Abstract
Human immunodeficiency virus protease activity can be regulated by reversible oxidation of a sulfur-containing amino acid at the dimer interface. We show here that oxidation of this amino acid in human immunodeficiency virus type 1 protease prevents dimer formation. Moreover, we show that human T-cell leukemia virus type 1 protease can be similarly regulated through reversible glutathionylation of its two conserved cysteine residues. Based on the known three-dimensional structures and multiple sequence alignments of retroviral proteases, it is predicted that the majority of retroviral proteases have sulfur-containing amino acids at the dimer interface. The regulation of protease activity by the modification of a sulfur-containing amino acid at the dimer interface may be a conserved mechanism among the majority of retroviruses.
We previously demonstrated that oxidative modification of cysteine 95 in human immunodeficiency virus type 1 (HIV-1) protease and methionine 95 in HIV-2 protease inhibits protease activity (2, 3). The activity of these oxidized proteases can be restored with the use of cellular enzymes, thioltransferase (TTase) for HIV-1 and methionine sulfoxide reductase for HIV-2. TTase is found within HIV-1 virions, supporting a possible role for this enzyme in activating protease activity during virus budding (3). In this study we addressed the mechanism by which oxidation leads to the inhibition of protease activity and investigated the potential role of oxidation in the inhibition of the protease from human T-cell leukemia virus type 1 (HTLV-1), a transforming retrovirus. We also analyzed other retrovirus protease sequences and structures to explore whether they might be similarly regulated.
Oxidation prevents retroviral protease dimer formation.
Residue 95 of the HIV-1 and HIV-2 proteases is located at the dimer interface. For HIV-1 protease, the interface interaction contributes close to 75% of the free energy of dimer stabilization (17). To determine if oxidation of residue 95 inhibited protease activity by preventing dimer formation, we ran analytical ultracentrifugation on a previously described autoproteolysis-resistant form of the HIV-1 protease (10, 24) modified with glutathione at cysteine 95. The protease mutations were introduced by using PCR as described previously (20) and included the Q8K, L33I, L63I, and C67A mutations, which are known to substantially reduce autoproteolysis (10, 24). The recombinant protease, called KIIA protease, was glutathionylated at cysteine 95 with 100 mM glutathione in 250 mM Tris-HCl, pH 7.8, and 6.0 M guanidine at 37°C for 4 h. The modified protease, KIIA-glut, was then purified by reversed-phase high-performance liquid chromatography (RP-HPLC) by using a 3-ml Resource reversed-phase chromatography column from Amersham Biosciences (Piscataway, N.J.), and its molecular mass was verified by mass spectrometry as described previously (3). KIIA-glut protease (eluting in approximately 5 ml and >95% pure) was refolded by dialysis against 4 liters of 50 mM sodium acetate buffer, pH 4.0, with 1 mM EDTA and then concentrated by using a Centriplus 10 device from Amicon (Bedford, Mass.). Unmodified KIIA protease was prepared by the same methods. Protease activity was assessed before and after treatment with 1 mM tris(2-carboxyethyl)phosphine HCl (TCEP) obtained from Calbiochem (La Jolla, Calif.) as described previously (3).
