Abstract
The 66-kDa leader proteinase (L-Pro) of the Beet yellows virus (BYV) possesses a nonconserved N-terminal domain and a conserved, papain-like C-terminal domain. Previous work revealed that the N-terminal domain functions in RNA amplification, whereas the C-terminal domain is required for autoproteolysis. Alanine-scanning mutagenesis was applied to complete the functional analysis of L-Pro throughout the virus life cycle. This analysis indicated that the C-terminal domain of L-Pro, in addition to being required for proteolysis, also functions in RNA amplification and that these two functions are genetically separable. Examination of the role of L-Pro in BYV cell-to-cell movement revealed that none of the 20 examined replication-competent mutants was movement defective. In contrast, six of the L-Pro mutations affected the long-distance transport of BYV to various degrees, whereas three mutations completely abolished the transport. Because these mutations were located throughout the protein molecule, both domains of L-Pro function in virus transport. We conclude that in addition to previously identified functions of L-Pro, it also serves as the BYV long-distance transport factor.
The papain-like leader proteinases provide a particularly illuminating example of the multifunctional viral proteins. These proteinases were found in a number of evolutionarily diverse lineages of the animal, fungal, and plant positive-strand RNA viruses (8, 12, 13, 20, 35). In addition to the primary role in processing of the viral polyproteins, these proteinases were implicated in genome replication, synthesis of the subgenomic mRNAs, virus spread, and various aspects of virus-host interactions (6, 26, 33, 34). Among the plant viruses, the papain-like leader proteinases in the members of two viral families, Potyviridae and Closteroviridae, were characterized (15, 31). The potyviral helper component proteinase (HC-Pro) and the closteroviral leader proteinase (L-Pro) share a two-domain structure with the N-terminal, nonproteolytic domain and the C-terminal, papain-like domain. HC-Pro functions in self-processing, genome amplification, long-distance transport, and aphid transmission (4, 7, 19, 28). At least some of these activities are due to the ability of HC-Pro to suppress RNA silencing, a host defense response that targets viral RNA for sequence-specific degradation (17, 18).
L-Pro of the Beet yellows virus (BYV), a prototype Closterovirus (10), is encoded in a 5′-proximal part of BYV open reading frame 1a (Fig. 1). Previous work implicated L-Pro in self-processing and genome amplification and suggested that the functional profile of L-Pro overlaps that of the potyviral HC-Pro (1, 11, 26). A more recent study demonstrated that although L-Pro is not essential for basal-level RNA replication, deletion of the N-terminal domain resulted in a 1,000-fold reduction of BYV RNA accumulation (23). In addition, a short RNA element indispensable for genome replication was identified within the 5′-terminal region of the L-Pro open reading frame.
FIG. 1.
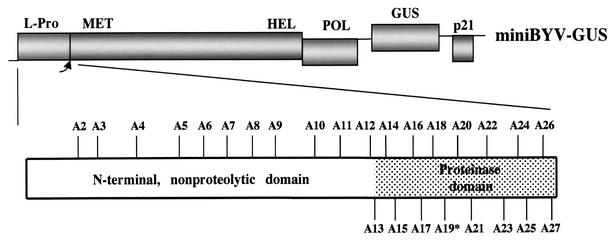
Mutations introduced into L-Pro of BYV. The diagram on the top shows a genetic map of the mini-BYV variant, with the boxes representing coding regions. MET, HEL, and POL, methyltransferase, RNA helicase, and RNA polymerase domains of the BYV replicase, respectively. The diagram at the bottom shows the approximate positions of the mutations introduced into the N-terminal (23) and C-terminal proteinase domains of L-Pro. The mutant A19, in which catalytic Cys was replaced with Ala, is marked by an asterisk.
Interestingly, some of the closteroviruses, such as Citrus tristeza virus (CTV), encode a tandem of leader proteinases, L1 and L2 (16). Comparative analysis of the closteroviral and potyviral proteinases by using a gene swapping approach indicated that BYV L-Pro and CTV L1 belong to the same functional class, whereas CTV L2 belongs to another (24). This analysis also demonstrated that potyviral HC-Pro cannot functionally replace BYV L-Pro, suggesting that these proteinases are mechanistically distinct. Moreover, it was found that the homologous, papain-like domains of L-Pro, L1, L2, and HC-Pro are functionally specialized. Although each of these domains efficiently processed chimeric polyprotein, only that of L1 was capable of partial rescue of the proteinase function in RNA amplification (24). A similar approach was used to compare leader proteinases from a broader range of plant, fungal, and animal viruses (25). Surprisingly, it was found that the replacement of BYV L-Pro with the leader proteinase of Equine arteritis virus resulted in a replication-competent BYV-Equine arteritis virus chimera. Further analysis of this chimera revealed defects in its invasiveness. These results suggested that the authentic BYV L-Pro is required for the ability of the virus to establish infection in the inoculated plant tissue (25).
