Abstract
Hepatitis C virus (HCV) genotype 1 (subtypes 1a and 1b) is responsible for the majority of treatment-resistant liver disease worldwide. Thus far, efficient HCV RNA replication has been observed only for subgenomic and full-length RNAs derived from genotype 1b isolates. Here, we report the establishment of efficient RNA replication systems for genotype 1a strain H77. Replication of subgenomic and full-length H77 1a RNAs required the highly permissive Huh-7.5 hepatoma subline and adaptive amino acid substitutions in both NS3 and NS5A. Replication could be detected by RNA quantification, fluorescence-activated cell sorting, and metabolic labeling of HCV-specific proteins. Replication efficiencies were similar for subgenomic and full-length RNAs and were most efficient for HCV RNAs lacking heterologous RNA elements. Interestingly, both subtype 1a and 1b NS3 adaptive mutations are surface exposed and present on only one face of the NS3 structure. The cell culture-adapted subtype 1a replicons should be useful for basic replication studies and for antiviral development. These results are also encouraging for the development of adapted replicons for the remaining HCV genotypes.
Persistent infection with hepatitis C virus (HCV) is one of the primary causes of chronic liver disease. Progression to chronic active hepatitis with cirrhosis occurs in ∼20 to 30% of infected individuals, and HCV-associated liver disease is now the leading cause of liver transplantation in the United States (7). Genotypes 1a and 1b, the most prevalent worldwide, have the poorest rates of response to the present treatment regimen, a combination of pegylated alfa interferon 2b with ribavirin (4, 5, 18).
HCV, a member of the family Flaviviridae, is a small enveloped virus whose genome is a 9.6-kb single-stranded RNA with positive polarity consisting of a 5′ nontranslated region (NTR), a large open reading frame encoding the virus-specific proteins, and a 3′ NTR (reviewed in references 1, 15, and 21). The 5′ NTR contains an internal ribosome entry site (IRES) mediating translation of a single polyprotein of ∼3,000 amino acids with the structural proteins (C, E1, and E2) located in the N terminus and the nonstructural proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) encoded in the remainder. The NS3-5B coding region is sufficient for RNA replication in cell culture (17), and these proteins are presumed to function as components of the HCV replicase. The NS3 protein possesses serine protease and nucleoside triphosphatase-helicase activities, NS4A is a cofactor for the NS3 serine protease, and NS5B is the RNA-dependent RNA polymerase. The functions of NS4B and NS5A remain elusive, although NS5A, a phosphorylated protein, has been a target for adaptive mutation, allowing efficient initiation of HCV replication in vitro (2, 9, 13). Amino acid substitutions in the NS3, NS4B, and NS5B proteins can also enhance replication to various degrees (9, 13, 16).
Initially, only the genotype 1b Con1 RNA sequence was replication competent in the human hepatoma cell line Huh-7, and adaptive mutations in the HCV-encoded proteins were required for efficient HCV replication (2, 9, 13, 16). More recently, a second genotype 1b isolate, HCV-N, was reported to replicate in Huh-7 cells; however, unlike the Con1 strain, the HCV-N infectious clone replicated in the absence of additional cell culture-adaptive mutations (10). Attempts to extend this system to other genotypes have been largely unsuccessful (2, 9, 10, 14). Despite the ability of RNA transcripts from the genotype 1a H77 infectious clone of HCV to initiate a robust replication cycle in chimpanzees after direct intrahepatic inoculation (11), selectable subgenomic replicons based on this consensus clone failed to confer antibiotic resistance in a variety of hepatoma cells, including the Huh-7 cell line (2).
Recently, a Huh-7 subline, Huh-7.5, that was highly permissive for Con1 subgenomic and full-length RNA replication was isolated (3). This subline facilitated the selection of G418-resistant colonies supporting replication of H77-derived subgenomic replicons carrying a mutation in NS5A shown to enhance replication of Con1 replicons (3). Analysis of replicating HCV RNAs in these cell clones revealed additional adaptation in the NS3 coding region that increased the replicative capacities of subgenomic and full-length H77 RNAs to comparable levels.
MATERIALS AND METHODS
Cell cultures.
Huh-7.5 cell monolayers (3) were propagated in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 0.1 mM nonessential amino acids (DMEM- 10% FBS). For cells supporting subgenomic replicons, 750 μg of G418 (Geneticin; Gibco-BRL)/ml was added to the culture medium.