Analytical ultracentrifugation of KIIA-glut was performed on a Beckman Optima XL-A analytical ultracentrifuge with an An-60 Ti rotor and standard double-sector centerpiece cells. For equilibrium measurements, samples (90 μl) were centrifuged for 14 to 20 h at either 22,500 rpm (at 20°C) for unmodified KIIA protease or 20,000 rpm (at 10°C) for KIIA-glut. KIIA protease was analyzed in 50 mM sodium acetate, pH 4.0, containing 1 mM dithiothreitol, while KIIA-glut was analyzed in the same buffer without dithiothreitol but with a 2 M excess of the HIV-1 protease inhibitor KNI-272 to prevent any possible autoproteolysis (22). At sedimentation equilibrium, the unmodified KIIA protease was clearly dimeric, with a determined relative molecular weight of 21,300, almost exactly twice that of the predicted monomer (10,778.4) (Fig. 1A). By comparison, KIIA-glut had a determined molecular weight of 12,000, which is close to the predicted molecular weight of the monomeric protein modified with glutathione (11,051) (Fig. 1B). The enzymatic activity of KIIA-glut was inhibited more than 95% compared to that of the chemically reduced protease (Fig. 1B, inset). Mass spectroscopy of the samples before and after centrifugation with an Agilent 1100 Series liquid chromatography-mass spectrometry system confirmed their structural integrity (data not shown). The conformation of the proteins was probed by far-UV circular dichroism (Fig. 1C). The native protein exhibits a fairly weak negative ellipticity centered at about 215 to 216 nm, which is typical of β-sheet structure and consistent with the X-ray model (β-sheet, 45 to 50%; protein data bank identification number 1A94). Glutathionylation of cysteine 95 results in an increase in negative ellipticity, now maximal at ∼200 nm, which is characteristic of unfolded protein (23). This interpretation is further supported by an analogous spectral shift induced in the control protein by denaturation with guanidine hydrochloride (the spectral region shown in Fig. 1C is limited by the very high absorbance of guanidine-HCl in the far-UV region). These results indicate that the modified protein is monomeric and at least partially unfolded, which is consistent with results from previous work in which the unfolding of the protease was best described by a two-state model where folded dimers were in equilibrium with unfolded monomers (5). A similar sedimentation equilibrium experiment also indicated that HIV-2 protease behaves as a monomer following oxidation of the methionine residues to methionine sulfoxide (data not shown), a modification that inhibits HIV-2 protease activity (2).
FIG. 1.
Equilibrium analytical ultracentrifugation and circular dichroic spectra of HIV-1 protease mutant (KIIA) (A) and HIV-1 protease mutant glutathionylated at cysteine residue 95 (KIIA-glut) (B). The absorbance gradients in the centrifuge cell after the sedimentation equilibrium was attained at 22,500 rpm (A) or 20,000 rpm (B) are shown in the lower panels. The open circles represent the experimental values, and the solid lines represent the results of fitting to a single ideal species. The best fit for the data shown in panel A yielded a molecular weight of 21,300 and that for the data shown in panel B yielded a molecular weight of 12,000. The corresponding upper panels show the differences in the fitted and experimental values as a function of radial position (residuals). The residuals of these fits were random, indicating that the single species model is appropriate for the analyses. (B, inset) Protease activity for the glutathionylated protease in the absence and presence of 1 mM TCEP reducing agent. (C) Circular dichroic spectra of the following HIV-1 protease forms: KIIA mutant protease (solid line), KIIA mutant protease glutathionylated at cysteine 95 (dashed line), and mutant KIIA treated with 4 M guanidine-HCl (Gdn-HCl; dotted line). Spectra (average of four scans) were recorded in 50 mM sodium acetate, pH 4.0, and are shown in far-UV region to limits of reliable detection as monitored by photomultiplier voltage (usually less than 600 V). The units of the ordinates are mean residue ellipticity (θmwr) and have the dimension degrees per square centimeter per decamole. θmwr can be converted to the molar circular dichroism absorption coefficient Δe (units per molar per centimeter) by multiplication by Na and division by 3,298, where Na is the number of residues.
Regulation of HTLV-1 protease activity through reversible oxidation.