Similar to that of other plant viruses, spread of BYV in plants is a two-phase process that involves relatively slow translocation from cell to cell and much faster long-distance transport via the vascular system (5, 21). The cell-to-cell movement of BYV requires functions of five virus genes (2). One of these genes codes for p6, a typical movement protein. The products of the remaining four genes assemble the filamentous virions that carry a tail-like movement device (2, 3, 27). Interestingly, the five movement-associated genes form a quintuple gene block, which is conserved among diverse members of the family Closteroviridae (2). This fact suggests that the cell-to-cell movement function is also conserved throughout this family. In those phloem-limited closteroviruses, which are incapable of movement between leaf mesophyll cells, this function could be involved in virus spread between the phloem cells. In addition to proteins that are required for both assembly and movement, the virion tails harbor p20, which is not essential for either of those processes. A recent study demonstrated that this nonconserved protein is specifically required for virus long-distance transport (29).
Here we employed reverse genetics to complete the functional analysis of the BYV L-Pro. We further explored the relationship of L-Pro to genome amplification and virus transport. We confirm that the papain-like domain of L-Pro plays a role in RNA amplification that is separable from its primary role in polyprotein processing. We also reveal a novel function of L-Pro in virus long-distance transport. Thus, BYV provides the first example of a plant virus whose spread through the vascular system requires at least two protein factors, L-Pro and p20. The unique and complex functional profile of the BYV L-Pro highlights evolutionary plasticity of the viral leader proteinases that provide a structural platform for diverse biological activities.
MATERIALS AND METHODS
Engineering of mutant BYV variants.
Generation of the mutations A1 to A12, which targeted the N-terminal, nonproteolytic L-Pro domain, was described previously (23). The same procedures were used to introduce 15 additional alanine-scanning mutations (A13 to A27) into the BYV genome region that encodes a C-terminal, proteolytic domain of L-Pro (Fig. 1). Most of these mutations resulted in replacement of the charged or polar amino acid residues with Ala (Table 1). The exceptions were mutations A19 and A20, in which catalytic Cys509 and a neighboring Cys517 were replaced with Ala. Each mutation was confirmed via nucleotide sequencing and introduced into plasmids pBYV-GUS-p21 (14) and pBYV-GFP (27), which correspond to BYV variants mini-BYV and BYV-GFP, respectively. In addition, 20 mutations (Table 2) were introduced into the p35S-BYV-GFP plasmid (29).
TABLE 1.
Mutation analysis of the proteinase domain of BYV L-Prolegend
| Classa | BYV variant | Mutation | Processing efficiencyb | GUS activityb |
|---|---|---|---|---|
| I | A14 | R456A | 100 ± 8 | 100 ± 13 |
| A15 | E470A | 67 ± 3 | 87 ± 10 | |
| A16 | Q481A | 95 ± 5 | 95 ± 12 | |
| A17 | D492A | 93 ± 4 | 125 ± 17 | |
| A21 | Q521A | 99 ± 8 | 102 ± 15 | |
| A23 | K543A | 95 ± 6 | 114 ± 13 | |
| A24 | H556A | 72 ± 5 | 95 ± 10 | |
| A25 | R559A | 81 ± 6 | 86 ± 10 | |
| A27 | S578A | 104 ± 5 | 92 ± 11 | |
| II | A18 | R503A | 92 ± 7 | 43 ± 3 |
| A20 | C517A | 54 ± 4 | 27 ± 6 | |
| A22 | D529A | 58 ± 4 | 2 ± 1 | |
| A26 | D571A | 59 ± 3 | 3 ± 1 | |
| III | A13 | D446A | 97 ± 7 | <0.001 |
| A19 | C509A | UDc | <0.001 |
The mutant variants are divided into classes according to the levels of GUS activity. These levels are ≥86% for class I, ≤2% for class II, and <0.001% for
class III.
Expressed as percentages of the levels found for the wild type. Means and standard deviations are shown.
UD, undetectable.
TABLE 2.