Plasmid construction.
Standard recombinant DNA technology was used to construct and purify all plasmids. Primed DNA synthesis was performed with KlenTaqLA DNA polymerase (W. Barnes, Washington University, St. Louis, Mo.), and regions amplified by PCR were confirmed by automated nucleotide sequencing. Plasmid DNAs for in vitro transcription were prepared from large-scale bacterial cultures and purified by centrifugation in CsCl gradients.
All nucleotides and amino acid numbers refer to the location within the H77 full-length HCV genome p90/HCVFLlongpU (11), commencing with the core coding region. The selectable replicon, pHCVrep13/Neo (H/SG-Neo), and the derivative, pHCVrep13(S2204I)/Neo [H/SG-Neo (I)] (Fig. 1), containing the NS5A mutation, S2204I, have been described (2). For these subgenomic constructs, NS5B polymerase-defective derivatives were generated carrying a triple-amino-acid substitution changing the Gly-Asp-Asp (GDD) motif in the active site to Ala-Ala-Gly (AAG) (12), and throughout this report they are referred to as pol−. All plasmids were engineered to carry an HpaI runoff site for RNA transcription, generating RNA transcripts containing one additional U nucleotide at the 3′ terminus. The Con1-derived constructs used in this study have been described previously (3).
FIG. 1.
Schematic representation of HCV RNAs used in this study. The 5′ and 3′ NTR structures are shown, and open reading frames are depicted as open boxes with the polyprotein cleavage products indicated. The first 21 amino acids of the core coding region (solid box), the neo gene (Neo; shaded box), and the EMCV IRES (EMCV; solid line) are illustrated. The nomenclature adopted for each construct is displayed on the right, and throughout this report, the HCV RNAs are prefaced by either H or Con1 to indicate H77- or Con1-derived sequences, respectively.
The plasmid pHCVrep90/A1226D+S2204I [H/SG-Neo (D+I)] (Fig. 1), containing the mutation A1226D in NS3, was constructed by ligating the DraIII-XhoI fragment of a reverse transcription (RT)-PCR product amplified from total RNA isolated from cell clone H-2 using the primers 2424 (5′ GCAGCCTGTTGACAGGCAATA 3′) and 2419 (5′ CATAATAATTTGTGACGAGTG 3′), together with the KpnI-DraIII and XhoI-NsiI fragments from pHCVrep13(S2204I)/Neo, into pHCVrep13(S2204I)/Neo cleaved with KpnI and NsiI. To engineer the mutation P1496L in pHCVrep13(S2204I)/Neo, thus creating pHCVrep90/P1496L+S2204I [H/SG-Neo (L+I)] (Fig. 1), nucleotides 4485 to 4861 in NS3 were PCR amplified from pHCVrep13(S2204I)/Neo using the mutant primer 2616 (5′ ACTCAGCCGGAGGGGCGCTCCCCCGGTGCCACAAATCTGTAGATGCCTAGCTTCCCCCTGCCAGTCCTGCC 3′) and oligonucleotide 2617 (5′ CTATCCCCCTCGAGGTGATCAAG 3′). The PCR product was digested with XhoI and BglI and, together with the BglI-BamHI and BamHI-EcoRI fragments from pHCVrep13(S2204I)/Neo, was inserted between the XhoI-EcoRI sites of pHCVrep13(S2204I)/Neo.
The replicons pNTR-EMCV/HCVrep90(A1226D+S2204I) and pNTR-EMCV/HCVrep90(P1496L+S2204I) [H/SG-5′HE (D+I) and H/SG-5′HE (L+I)] (Fig. 1) containing the mutations A1226D and P1496L, respectively, were constructed by inserting the KpnI-EcoRI fragment from either pHCVrep90/A1226D+S2204I or pHCVrep90/P1496L+S2204I between the KpnI and EcoRI sites of pNTR-EMCV/HCVrep13(S2204I) (K. J. Blight, unpublished data).
The plasmid p90/HCVFL(S2204I), containing the full-length H77 genome with S2204I in NS5A and an HpaI runoff site, was constructed from p90/HCVFLlongpU (K. J. Blight, unpublished data). The derivatives, p90/FL(A1226D+S2204I) and p90/FL(P1496L+S2204I) [H/FL (D+I) and H/FL (L+I)] (Fig. 1), containing the mutations A1226D and P1496L, were generated by replacing the AvrII-MluI portion of p90/HCVFL(S2204I) with the AvrII-NsiI fragment from either pHCVrep90/A1226D+S2204I or pHCVrep90/P1496L+S2204I and the NsiI-MluI fragment from pHCVrep13(S2204I)/Neo.