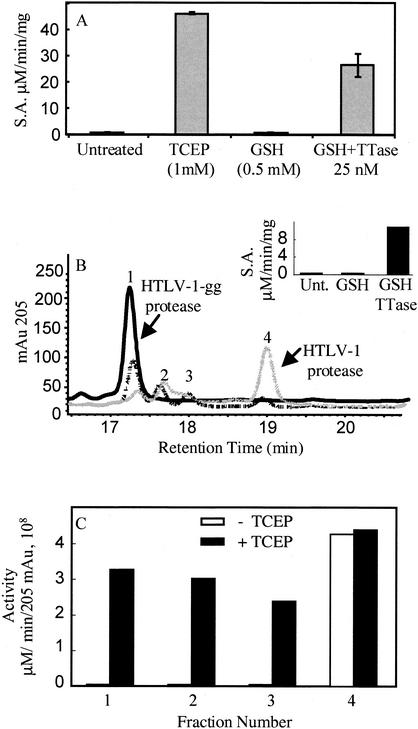
To determine if the regulation of protease activity through reversible oxidation extends beyond the lytic lentiviruses, we investigated the protease from the transforming deltaretrovirus HTLV-1. This protease contains two conserved cysteine residues at positions 90 and 109 that are not required for protease activity (8). A recently published molecular model of HTLV-1 protease (19) places cysteine 109 at the start of a C-terminal β strand of the predicted dimer interface region. To evaluate the effect of cysteine oxidation on HTLV-1 protease activity, we used a recombinant HTLV-1 protease containing the L40I mutation that confers resistance to autoproteolysis (a gift from John Louis) (8). We glutathionylated both cysteines of the HTLV-1 protease under the same conditions described previously for the KIIA protease. The fully glutathionylated protease was purified by RP-HPLC with a Vydac C18 column (2.1 by 50 mm). A 5 to 35% gradient of acetonitrile in 0.01% trifluoroacetic acid (TFA) was applied over 5 min, followed by a 35 to 65% gradient over 15 min. Mass spectrometry analysis performed as described previously (3) confirmed that the purified glutathionylated form of the L40I protease, which eluted almost 2 min earlier than the unmodified protease, was 610 atomic mass units (amu) larger than the unmodified protease (HTLV-1gg, 14,082.0 ± 0.8 amu). This increase in mass is consistent with the addition of two glutathione moieties. HTLV-1gg protease was analyzed for activity by using the peptide substrate APQVLPVMHP as described previously (8) and was found to be completely inactive (Fig. 2A). However, the activity of the HTLV-1gg protease was readily restored by treatment with TCEP (Fig. 2A). TCEP treatment also changed the RP-HPLC elution time back to that observed for the unmodified protease (data not shown). These results confirmed that HTLV-1 protease could be reversibly inactivated through oxidation of the cysteine residues.
FIG. 2.
Restoration of protease activity following treatment of glutathionylated HTLV-1 protease with TTase. (A) Activity of glutathionylated HTLV-1 protease. Shown are the means and standard deviations for results from three independent samples of HTLV-1gg run under the following conditions: untreated, treated with TCEP (1 mM), treated with glutathione (GSH; 0.5 mM), and treated with 0.5 mM glutathione in the presence of 25 nM TTase. S.A., specific activity. (B) Glutathionylated HTLV-1 protease (10 μl containing 1.25 μg of HTLV-1gg) was diluted 1:50 in 490 μl of refolding buffer (without NP-40 detergent) containing no treatment, 0.5 mM reduced glutathione, 0.5 mM reduced glutathione in the presence of 50 nM TTase, or 1 mM TCEP. Following a 15-min incubation, an aliquot of the sample was analyzed for protease activity. Solid guanidine was added to the remainder of the sample (495 μl) to bring it to 6 M guanidine, and then the sample was acidified with 25 μl of 10% TFA. RP-HPLC analysis was used to evaluate the extent of deglutathionylation of the protease. RP-HPLC chromatograms are shown overlaid for the glutathionylated HTLV-1 protease (HTLV-1gg) incubated in buffer for 15 min in the absence of any treatment (black solid line), in the presence of 0.5 mM reduced glutathione (dotted line), and in the presence of 0.5 mM reduced glutathione with 50 nM TTase (gray solid line). mAu 205, absorbance at 205 nm. (Inset) Protease activity for samples corresponding to those for which results of RP-HPLC analysis are shown in panel B. Unt., untreated; GSH, glutathionylated protease treated with 0.5 mM glutathione; GSH/TTase, glutathionylated protease treated with glutathione and 50 nM TTase. The activity of the fully reduced TCEP-treated sample was 12.1 μM/min/mg in phosphate-buffered saline containing 10% glycerol and 1 mM EDTA (data not shown). (C) Glutathionylated HTLV-1 protease was treated with 5 mM reduced glutathione, and fractions corresponding to peaks 1 to 4 in panel B were