Systemic infectivity of BYV-GFP and its mutant variants
| Classa | BYV variant | No. of infected plants/no. of inoculated plants | Systemic infectivityb | Time of symptom appearance (wk) |
|---|---|---|---|---|
| I | BYV-GFP | 16/18 | 89 | 3 |
| A3 | 13/18 | 72 | 6 | |
| A4 | 11/18 | 61 | 3 | |
| A6 | 14/18 | 78 | 3 | |
| A8 | 13/18 | 72 | 3 | |
| A10 | 14/18 | 78 | 3 | |
| A11 | 16/18 | 89 | 3 | |
| A14 | 12/18 | 67 | 3 | |
| A16 | 14/18 | 78 | 3 | |
| A18 | 11/18 | 61 | 3 | |
| A23 | 11/18 | 61 | 3 | |
| A27 | 15/18 | 83 | 3 | |
| II | A2 | 1/18 | 6 | 3 |
| A9 | 8/18 | 44 | 3 | |
| A12 | 9/18 | 50 | 3 | |
| A15 | 4/18 | 22 | 5 | |
| A21 | 9/18 | 50 | 4 | |
| A25 | 7/18 | 39 | 4 | |
| III | A5 | 0/18 | 0 | NAc |
| A7 | 0/18 | 0 | NA | |
| A17 | 0/18 | 0 | NA |
The BYV-GFP variants are arbitrarily divided into classes according to efficiency of systemic infection. These efficiencies are ≥61% for class I, ≤50% for class II, and zero for class III.
Expressed as percentages of the infected versus inoculated plants.
NA, not applicable.
In vitro translation and protoplast transfection assays.
The pBYV-GUS-p21 variants were linearized by using XbaI (for in vitro analysis) or SmaI (for protoplast experiments) and transcribed with SP6 RNA polymerase. The resulting capped RNA transcripts were translated by using wheat germ extracts (Promega) and [35S]cysteine (Amersham/Pharmacia Biotech) or rabbit reticulocyte lysates (Promega) and a non-isotope-labeled amino acid mixture. Alternatively, the transcripts were transfected into protoplasts derived from a suspension culture of Nicotiana tabacum cells. The efficiency of the in vitro proteolysis and accumulation of β-glucuronidase (GUS) activity in protoplasts were determined from means of four independent experiments as described previously (25). It was demonstrated previously that the level of the GUS activity provided an accurate integral measure of BYV genome amplification, transcription, and translation (23, 25).
Plant inoculations.
The pBYV-GFP-based variants were manually inoculated into leaves of Claytonia perfoliata, and the number and diameter of the resulting fluorescent infection foci were determined at 8 days postinoculation (27). At least 20 leaves were inoculated in three independent experiments for each variant. The total number of infection foci counted varied from ∼200 to ∼300 for each of the tested BYV-GFP variants. To test the systemic infectivities of the selected 20 variants (Table 2), corresponding plasmids of the p35S-BYV-GFP series were mobilized into Agrobacterium tumefaciens strain C58 by electroporation. The resulting bacterial isolates were infiltrated into leaves of young Nicotiana benthamiana (six- to eight-leaf stage) plants. The upper, noninoculated leaves of these plants were monitored visually for symptom development, whereas epifluorescence microscopy (27) and a Spot camera (Diagnostic Instruments, Inc.) were used to document accumulation of the virus-expressed green fluorescent protein (GFP).
Immunoblot analyses.
Total plant extracts were prepared by grinding 1 g of tissue in liquid nitrogen and adding 5 ml of buffer containing 75 mM Tris-HCl (pH 6.8), 8 M urea, 4.5% sodium dodecyl sulfate, and 100 mM dithiothreitol. For fractionation experiments, the extracts were prepared in the same way except that the buffer contained 100 mM Tris-HCl (pH 8.0), 10 mM EDTA, and 5 mM dithiothreitol. The extracts were clarified by centrifugation at 1,000 × g for 10 min. The supernatant was centrifuged at 30,000 × g, yielding S30 supernatant and pellet, which was resuspended in the same buffer with the addition of 2% Triton X-100. The suspension was subjected to additional cycle of fractionation at 30,000 × g, yielding Triton X-100-treated S30 supernatant and P30 pellet. The extracts and other samples were analyzed by immunoblotting (3) with antiserum at 1:1,000 dilution. This antiserum was generated against a synthetic 23-residue, C-terminal oligopeptide of L-Pro (SLFHCDVASAFSSPFYSLPRFIG) by Genemed Synthesis, Inc. (San Francisco, Calif.).
RESULTS
Mutation analysis of the proteinase domain.