The selectable biscistronic full-length HCV clone p90/FL-Neo(P1496L+S2204I) [H/FL-Neo (L+I)] (Fig. 1) was assembled in a two-step cloning procedure. First, in a four-piece ligation reaction, the XbaI-BsaI fragment from pHCVrep13(S2204I)/Neo, the BsaI-AatII fragment from pHCVBMFL(S2204I)/Neo (3), and the AatII-XhoI fragment from p90/HCVFL(S2204I) were cloned between the XbaI and XhoI sites in pSL1180 (Pharmacia), generating the intermediate plasmid pSL1180/90NTR-C12-Neo-EMCV-CXho. Second, the XbaI-KpnI portion of p90/FL(P1496L+S2204I) was replaced with the XbaI-XhoI fragment excised from pSL1180/90NTR-C12-Neo-EMCV-CXho, together with a XhoI-KpnI-digested PCR product generated by extension of the overlapping oligonucleotides 2411 (5′ CCACCTCGAGGTAGACGTCAGCCTATCCCCAAGGCACGTCGGCCCGAGG 3′) and 2412 (5′ CCAGTGGTACCCGGGCTGAGCCCAGGTCCTGCCCTCGGGCCGACGTGC 3′).
To delete the E1-p7 coding region from p90/HCVFL(S2204I), the KpnI-MslI portion of a PCR product amplified from p90/HCVFL(S2204I) with the forward primer 2749 (5′ GCCCGGGTACCCTTGGCCCCTCT 3′) and the reverse primer 2748 (5′ CCAGAGCACCTCCGTGTCCAGGGCTGAAGCGGGCACGGTCAGGCA 3′) and the MslI-BsrGI fragment from p90/HCVFL(S2204I) were ligated between the KpnI and BsrGI sites of p90/HCVFL(S2204I). The P1496L mutation and an HpaI runoff site were engineered between the AvrII and MluI sites in this construct, named p90/FLΔE1-p7(S2204I), by inserting the AvrII-NsiI fragment from pHCVrep90/P1496L+S2204I, together with the NsiI-MluI fragment from pHCVrep13(S2204I)/Neo, thus creating p90/FLΔE1-p7(P1496L+S2204I) [H/ΔE1-p7 (L+I)] (Fig. 1).
RNA transcription and electroporation of cultured cells.
Plasmid DNAs containing H77 and Con1 sequences were linearized with HpaI and ScaI, respectively. In vitro transcription was performed as described previously (3). DNase-treated RNA transcripts (0.5 to 1 μg) were electroporated into 5 × 106 Huh-7.5 cells using a BTX ElectroSquarePorator essentially as described previously (3). The transfected cells were plated in (i) 150-mm-diameter dishes for selection of G418-resistant colonies, (ii) 100-mm-diameter dishes for determining the efficiency of G418-resistant colony formation and fluorescence-activated cell sorting (FACS) analysis, (iii) 35-mm-diameter wells for quantifying HCV RNA and for metabolic labeling experiments, or (iv) eight-well chamber slides (Becton Dickinson) for immunofluorescence studies. For dishes requiring G418 selection, 48 h after the cells were plated, the medium was replaced with DMEM- 10% FBS supplemented with 1 mg of G418/ml. Three weeks later, the resultant foci were either isolated and expanded for further analysis or fixed with 7% formaldehyde, stained with 1% crystal violet in 50% ethanol, and counted to determine G418 transduction efficiency (3).
RNA extraction, RT-PCR amplification, and quantification of HCV RNA.
Total cellular RNA was isolated from cell monolayers using TRIZOL reagent (Gibco-BRL) according to the manufacturer's protocol and precipitated with isopropanol. The RNA pellet was washed in 80% ethanol and resuspended in H2O. The NS3-5B coding region was amplified from total cellular RNA extracted from G418-resistant cell clones in four overlapping fragments by RT-PCR. Approximately 105 molecules of HCV RNA were mixed with 5 pmol of HCV-specific primer, and the primer was extended at 43.5°C for 1 h in a 5-μl reaction mixture containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, 0.5 mM (each) deoxynucleoside triphosphate, 4 U of RNasin (Promega), and 50 U of Superscript II reverse transcriptase (Gibco-BRL). The cDNAs were then amplified with KlenTaqLA DNA polymerase using 35 cycles of 95°C for 30 s, 55 to 60°C for 30 s, and 68°C for 4 min. The PCR products were recovered from preparative low-melting-point agarose gels by phenol extraction, and ∼40 ng of purified PCR product was directly sequenced with an ABI 9600 automatic DNA sequencer.