collected, dried down by lyophilization, resuspended in 30 μl of 50% acetonitrile-0.05% TFA, and assayed for protease activity (30 min) following a 1:25 dilution in refolding buffer with or without 1 mM TCEP. Protease specific activity was calculated based on the absorbance at 205 nm (205 mAu) corresponding to each fraction collected by RP-HPLC. The fractions corresponding to each peak were also analyzed by electrospray mass spectrometry as described in the text. Fraction 1, HTLV-1 protease glutathionylated at cysteine 90 and cysteine 109; fraction 2, HTLV-1 protease with internal disulfide bond; fraction 3, a mixture of HTLV-1 protease with glutathione bound to cysteine 90 or cysteine 109; fraction 4, wild-type HTLV-1 protease.
TTase restores HTLV-1gg protease activity.
TTase is a ubiquitous and specific glutathionyl transferase that was shown previously to reverse glutathionylation of HIV-1 protease and restore activity (3). Treating HTLV-1gg with 0.5 mM glutathione in refolding buffer alone (8) was not sufficient to restore protease activity (Fig. 2A). However, treatment of the HTLV-1gg protease with 0.5 mM glutathione in the presence of TTase (25 nM) for 30 min restored more than 50% of the protease activity (Fig. 2A). To determine if TTase treatment removed the glutathione from HTLV-1gg, we analyzed the protease by RP-HPLC following treatment with TTase in phosphate-buffered saline containing 10% glycerol and 1 mM EDTA. This buffer was required since the NP-40 detergent in the standard refolding buffer (8) coeluted with the HTLV-1 protease. Treatment of the HTLV-1gg protease with TTase (50 nM) resulted in almost complete restoration of protease activity relative to the activity of the TCEP-treated protease (Fig. 2B, inset) and, at the same time, converted a majority of the enzyme back to the wild-type form, as determined by RP-HPLC and mass spectrometry analysis (Fig. 2B). Glutathione treatment alone did not restore activity or convert significant amounts of the enzyme back to wild-type protease (Fig. 2B). The fact that TTase converted HTLV-1gg back to the wild-type form indicates that both of the glutathionylated cysteine residues were substrates for human TTase.
Two other closely eluting peak fractions were also generated following TTase treatment of HTLV-1gg (Fig. 2B, peaks 2 and 3). The four peak fractions were individually collected from RP-HPLC and assayed for protease activity in the presence and absence of a reducing agent. In the absence of a reducing agent, the fractions for peaks 1, 2, and 3 were inactive while that for peak 4, representing the unmodified protease, was active (Fig. 2C). However, in the presence of the reducing agent TCEP, HTLV-1 protease activity was detected in all fractions, indicating that fractions for all peaks contained intact HTLV-1 protease (Fig. 2C). The peak 1 fraction was already characterized as the diglutathionylated form of the HTLV-1 protease. To further characterize the fractions corresponding to peaks 2 and 3, the fractions were subjected to electrospray mass spectrometry on a Finnigan TSQ 700 by direct infusion. The peak 2 fraction had a mass of 13,474 amu, essentially the same as that calculated for the native HTLV-1 protein. However, the lack of activity for this peak fraction in the absence of a reducing agent (see Fig. 2C) indicates that it most likely represents the wild-type HTLV-1 protease containing an intramolecular disulfide bond. The difference in mass would be only 2 amu, a difference within the error of the spectrometer for this m/z range. To further characterize these fractions, the fractions corresponding to peaks 2 and 3 were trypsinized and the resultant peptides were identified by mass spectrometry. Samples (approximately 5 μg) were dried down and then resuspended in 4 μl of acetonitrile followed by the addition of 16 μl of 200 mM ammonium bicarbonate, pH 8.0. Then 0.5 μl of a 20-μg/ml concentration of trypsin from Promega (Madison, Wis.) was added, and the mixture