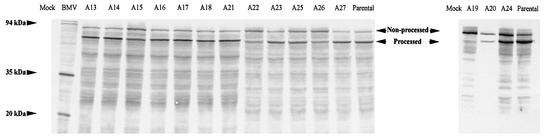
Our previous gene-swapping experiments suggested that in addition to the primary function in polyprotein processing, the proteinase domain of the L-Pro plays a role in RNA amplification (24, 25). To further examine this possibility, we employed alanine-scanning mutagenesis (Fig. 1). Fifteen mutants were generated and numbered A13 to A27, in continuation of the previously described 12 mutants that targeted the N-terminal domain (23). The processing activity of each mutant was assayed in a cell-free translation system. As seen from Fig. 2 and Table 1, all but one mutant were capable of processing polyprotein to various degrees. As expected, the A19 mutant lost the proteinase activity due to replacement of the catalytic Cys residue (1).
FIG. 2.
Autocatalytic processing of the wild-type and mutant BYV polyproteins upon translation in the wheat germ extracts. Mock, translation with no added mRNA (negative control); BMV, translation of Brome mosaic virus RNA (positive control); Parental, translation of RNA that encodes a wild-type L-Pro; A13 to A27, translation of RNAs that harbor corresponding mutant variants of L-Pro. The positions and molecular masses of translation products of BMV RNAs 2, 3, and 4 are shown at the left. The arrowheads between the two autoradiograms indicate positions of the nonprocessed polyprotein and processed L-Pro.
The RNA amplification levels of the mutants were measured in protoplast transfection experiments with a mini-BYV variant tagged by insertion of the reporter GUS gene (14). As demonstrated previously, GUS assays provide a sensitive and accurate measure of the accumulation of viral RNA (23-25). According to the levels of GUS accumulation, all mutants were arbitrarily separated into three classes (Table 1). Class I included nine mutants with GUS levels not significantly different from that of the parental variant (86 to 114%). The GUS levels found in the four mutants in class II were from 2 to 43% of the parental level. Finally, the two class III mutants failed to produce any detectable GUS activity.
Two major conclusions can be drawn from these analyses. First, these data reconfirm that the processing by L-Pro is a prerequisite for genome replication. Indeed, the point mutation A19, which eliminated the proteinase activity of L-Pro, was also replication incompetent. Second, several mutations had differential effects on polyprotein processing and RNA amplification as revealed by GUS accumulation. In particular, class II mutants A20, A22, and A26 had very similar processing rates, whereas their GUS activity levels varied by a factor of ∼10. Although the processing efficiency of the class II mutant A18 was very similar to that of the parental variant, the corresponding GUS activity level was reduced by a factor of ∼2.5 Finally, class III mutant A13 failed to replicate and produce any detectable GUS activity despite having a normal ability to process the polyprotein. This lack of correlation between processing efficiency and RNA amplification (as measured by GUS expression) provides evidence for genetically separable roles of the proteinase domain in each of these processes.
Cell-to-cell movement of the replication-competent L-Pro mutants.
As suggested by previous analyses of the chimeric BYV variants (24, 25), L-Pro could be indirectly involved in mediating virus movement from cell to cell. To further investigate this possibility, we studied 20 alanine-scanning mutants that were found to replicate efficiently in transfected protoplasts. These included mutants A2 to A12, described previously (23), as well as eight class I mutants and class II mutant A18 characterized in this study (Fig. 1 and Table 1). Each of the mutations was introduced into BYV-GFP, and the resulting variants were inoculated into leaves of C. perfoliata. The use of BYV-GFP and this host plant species was demonstrated to provide the most sensitive model system for quantification of BYV cell-to-cell movement (27).
Under these experimental conditions, parental BYV-GFP produced 14 ± 6 infection foci per leaf. The mean diameter of these foci was 4.9 ± 3.2 cells. The specific infectivity of each mutant (number of foci per leaf) was similar to that of parental BYV-GFP, varying from 9 ± 2 to 15 ± 6. Likewise, the mean size of the foci generated by the mutants was not significantly different from that of foci generated by BYV-GFP, varying from 4.1 ± 2.7 to 4.9 ± 2.2 cells. Since the characterized mutations were located throughout the L-Pro molecule, neither the N-terminal domain nor the proteinase domain of L-Pro played a significant role in BYV cell-to-cell movement. It cannot be excluded, however, that none of the tested point mutations resulted in major changes in the L-Pro structure required to affect its putative function in cell-to-cell movement.
L-Pro is required for long-distance transport of BYV.
Typically, the three principal phases in the life cycle of a plant virus are cell-autonomous replication, cell-to-cell movement, and long-distance transport. In order to complete the functional profiling of L-Pro, we examined its possible involvement in the long-distance transport of BYV. To this end, 20 alanine-scanning mutants that were competent in replication and intercellular spread in C. perfoliata were agroinoculated into the BYV systemic host N. benthamiana (29). We did not use C. perfoliata for these experiments because its systemic infection by BYV requires aphid transmission (32), whereas this plant species is not amenable to agroinoculation. Thus, the peculiarities of BYV biology make it impractical to use the same host species for examination of both cell-to-cell movement and long-distance transport. However, despite the difference in susceptibility, the rate of and genetic requirements for BYV cell-to-cell movement are similar in C. perfoliata and N. benthamiana (27). This fact validates utilization of the two-host model system for characterization of the BYV infection cycle.