HCV-specific RNA levels in total cellular RNA preparations extracted from transfected Huh-7.5 cell monolayers were quantified using primers specific for the 5′ NTR in a real-time RT-PCR assay as previously described (3).
Protein detection.
Transfected cell monolayers were removed from the 100-mm-diameter dishes, and a single-cell suspension was prepared for FACS analysis as described previously (3). Briefly, cells were resuspended at 2 × 106 per ml and fixed for 20 min at room temperature (rt) in a final concentration of 2% paraformaldehyde in phosphate-buffered saline (PBS). The fixed cells were washed twice in PBS, resuspended at 2 × 106 per ml, permeabilized for 20 min at rt in 0.1% saponin- PBS, and stained for 1 h at rt with an HCV-specific monoclonal antibody to NS3 (10E5/24; 1/500 dilution) kindly provided by Raffaele De Francesco, Istituto di Ricerche di Biologia Molecolare, Rome, Italy (19). Bound monoclonal antibody was detected by incubation for 1 h at rt with anti-mouse immunoglobulin G (IgG) conjugated to Alexa 488 (Molecular Probes) diluted 1:1,000 in 3% FBS- 0.1% saponin- PBS. The stained cells were washed three times with 0.1% saponin- PBS and resuspended in FACSflow buffer (BD Biosciences) prior to analysis using a FACSCalibur (BD Biosciences).
For metabolic labeling experiments, cell monolayers in 35-mm-diameter wells were incubated for 10 to 12 h in methionine- and cysteine-deficient minimal essential medium containing 1/40 the normal concentration of methionine, 5% dialyzed FBS, and Express 35S protein-labeling mix (140 μCi/ml; NEN). Cell lysis and immunoprecipitation under denaturing conditions using HCV-positive patient serum, JHF (recognizing NS3, NS4B, and NS5A), and Pansorbin cells have been described previously (3).
RESULTS
Replication of H77-derived subgenomic replicons.
Con1 and HCV-N, both genotype 1b, are the only two isolates reported to replicate in Huh-7 cells. To test the abilities of Con1 adaptive mutations to enhance replication of genotype 1a-derived replicons, we incorporated the highly adaptive Ser-to-Ile substitution in NS5A (S2204I) into subgenomic replicons derived from a chimpanzee-infectious H77 genotype 1a cDNA clone (11). Although modeled after the Con1 selectable replicons, these genotype 1a-derived replicons failed to replicate in Huh-7 cells (2). The lack of replication might have been due to the Huh-7 cellular environment, requirements for different or additional adaptive mutations, or a combination of these factors. Recently, a Huh-7 subline (Huh-7.5) was established by “curing” a cell clone containing a Con1 subgenomic replicon by prolonged treatment with alpha interferon (3). A majority of these cells (>70%) are permissive for HCV RNA replication after transfection with Con1 replicons harboring certain adaptive mutations (e.g., S2204I) (3). The increased permissiveness of the Huh-7.5 cell line for Con1 replication led us to reexamine the replicative abilities of H77 subgenomic replicons in these cells.