was incubated at 37°C for 4 h. The tryptic digests of the purified fractions were diluted twofold into a solution consisting of 50% acetonitrile-0.2% formic acid. The digests were independently analyzed by an electrospray quadrupole time of flight mass spectrometer (QTOF Ultima Global; Micromass, Manchester, United Kingdom) with a Micromass nanospray source. These solutions were directly infused into the Z-spray electrospray ionization source at 1 μl/min with all parameters tuned to optimize the sensitivity and obtain mass resolution greater than 10,000. Fragmentation of precursor peptide ions of interest was performed by producing collisionally induced dissociation with argon as the collision gas by increasing the gas pressure by 5 × 10−6 torr. Data were analyzed by using the MassLynx software package, version 3.5, and Protein Prospector (Web version at http://prospector.ucsf.edu). The peptide mass corresponding to the doubly and triply charged ions for the disulfide-linked peptide was easily detected in the peak 2 fraction (expected, 1632.35 [M + 2H]; obtained, 1632.53 [M + 2H]), confirming the presence of HTLV-1 protease with an internal disulfide bond. Therefore, peak 2 represents HTLV-1 protease containing an internal disulfide bond that causes reversible inactivation.
The fraction corresponding to peak 3 in Fig. 2B consisted predominantly of monoglutathionylated HTLV-1 protease with a mass of 13,778 ± 0.8 Da (calculated 13,779). To analyze the structure of the protease in the peak 3 fraction, tryptic digests were analyzed. Peptides corresponding to both glutathionylated cys 90 and cys 109 were detected. This included the glutathionylated peptide containing residues 82 to 95 with m/z 1,248.04 (calculated 1,248.8) and the glutathionylated peptide containing residues 104 to 119 with m/z 2,081.02 (calculated 2,080.9). Therefore, the fraction corresponding to peak 3 represents a mixture of the monoglutathionylated forms of the HTLV-1 protease.
Comparisons of retroviral protease structures and sequence alignments provide evidence for the presence of a general redox regulatory mechanism.
Based on the finding that the protease from HTLV-1, a deltaretrovirus, could be regulated by reversible oxidation in a manner similar to that for the lentiviral proteases, we decided to explore the potential for this mechanism in other retroviral proteases. Inspection of the three-dimensional structures for the available retroviral proteases revealed the presence of a sulfur-containing amino acid at the dimer interface in five out of six structures (Fig. 3). Using the sequences from these structures, we aligned the sequences for proteases whose structures have not been solved in an effort to predict their interface region by using the ClustalW method within the MacVector sequence analysis software package (Genetics Computer Group, Madison, Wis.). We found that nearly all the retroviral proteases have one or more sulfur-containing amino acids present at or near the predicted dimer interface region (Fig. 4). The major exception is the gammaretrovirus family of proteases, where none of the sequences have this property, although there is a conserved cysteine within this group of proteases (Fig. 4).
FIG. 3.
Space-filling models for retroviral proteases indicating the presence of oxidizable amino acids at the dimer interface. RASMOL (13) was used to obtain the graphical depictions of the X-ray structures. The cysteine and methionine residues located within the dimer interface are shaded in black. Coordinates for the protein structures can be accessed at www.rcsb.pdb/index.html by using the following protein data bank identification numbers: HIV-1 protease, 1ODW; HIV-2 protease, 1JLD; simian immunodeficiency virus (SIV) protease, 1TCW; Rous sarcoma virus (RSV) protease, 2RSP; and feline immunodeficiency virus (FIV) protease, 2FIV. Coordinates for equine infectious anemia virus (EIAV) protease were from reference 6 and were kindly provided by Alex Wlodawer.