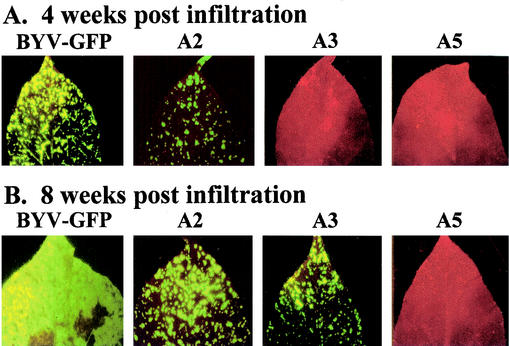
The competence of the virus in long-distance transport, which is synonymous with the systemic infectivity, was assessed by appearance of symptoms and GFP expression in the upper, noninoculated leaves. Approximately 90% of the plants inoculated with parental BYV-GFP exhibited symptoms of systemic infection and GFP expression in upper leaves by 3 to 4 weeks postinoculation (wpi) (Table 2). The patterns of systemic infection for 10 mutants were not significantly different from that of BYV-GFP. By 4 wpi, they systemically infected more than 60% of the inoculated plants (Table 2). Mutant A3 exhibited delayed systemic spread; it was detected in the upper leaves only at 6 wpi. Systemic transport of the six mutants in class II in Table 2 was affected to various degrees; they were able to systemically infect from only 6% (A2) to 50% (A12 and A21) of the inoculated plants. Strikingly, the remaining three mutants, A5, A7, and A17, failed to establish systemic infection in any of the inoculated plants, as determined by the lack of symptoms or GFP accumulation (Table 2). Selective immunoblot analysis of the symptomless plants with anti-BYV serum confirmed the absence of the virus (not shown).
These results are further illustrated in Fig. 3, which shows the spread of the green fluorescent areas in the upper leaves of plants inoculated with parental BYV-GFP and with a subset of L-Pro mutants. By 8 wpi, most of the plants inoculated with BYV-GFP were dead or severely necrotic. Those few live, systemically infected leaves, including one shown in Fig. 3B, were almost entirely colonized by GFP-producing virus. In contrast, the L-Pro mutants with delayed or reduced systemic transport exhibited only limited invasion into the leaf tissues and relatively mild visual symptoms (exemplified by mutants A2 and A3 in Fig. 3). As indicated by the lack of detectable GFP fluorescence throughout the observation period, mutants A5, A7, and A17 were unable to invade the noninoculated leaves (exemplified by mutant A5 in Fig. 3).
FIG. 3.
Systemic transport of the parental BYV-GFP and selected mutant variants. The noninoculated upper leaves of the N. benthamiana plants are shown. The leaves were photographed with an epifluorescence microscope at 4 (A) or 8 (B) weeks postinfiltration. The virus-infected areas are green due to GFP fluorescence, whereas the red color corresponds to noninfected, autofluorescent areas.
Taken together, these results demonstrate that L-Pro is required for systemic transport of BYV. Since transport-defective mutations were identified in both the N-terminal and proteinase domains, this additional function involves each of the principal domains of L-Pro. It should be emphasized that the transport function of L-Pro is genetically separable from its functions in processing and RNA amplification.
Distribution of L-Pro in virus-infected plants.
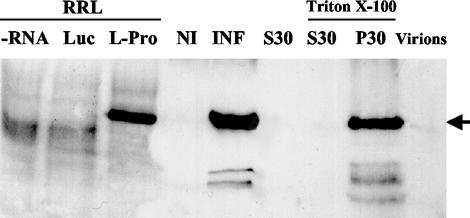
In a recent study we identified a 20-kDa BYV protein (p20) as a long-distance transport factor (LTF) and found that p20 is associated with the BYV virions (29). Because of that, we were interested in determining if L-Pro is also capable of binding virions. To this end, we generated polyclonal serum against the synthetic oligopeptide that corresponded to the 23 C-terminal residues of L-Pro and tested its specificity. As seen in Fig. 4 (lanes L-Pro and INF), this antibody readily detected the L-Pro produced in a cell-free system and in BYV-infected plants. The identical electrophoretic mobility of these forms of L-Pro indicates its correct processing and apparent lack of posttranslational modification.