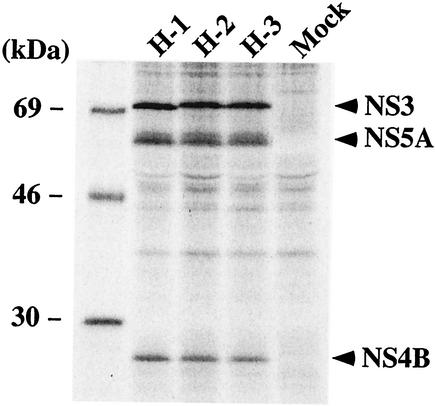
Two G418-selectable replicons, H/SG-Neo and H/SG-Neo (I) (Fig. 1), containing wild-type sequence and S2204I in NS5A, respectively, were evaluated for the ability to replicate compared to an NS5B polymerase-defective replicon, pol−. In vitro-synthesized RNA was electroporated into Huh-7.5 cells, and G418 selection was imposed 48 h later. After 3 weeks, a few G418-resistant colonies were visible for Huh-7.5 cells transfected with H/SG-Neo (I), but not with H/SG-Neo and H/SG-Neo (pol−). From two independent electroporations, 16 individual cell colonies were isolated, and HCV replication was verified by metabolically labeling cell monolayers and immunoprecipitation of HCV antigens. After the separation of labeled proteins by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), NS3, NS4B, and NS5A were detected in each cell clone, but not in the parental Huh-7.5 cells. Data for three of these clones are shown in Fig. 2. Interestingly, the migration of NS3 differed depending on the clone, suggesting that a modification(s) within NS3 may have occurred.
FIG. 2.
Detection of HCV proteins in cell clones supporting H77 subgenomic replication. Two days postseeding, monolayers of the cell clones H-1, H-2, and H-3 and parental Huh-7.5 cells (Mock) were incubated for 10 h in the presence of [35S]methionine and [35S]cysteine. The labeled cells were lysed and immunopreciptated with HCV-positive human serum (JHF, anti-NS3, NS4B, and NS5A), and the labeled proteins were separated by SDS- 10% PAGE. The mobilities of molecular-mass standards are indicated on the left, and the migrations of NS3, NS4B, and NS5A are shown on the right.
Identification of adaptive mutations.
The low frequency of cells supporting H77 replication suggested that adaptive mutations were generated, allowing replication at a level sufficient to confer resistance to G418. To determine if subgenomic RNAs within individual cell clones had acquired additional mutations, the NS3-5B coding region was amplified by RT-PCR from total cellular RNA isolated from three clones (H-1, H-2, and H-3 [Fig. 2]) and sequenced. The S2204I change in NS5A was maintained, but in addition, a single-amino-acid substitution in the helicase domain of NS3 was found in each clone. For one clone (H-1), a Pro-to-Leu substitution at locus 1496 (P1496L) was identified, while in the other two clones, H-2 and H-3, Ala at position 1226 was mutated to Asp (A1226D). For an additional nine cell clones, the NS3-4A coding region was amplified, and sequence analysis of these RT-PCR products identified the mutation P1496L in seven clones and A1226D in the remainder. In addition to P1496L, one replicon (H-6) encoded a second NS3 mutation (V1355I). Due to the conservative nature of this substitution, we did not investigate the effect of the mutation on HCV replication. Thus, of the 12 independent cell clones sequenced at the population level, only mutations localized within the helicase domain of NS3 were identified.
Adaptive values of mutations identified in NS3.
To analyze whether A1226D and P1496L in NS3 conferred an adaptive phenotype, these mutations were independently introduced into H/SG-Neo (I) (Fig. 1) to produce H/SG-Neo (D+I) and H/SG-Neo (L+I), respectively. Their replication efficiencies were compared in Huh-7.5 cells by measuring the number of cells transduced to G418 resistance. The frequency of Huh-7.5 cells able to support H/SG-Neo (L+I) replication was ∼30,000-fold higher than for H/SG-Neo (I), while H/SG-Neo (D+I) demonstrated an ∼800-fold increase in the number of G418-resistant colonies (Fig. 3A). However, the replicative capacities of H/SG-Neo (L+I) and H/SG-Neo (D+I) were consistently lower than that of Con1/SG-Neo (I) (∼7- and ∼34-fold, respectively) (Fig. 3A).
FIG. 3.
Colony-forming abilities of H77 subgenomic RNAs containing mutations in NS3. (A) Huh-7.5 cells were electroporated with 0.5 μg (each) of the subgenomic replicons H/SG-Neo (L+I), H/SG-Neo (D+I), Con1/SG-Neo (I), and H/SG-Neo (pol−). Forty-eight hours later, the cells were subjected to G418 selection, and the resulting colonies were fixed and stained with crystal violet. Representative dishes after 2.5 × 104 cells were plated are illustrated. The percentage below each dish refers to the calculated G418 transduction efficiency of the replicon that was determined by serially titrating transfected cells from 2 × 105 to 1 × 103 cells per 100-mm-diameter dish, together with feeder cells electroporated with the pol− replicon. The resulting G418-resistant foci were counted for at least three cell densities, and the relative G418 transduction efficiency was expressed as a percentage after dividing the number of colonies by the number of electroporated Huh-7.5 cells initially plated. (B) One microgram (each) of the subgenomic RNAs H/SG-Neo (L+I), H/SG-Neo (L), H/SG-Neo (D), and H/SG-Neo (pol−) was transfected into Huh-7.5 cells, and after 3 weeks of G418 selection, the transduction efficiency was determined as described for panel A. Dishes seeded with 2 × 105 electroporated cells are depicted, with the relative transduction efficiencies shown below.