FIG.4.
ClustalW alignment of retroviral protease sequences from the different genera of retroviruses indicating the predicted dimer interface region. Full-length protease sequences obtained from the Entrez protein database were first aligned by using the ClustalW method within the MacVector sequence analysis software package (Genetics Computer Group). For clarity, only N- and/or C-terminal regions containing the predicted dimer interface regions of interest are shown. For the N terminus, the sequence up to the active site DTG/DSG is included. The active site aspartate in each protease sequence is indicated by an asterisk. For the C terminus, the sequence is shown starting from the highly conserved GR region up to the predicted interface region. For the gammaretroviruses, the conserved cysteine is included in the C-terminal region and is indicated by the arrow. The black bars indicate the sequences predicted to be involved in the dimer interface by comparison to the dimer interface amino acids known for HIV-1 protease. The cysteine and methionine residues located at or very near the dimer interface amino acids are indicated in bold. For HTLV-2 and bovine leukemia virus (BLV), an amino terminal region for the predicted dimer interface is not found by the multiple alignments with the full-length protease sequences. In the following list, the accession numbers are in parentheses following the virus type. Lentiviruses: HIV-1 (K02013); HIV-2 (M15390); SIV, simian immunodeficiency virus (AY033233); FIV, feline immunodeficiency virus (M25381); BIV, bovine immunodeficiency virus (M32690); CAEV, caprine arthritis encephalitis virus (M33677); VISNA, visna/maedi virus (M60609); EIAV, equine infectious anemia virus (M16575). Alpharetroviruses: ALV, avian leukosis virus (M37980); RSV Pr, RSV Prague C (J02342); RSV SRB, RSV Schmidt-Ruppin B (AF052428); RSV SRD, RSV Schmidt-Ruppin D (D10652). Betaretroviruses: M-PMV, Mason-Pfizer monkey virus (M12349); SRV-1, simian retrovirus type 1 (M11841); SRV-2, simian retrovirus type 2 (M16605); SMRV, squirrel monkey retrovirus (M23385); JSRV, jaagsiekte sheep retrovirus (M80216); MMTV, mouse mammary tumor virus (M15122). Deltaretroviruses: HTLV-1 (D13784); HTLV-2 (M10060); STLV-3, simian T-lymphotropic virus type 3 (Y07616); BLV (AF162285). Gammaretroviruses: Fr-MLV, friend murine leukemia virus (M93134); Rad-LV, radiation leukemia virus (K03363); SNV, spleen necrosis virus (M54933); Mo-MLV, Moloney murine leukemia virus (J02255); FeLV, feline leukemia virus (M18247); GALV, gibbon ape leukemia virus (M26927). Spumaviruses: CFV, chimpanzee foamy virus (U94514); SFV-3, simian foamy virus type 3 (M74895); SFV-1, simian foamy virus type 1 (X54482); FFV, feline foamy virus (Y08851); CFV/HU, chimpanzee/human foamy virus (Y08851); BFU, bovine foamy virus (U94514).
Mechanisms that can prevent the premature activation of retroviral proteases within the cytoplasm of infected cells could be beneficial to viral production, since the overexpression of protease activity can lead to cellular toxicity and/or loss of viral particle formation (7, 9). Studies have shown that a number of critical cellular proteins can be cleaved by viral proteases when the proteases are overexpressed in cells (1, 12, 14-16, 18, 21). The oxidation of sulfur-containing amino acids at the dimer interface that can prevent dimer formation and activity provides a mechanism by which protease activity may be regulated. We have shown previously that HIV-1-infected cells cultured in the presence of protease inhibitors produce immature viral particles containing inactivated forms of the protease that can be reactivated with a reducing agent (4). In addition, Parker and Hunter have demonstrated that immature Mason-Pfizer monkey virus capsids isolated from infected cells can undergo proteolytic processing following the addition of a reducing agent to the particles (11). This provides a mechanism by which Mason-Pfizer monkey virus can prevent processing from occurring in the cytoplasm until virus budding. Therefore, protease oxidation occurs in infected cells and may be one mechanism to delay polyprotein processing within infected cells (25). The results here provide evidence that the ability to regulate retroviral protease activity through reversible oxidation is evolutionarily conserved and may be an important biological mechanism.