FIG. 4.
Immunoblot analysis of BYV L-Pro with anti-L-Pro serum. RRL, non-isotope-labeled products of the translation in the rabbit reticulocyte lysates. Triton X-100, fractions that were treated with 2% Triton X-100. Lanes: −RNA, no exogeneous RNA added; Luc, translation of luciferase mRNA; L-Pro, translation of BYV L-Pro mRNA; NI and INF, total protein extracts from the noninfected and BYV-infected plants, respectively; S30 and P30, supernatant and pellet, respectively, after extract centrifugation at 30,000 × g; Virions, isolated BYV virions.
Interestingly, immunoblot analysis revealed that the amount of L-Pro present in the isolated BYV virions is beyond the limit of detection (Fig. 4, lane Virions). This result is compatible with at least three possibilities: (i) a very low number of L-Pro molecules in virions, (ii) transient or weak association between L-Pro and virions, or (iii) a lack of such an association. Although none of these possibility can be excluded with the existing data, the first one seems less likely because we have previously identified two minor virion components, Hsp70h (22) and p20 (29). On the other hand, fractionation of the protein extracts derived from BYV-infected plants showed that virtually all of the L-Pro is present in the 30,000 × g pellet. Because treatment of this pellet with Triton X-100 did not result in solubilization of L-Pro (Fig. 4, lane P30), it can be concluded that L-Pro is not associated with the cell membranes. This result is in agreement with the previous detection of the GFP-L-Pro fusion protein in cytoplasmic inclusion bodies (25).
DISCUSSION
The major objective of this study was to define the complete functional profile of the BYV L-Pro. Early work revealed that L-Pro possesses two principal domains, a nonconserved N-terminal domain and a conserved C-terminal domain that is related to papain-like proteinases (1). To assess the functional significance of these putative domains, we conducted an extensive genetic study using alanine-scanning mutagenesis and gene swapping. This study revealed that L-Pro provides an illuminating example of a multifunctional viral protein that enables progression of virus infection through its major phases.
Initial analyses showed that the C-terminal domain is both essential and sufficient for autocatalytic release of L-Pro from the viral polyprotein (23, 26). The subsequent gene-swapping experiments suggested that in addition to proteolysis per se, this domain is required for efficient RNA amplification (24, 25). The extensive mutational analysis presented here supported this suggestion and indicated that the roles played by C-terminal domain in proteolysis and genome amplification are genetically separable. A previous study of the N-terminal domain revealed that although it is not essential for basal-level replication, its deletion results in a dramatic reduction in RNA accumulation (23). It is yet to be determined if the functions of the two principal L-Pro domains in genome amplification are independent or interdependent. The dramatic phenotype of mutant A13, which is competent in proteolysis but deficient in RNA amplification, seems to support the latter possibility. Since this mutation is located near the junction of L-Pro domains, it could abolish L-Pro function by affecting the relative orientation of two domains.
Our finding of a novel function of L-Pro in BYV long-distance transport provides an example of activity that requires an entire L-Pro molecule. Indeed, the mutations that affect virus transport were found in each of the L-Pro domains. L-Pro, however, is not the only BYV protein that plays the role of an LTF. Very recently we demonstrated that p20 is also required for systemic spread of BYV (29). Although the defects in virus spread due to mutations in L-Pro and p20 are phenotypically similar, the underlying mechanisms could be different for the following reasons. First, p20 is tightly associated with the BYV virions (29), whereas L-Pro is not. Even though we cannot exclude transient or weak binding of L-Pro to virions, the nature of this interaction is certainly different from that of p20. Second, the regulation and functional profiles of the two BYV LTFs are very distinct. L-Pro is expressed via translation of the genomic RNA from the very onset of infection, whereas p20 is translated from a subgenomic mRNA only late in infection (14). Furthermore, p20 is not involved in the amplification of the BYV RNA (26), whereas L-Pro acts as a strong enhancer of this process (23).
Is it possible to reconcile the major roles played by L-Pro in BYV replication and transport? Since long-distance transport involves virus passage through and replication within the phloem (21), it seems plausible that L-Pro provides a specific activity required for efficient replication of BYV in this plant tissue. The ability of some mutants (e.g., A2 and A15 [Table 2]) to establish a productive infection in only a fraction of the inoculated plants suggests interference of L-Pro with the host defense response, the extent of which may vary between the individual plants. Because these same mutants did not exhibit defects in their ability to invade the leaf mesophyll tissue and to move from cell to cell, this putative defense response could be specific to phloem tissue.
