA-Rta and siRNA-45, respectively (Fig. 4C). Again, siRNA targeted to an unrelated transcript, KSHV K8, did not show inhibition on virus replication, indicating the specificity of siRNA-induced inhibition of gammaherpesvirus replication. The prolonged (6-day) inhibition of gammaherpesvirus replication by RNAi during both cotransfection and de novo infection indicates the effectiveness of the RNAi mechanism. From these results, we conclude that RNAi is capable of blocking gammaherpesvirus replication and that both Rta and ORF 45 are required for efficient virus replication. This is the first report of RNAi's capability of inhibiting herpesvirus replication. The mechanism of ORF 45 function in virus replication is still unclear. However, inhibition of virus replication in the presence of siRNA-45 suggests that its function affects a process involved in virus replication and/or assembly.
These data indicate that RNAi can provide an antigammaherpesvirus response in mammalian cells. Using the efficient RNAi method, we can identify essential genes during gammaherpesvirus replication. These essential genes, i.e., Rta and ORF 45, are potential targets for antiviral therapy. siRNAs stably expressed from plasmid to target multiple essential viral genes may represent a powerful combination therapy for herpesvirus-related disorders.
Acknowledgments
We thank Jiuyong Xie and Ting-Ting Wu for helpful discussions, and Helen Brown, Eric Bortz, Xudong Li, Wendy Wong, and Wendy Aft for critical editing of the manuscript.
This work was supported by NIH grants CA83525, CA91791, and DE14153 and by the STOP Cancer Foundation (R.S.). Q.J. was supported by a Universitywide AIDS Research Program of the University of California grant, F00-LA-016.
REFERENCES
- 1.Ahn, J. W., K. L. Powell, P. Kellam, and D. G. Alber. 2002. Gammaherpesvirus lytic gene expression as characterized by DNA array. J. Virol. 76:6244-6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baglioni, C., and T. W. Nilsen. 1983. Mechanisms of antiviral action of interferon. Interferon 5:23-42. [PubMed] [Google Scholar]
- 3.Caplen, N. J., S. Parrish, F. Imani, A. Fire, and R. A. Morgan. 2001. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. USA 98:9742-9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 5.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 6.Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver, and C. C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806-811. [DOI] [PubMed] [Google Scholar]
- 7.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 8.Hannon, G. J. 2002. RNA interference. Nature 418:244-251. [DOI] [PubMed] [Google Scholar]
- 9.Hardwick, J. M., P. M. Lieberman, and S. D. Hayward. 1988. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J. Virol. 62:2274-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang, L. M., M. F. Chao, M. Y. Chen, H. Shih, Y. P. Chiang, C. Y. Chuang, and C. Y. Lee. 2001. Reciprocal regulatory interaction between human herpesvirus 8 and human immunodeficiency virus type 1. J. Biol. Chem. 276:13427-13432. [DOI] [PubMed] [Google Scholar]
- 11.Jacque, J. M., K. Triques, and M. Stevenson. 2002. Modulation of HIV-1 replication by RNA interference. Nature 418:435-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenner, R. G., M. M. Alba, C. Boshoff, and P. Kellam. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol. 75:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kennerdell, J. R., and R. W. Carthew. 1998. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95:1017-1026. [DOI] [PubMed] [Google Scholar]
- 14.Lee, N. S., T. Dohjima, G. Bauer, H. Li, M. J. Li, A. Ehsani, P. Salvaterra, and J. Rossi. 2002. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 20:500-505. [DOI] [PubMed] [Google Scholar]
- 15.Li, H., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319-1321. [DOI] [PubMed] [Google Scholar]
- 16.Li, W. X., and S. W. Ding. 2001. Viral suppressors of RNA silencing. Curr. Opin. Biotechnol. 12:150-154. [DOI] [PubMed] [Google Scholar]
- 17.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mistrikova, J., and D. Blaskovic. 1985. Ecology of the murine alphaherpesvirus and its isolation from lungs of rodents in cell culture. Acta Virol. 29:312-317. [PubMed] [Google Scholar]