Acknowledgments
We thank Rod Levine for helpful discussions throughout the course of this work.
A portion of this study was supported, in part, by an NIH Intramural AIDS Antiviral Targeted Program Grant.
REFERENCES
- 1.Adams, L. D., A. G. Tomasselli, P. Robbins, B. Moss, and R. L. Heinrikson. 1992. HIV-1 protease cleaves actin during acute infection of human T-lymphocytes. AIDS Res. Hum. Retrovir. 8:291-295. [DOI] [PubMed] [Google Scholar]
- 2.Davis, D. A., F. M. Newcomb, J. Moskovitz, P. T. Wingfield, S. J. Stahl, J. Kaufman, H. M. Fales, R. L. Levine, and R. Yarchoan. 2000. HIV-2 protease is inactivated after oxidation at the dimer interface and activity can be partly restored with methionine sulphoxide reductase. Biochem. J. 346:305-311. [PMC free article] [PubMed] [Google Scholar]
- 3.Davis, D. A., F. M. Newcomb, D. W. Starke, D. E. Ott, J. M. Mieyal, and R. Yarchoan. 1997. Thioltransferase (glutaredoxin) is detected within HIV-1 and can regulate the activity of glutathionylated HIV-1 protease in vitro. J. Biol. Chem. 272:25935-25940. [DOI] [PubMed] [Google Scholar]
- 4.Davis, D. A., K. Yusa, L. A. Gillim, F. M. Newcomb, H. Mitsuya, and R. Yarchoan. 1999. The conserved cysteines of the human immunodeficiency type 1 protease are involved in regulation of polyprotein processing and viral maturation of immature virions. J. Virol. 13:1156-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grant, S. K., I. C. Deckman, J. S. Culp, M. D. Minnich, and I. S. Brooks. 1992. Use of protein unfolding studies to determine the conformational and dimeric stabilities of HIV-1 and SIV proteases. Biochemistry 31:9491-9501. [DOI] [PubMed] [Google Scholar]
- 6.Gustchina, A., J. Kervinen, D. J. Powell, A. Zdanov, J. Kay, and A. Wlodawer. 1996. Structure of equine infectious anemia virus proteinase complexed with an inhibitor. Protein Sci. 5:1453-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katz, R. A., and A. M. Skalka. 1994. The retroviral enzymes. Annu. Rev. Biochem. 63:133-173. [DOI] [PubMed] [Google Scholar]
- 8.Louis, J. M., S. Oroszlan, and J. Tozser. 1999. Stabilization from autoproteolysis and kinetic characterization of the human T-cell leukemia virus type 1 proteinase. J. Biol. Chem. 274:6660-6666. [DOI] [PubMed] [Google Scholar]
- 9.Luukkonen, B. G., E. M. Fenyo, and S. Schwartz. 1995. Overexpression of human immunodeficiency virus type 1 protease increases intracellular cleavage of Gag and reduces virus infectivity. Virology 206:854-865. [DOI] [PubMed] [Google Scholar]
- 10.Mildner, A. M., D. J. Rothrock, J. W. Leone, C. A. Bannow, J. M. Lull, I. M. Reardon, J. L. Sarcich, W. J. Howe, C. S. Tomich, C. W. Smith, et al. 1994. The HIV-1 protease as enzyme and substrate: mutagenesis of autolysis sites and generation of a stable mutant with retained kinetic properties. Biochemistry 33:9405-9413. [DOI] [PubMed] [Google Scholar]
- 11.Parker, S. D., and E. Hunter. 2001. Activation of the Mason-Pfizer monkey virus protease within immature capsids in vitro. Proc. Natl. Acad. Sci. USA 98:14631-14636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riviere, Y., V. Blank, P. Kourilsky, and A. Israel. 1991. Processing of the precursor of NF-kappa B by the HIV-1 protease during acute infection. Nature 350:625-626. [DOI] [PubMed] [Google Scholar]