Genetic requirements for long-distance transport vary between plant viruses. Some viruses, exemplified by Tobacco mosaic virus, require only a cell-to-cell movement protein and a capsid protein to spread systemically (9). Other plant viruses, such as potyviruses, code for an LTF, a protein with a specialized function in long-distance transport (7). BYV is unusual in that its long-distance transport requires the action of at least two LTFs, p20 and L-Pro. While the functional profile of L-Pro is unique, it partially overlaps that of the potyviral LTF HC-Pro. Both L-Pro and HC-Pro are leader proteinases that are required for efficient genome amplification. However, the amplification and long-distance transport activities of HC-Pro correlate with each other and are empowered by its ability to suppress RNA silencing (18). In contrast, we show here that the L-Pro functions in genome amplification and virus transport are genetically separable. These differences in the functional profiles point to mechanistic distinctions between HC-Pro and L-Pro that are underscored by the inability of HC-Pro to rescue L-Pro function (24). Furthermore, HC-Pro, but not L-Pro, is capable of suppressing RNA silencing that is induced by transient expression of the double-stranded RNA (30).
In conclusion, we demonstrated that the BYV L-Pro functions include polyprotein processing, genome amplification, virus invasion into plant tissues, and virus long-distance transport via the phloem. Thus, L-Pro provides activities that are essential for integration of the distinct phases of the viral life cycle from genome expression and amplification to virus dissemination through the infected host.
Acknowledgments
We thank Mary L. Powelson for critical reading of the manuscript.
This work was supported by grants from the U.S. Department of Agriculture (NRICGP 2001-35319-10875) and the National Institutes of Health (R1GM53190B) to V.V.D.
REFERENCES
- 1.Agranovsky, A. A., E. V. Koonin, V. P. Boyko, E. Maiss, R. Frotschl, N. A. Lunina, and J. G. Atabekov. 1994. Beet yellows closterovirus: complete genome structure and identification of a leader papain-like thiol protease. Virology 198:311-324. [DOI] [PubMed] [Google Scholar]
- 2.Alzhanova, D. V., Y. Hagiwara, V. V. Peremyslov, and V. V. Dolja. 2000. Genetic analysis of the cell-to-cell movement of beet yellows closterovirus. Virology 268:192-200. [DOI] [PubMed] [Google Scholar]
- 3.Alzhanova, D. V., A. Napuli, R. Creamer, and V. V. Dolja. 2001. Cell-to-cell movement and assembly of a plant closterovirus: roles for the capsid proteins and Hsp70 homolog. EMBO J. 20:6997-7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrington, J. C., and K. L. Herndon. 1992. Characterization of the potyviral HC-pro autoproteolytic cleavage site. Virology 187:308-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrington, J. C., K. D. Kasschau, S. K. Mahajan, and M. C. Schaad. 1996. Cell-to-cell and long-distance transport of viruses in plants. Plant Cell 8:1669-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chinsangaram, J., M. E. Piccone, and M. J. Grubman. 1999. Ability of foot-and-mouth disease virus to form plaques in cell culture is associated with suppression of alpha/beta interferon. J. Virol. 73:9891-9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cronin, S., J. Verchot, R. Haldeman-Cahill, M. C. Schaad, and J. C. Carrington. 1995. Long-distance movement factor: a transport function of the potyvirus helper component proteinase. Plant Cell 7:549-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawe, A. L., and D. L. Nuss. 2001. Hypoviruses and chestnut blight: exploiting viruses to understand and modulate fungal pathogenesis. Annu. Rev. Genet. 35:1-29. [DOI] [PubMed] [Google Scholar]
- 9.Dawson, W. O. 1992. Tobamovirus-plant interactions. Virology 186:359-367. [DOI] [PubMed] [Google Scholar]
- 10.Dolja, V. V. Beet yellows virus: the importance of being different. Mol. Plant Pathol., in press. [DOI] [PubMed]
- 11.Dolja, V. V., J. Hong, K. E. Keller, R. R. Martin, and V. V. Peremyslov. 1997. Suppression of potyvirus infection by coexpressed closterovirus protein. Virology 234:243-252. [DOI] [PubMed] [Google Scholar]