- 19.Moore, P. S., and Y. Chang. 1995. Kaposi's sarcoma findings. Science 270:15.. [DOI] [PubMed] [Google Scholar]
- 20.Ngo, H., C. Tschudi, K. Gull, and E. Ullu. 1998. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 95:14687-14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Novina, C. D., M. F. Murray, D. M. Dykxhoorn, P. J. Beresford, J. Riess, S. K. Lee, R. G. Collman, J. Lieberman, P. Shankar, and P. A. Sharp. 2002. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 8:681-686. [DOI] [PubMed] [Google Scholar]
- 22.Poole, L. J., Y. Yu, P. S. Kim, Q. Z. Zheng, J. Pevsner, and G. S. Hayward. 2002. Altered patterns of cellular gene expression in dermal microvascular endothelial cells infected with Kaposi's sarcoma-associated herpesvirus. J. Virol. 76:3395-3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajcani, J., D. Blaskovic, J. Svobodova, F. Ciampor, D. Huckova, and D. Stanekova. 1985. Pathogenesis of acute and persistent murine herpesvirus infection in mice. Acta Virol. 29:51-60. [PubMed] [Google Scholar]
- 24.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr virus, p. 2575-2628. In D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 25.Sarid, R., O. Flore, R. A. Bohenzky, Y. Chang, and P. S. Moore. 1998. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1). J. Virol. 72:1005-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharp, P. A. 2001. RNA interference—2001. Genes Dev. 15:485-490. [DOI] [PubMed] [Google Scholar]
- 27.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 28.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 29.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun, R., S. F. Lin, K. Staskus, L. Gradoville, E. Grogan, A. Haase, and G. Miller. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sunil-Chandra, N. P., S. Efstathiou, J. Arno, and A. A. Nash. 1992. Virological and pathological features of mice infected with murine gamma-herpesvirus 68. J. Gen. Virol. 73:2347-2356. [DOI] [PubMed] [Google Scholar]
- 32.van Berkel, V., K. Preiter, H. W. Virgin IV, and S. H. Speck. 1999. Identification and initial characterization of the murine gammaherpesvirus 68 gene M3, encoding an abundantly secreted protein. J. Virol. 73:4524-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaucheret, H., C. Beclin, T. Elmayan, F. Feuerbach, C. Godon, J. B. Morel, P. Mourrain, J. C. Palauqui, and S. Vernhettes. 1998. Transgene-induced gene silencing in plants. Plant J. 16:651-659. [DOI] [PubMed] [Google Scholar]
- 34.Virgin, H. W., IV, P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Virgin, H. W., IV, R. M. Presti, X. Y. Li, C. Liu, and S. H. Speck. 1999. Three distinct regions of the murine gammaherpesvirus 68 genome are transcriptionally active in latently infected mice. J. Virol. 73:2321-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wargelius, A., S. Ellingsen, and A. Fjose. 1999. Double-stranded RNA induces specific developmental defects in zebrafish embryos. Biochem. Biophys. Res. Commun. 263:156-161. [DOI] [PubMed] [Google Scholar]
- 37.Wianny, F., and M. Zernicka-Goetz. 2000. Specific interference with gene function by double-stranded RNA in early mouse development. Nat. Cell Biol. 2:70-75. [DOI] [PubMed] [Google Scholar]
- 38.Wu, T. T., L. Tong, T. Rickabaugh, S. Speck, and R. Sun. 2001. Function of Rta is essential for lytic replication of murine gammaherpesvirus 68. J. Virol. 75:9262-9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zalani, S., E. Holley-Guthrie, and S. Kenney. 1996. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. USA 93:9194-9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zamore, P. D., T. Tuschl, P. A. Sharp, and D. P. Bartel. 2000. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101:25-33. [DOI] [PubMed] [Google Scholar]
- 41.Zhu, F. X., T. Cusano, and Y. Yuan. 1999. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:5556-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu, F. X., S. M. King, E. J. Smith, D. E. Levy, and Y. Yuan. 2002. A Kaposi's sarcoma-associated herpesviral protein inhibits virus-mediated induction of type I interferon by blocking IRF-7 phosphorylation and nuclear accumulation. Proc. Natl. Acad. Sci. USA 99:5573-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]