- 13.Sayle, R. A., and E. J. Milner-White. 1995. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 20:374.. [DOI] [PubMed] [Google Scholar]
- 14.Shoeman, R. L., C. Kesselmier, E. Mothes, B. Honer, and P. Traub. 1991. Non-viral cellular substrates for human immunodeficiency virus type 1 protease. FEBS Lett. 278:199-203. [DOI] [PubMed] [Google Scholar]
- 15.Snasel, J., R. Shoeman, M. Horejsi, O. Hruskova-Heidingsfeldova, J. Sedlacek, T. Ruml, and I. Pichova. 2000. Cleavage of vimentin by different retroviral proteases. Arch. Biochem. Biophys. 377:241-245. [DOI] [PubMed] [Google Scholar]
- 16.Strack, P. R., M. W. Frey, C. J. Rizzo, B. Cordova, H. J. George, R. Meade, S. P. Ho, J. Corman, R. Tritch, and B. D. Korant. 1996. Apoptosis mediated by HIV protease is preceded by cleavage of Bcl-2. Proc. Natl. Acad. Sci. USA 93:9571-9576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Todd, M. J., N. Semo, and E. Freire. 1998. The structural stability of the HIV-1 protease. J. Mol. Biol. 283:475-488. [DOI] [PubMed] [Google Scholar]
- 18.Tomasselli, A., J. O. Hui, L. Adams, J. Chosay, D. Lowery, B. Greenberg, A. Yem, M. R. Deibel, H. Zurcher-Neely, and R. L. Heinrikson. 1991. Actin, troponin C, Alzheimer amyloid precursor protein and pro-interleukin 1B as substrates of the protease from human immunodeficiency virus. J. Biol. Chem. 266:14548-14553. [PubMed] [Google Scholar]
- 19.Tozser, J., G. Zahuczky, P. Bagossi, J. M. Louis, T. D. Copeland, S. Oroszlan, R. W. Harrison, and I. T. Weber. 2000. Comparison of the substrate specificity of the human T-cell leukemia virus and human immunodeficiency virus proteinases. Eur. J. Biochem. 267:6287-6295. [DOI] [PubMed] [Google Scholar]
- 20.Vallette, F., E. Mege, A. Reiss, and M. Adesnik. 1989. Construction of mutant and chimeric genes using the polymerase chain reaction. Nucleic Acids Res. 17:723-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ventoso, I., R. Blanco, C. Perales, and L. Carrasco. 2001. HIV-1 protease cleaves eukaryotic initiation factor 4G and inhibits cap-dependent translation. Proc. Natl. Acad. Sci. USA 98:12966-12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang, Y. X., D. I. Freedberg, T. Yamazaki, P. T. Wingfield, S. J. Stahl, J. D. Kaufman, Y. Kiso, and D. A. Torchia. 1996. Solution NMR evidence that the HIV-1 protease catalytic aspartyl groups have different ionization states in the complex formed with the asymmetric drug KNI-272. Biochemistry 35:9945-9950. (Erratum, 36:280, 1997.) [DOI] [PubMed]
- 23.Woody, R. W. 1995. Circular dichroism. Methods Enzymol. 246:34-71. [DOI] [PubMed] [Google Scholar]
- 24.Xie, D., S. Gulnik, E. Gustchina, B. Yu, W. Shao, W. Qoronfleh, A. Nathan, and J. W. Erickson. 1999. Drug resistance mutations can effect dimer stability of HIV-1 protease at neutral pH. Protein Sci. 8:1702-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72:4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]