- 12.Dougherty, W. G., and B. L. Semler. 1993. Expression of virus-encoded proteinases: functional and structural similarities with cellular enzymes. Microbiol. Rev. 57:781-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorbalenya, A. E., E. V. Koonin, and M. M. Lai. 1991. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett. 288:201-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagiwara, Y., V. V. Peremyslov, and V. V. Dolja. 1999. Regulation of closterovirus gene expression examined by insertion of a self-processing reporter and by Northern hybridization. J. Virol. 73:7988-7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karasev, A. V. 2000. Genetic diversity and evolution of closteroviruses. Annu. Rev. Phytopathol. 38:293-324. [DOI] [PubMed] [Google Scholar]
- 16.Karasev, A. V., V. P. Boyko, S. Gowda, O. V. Nikolaeva, M. E. Hilf, E. V. Koonin, C. L. Niblett, K. Cline, D. J. Gumpf, R. F. Lee, S. M. Garnsey, D. J. Lewandowski, and W. O. Dawson. 1995. Complete sequence of the citrus tristeza virus RNA genome. Virology 208:511-520. [DOI] [PubMed] [Google Scholar]
- 17.Kasschau, K. D., and J. C. Carrington. 1998. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95:461-470. [DOI] [PubMed] [Google Scholar]
- 18.Kasschau, K. D., and J. C. Carrington. 2001. Long-distance movement and replication maintenance functions correlate with silencing suppression activity of potyviral HC-Pro. Virology 285:71-81. [DOI] [PubMed] [Google Scholar]
- 19.Kasschau, K. D., and J. C. Carrington. 1995. Requirement for HC-Pro processing during genome amplification of tobacco etch potyvirus. Virology 209:268-273. [DOI] [PubMed] [Google Scholar]
- 20.Koonin, E. V., and V. V. Dolja. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28:375-430. [DOI] [PubMed] [Google Scholar]
- 21.Lucas, W. J., and S. Wolf. 1999. Connections between virus movement, macromolecular signalling and assimilate allocation. Curr. Opin. Plant Biol. 2:192-197. [DOI] [PubMed] [Google Scholar]
- 22.Napuli, A. J., B. W. Falk, and V. V. Dolja. 2000. Interaction between HSP70 homolog and filamentous virions of the Beet yellows virus. Virology 274:232-239. [DOI] [PubMed] [Google Scholar]
- 23.Peng, C. W., and V. V. Dolja. 2000. Leader proteinase of the beet yellows closterovirus: mutation analysis of the function in genome amplification. J. Virol. 74:9766-9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng, C. W., V. V. Peremyslov, A. R. Mushegian, W. O. Dawson, and V. V. Dolja. 2001. Functional specialization and evolution of leader proteinases in the family Closteroviridae. J. Virol. 75:12153-12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng, C. W., V. V. Peremyslov, E. J. Snijder, and V. Dolja. 2002. A replication-competent chimera of the plant and animal viruses. Virology 294:75-84. [DOI] [PubMed] [Google Scholar]
- 26.Peremyslov, V. V., Y. Hagiwara, and V. V. Dolja. 1998. Genes required for replication of the 15.5-kilobase RNA genome of a plant closterovirus. J. Virol. 72:5870-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peremyslov, V. V., Y. Hagiwara, and V. V. Dolja. 1999. HSP70 homolog functions in cell-to-cell movement of a plant virus. Proc. Natl. Acad. Sci. USA 96:14771-14776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pirone, T. P., and S. Blanc. 1996. Helper-dependent vector transmission of plant viruses. Annu. Rev. Phytopathol. 34:227-247. [DOI] [PubMed] [Google Scholar]
- 29.Prokhnevsky, A. I., V. V. Peremyslov, A. J. Napuli, and V. V. Dolja. 2002. Interaction between long-distance transport factor and Hsp70-related movement protein of beet yellows virus. J. Virol. 76:11003-11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reed, J. C., K. D. Kasschau, A. I. Prokhnevsky, K. Gopinath, G. P. Pogue, J. C. Carrington, and V. V. Dolja. Suppressor of RNA silencing encoded by beet yellows virus. Virology, in press. [DOI] [PubMed]
- 31.Revers, F., O. Le Gall, T. Candresse, and A. J. Maule. 1999. New advances in understanding the molecular biology of plant/potyvirus interactions. Mol. Plant-Microbe Interact. 12:367-376. [Google Scholar]
- 32.Russel, G. E. 1963. Isolation of individual strains of beet yellows virus. Nature 197:623-624. [Google Scholar]
- 33.Suzuki, N., B. Chen, and D. L. Nuss. 1999. Mapping of a hypovirus p29 protease symptom determinant domain with sequence similarity to potyvirus HC-Pro protease. J. Virol. 73:9478-9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tijms, M. A., L. C. van Dinten, A. E. Gorbalenya, and E. J. Snijder. 2001. A zinc finger-containing papain-like protease couples subgenomic mRNA synthesis to genome translation in a positive-stranded RNA virus. Proc. Natl. Acad. Sci. USA 98:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853-879. [DOI] [PubMed] [Google Scholar]