Next, we addressed whether these NS3 mutations could confer a replicative advantage in the absence of the NS5A S2204I mutation. G418-resistant colonies were selected after transfection of Huh-7.5 cells with the selectable replicon H/SG-Neo (L) carrying only the P1496L substitution but not the replicon containing A1226D [H/SG-Neo (D)] (Fig. 3B). The calculated G418 transduction efficiency for H/SG-Neo (L) was ∼0.008%, a 250-fold reduction compared to that of H/SG-Neo (L+I) (Fig. 3B), demonstrating that P1496L, together with S2204I, is required for efficient H77 subgenomic RNA replication. The low frequency of colonies generated after transfection of H/SG-Neo (L) suggests that additional adaptive mutations may have arisen, which we are investigating further.
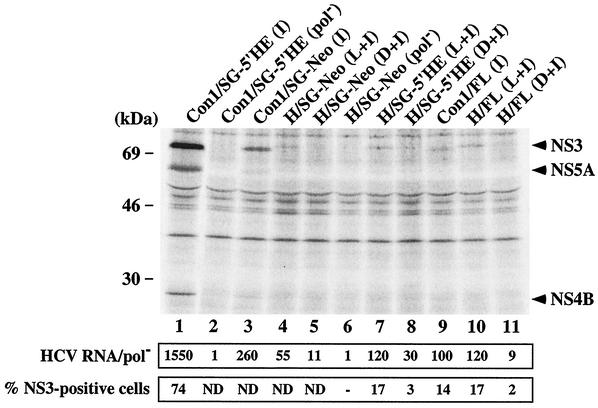
Previously, it was demonstrated that replication of Con1 subgenomic and genomic RNAs containing adaptive mutations could be monitored soon after RNA transfection of Huh-7.5 cells without the need for G418 selection (3). H77-derived subgenomic and full-length RNAs containing either P1496L or A1226D, together with S2204I, were transfected into Huh-7.5 cells; total RNA was extracted 96 h later, and HCV RNA levels were quantified by RT-PCR. The replication-defective replicon, H/SG-Neo (pol−), was transfected in parallel to allow discrimination between input RNA and that generated by productive replication. HCV RNA levels relative to the pol− control were higher for the subgenomic replicons (H/SG-Neo and H/SG-5′HE) containing P1496L and S2204I than for those harboring the A1226D and S2204I combination (Fig. 4; compare sample 4 to 5 and 7 to 8). The levels of H/FL (L+I) and H/FL (D+I) RNAs were ∼120- and ∼9-fold higher than that of pol− (Fig. 4, samples 10 and 11), consistent with the replicative ability of subgenomic RNAs carrying the same mutations. However, H77 subgenomic RNA levels were ∼5 to ∼13-fold lower than those observed for Con1-derived replicons [Con1/SG-Neo (I) and Con1/SG-5′HE (I)] (Fig. 4, compare sample 4 to 3 and 7 to 1), whereas H77 and Con1 full-length replication rates were comparable (Fig. 4, samples 9 and 10).
FIG. 4.
Detection of HCV proteins and RNA in Huh-7.5 cells transiently transfected with subgenomic and full-length HCV RNAs. (Top) Ninety-six hours after RNA transfection of Huh-7.5 cells, the monolayers were labeled with 35S protein-labeling mixture and lysed, and NS3, NS4B, and NS5A were analyzed by immunoprecipitation, SDS- 10% PAGE, and autoradiography. The positions of the molecular-mass standards are given on the left, and HCV-specific proteins are indicated on the right. (Middle) Total cellular RNA was extracted 96 h posttransfection, and HCV RNA levels were quantified as described in Materials and Methods. The ratio of HCV RNA to the pol− negative control is shown (HCV RNA/pol−). (Bottom) Ninety-six hours after transfection, cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% saponin, stained for HCV NS3, and analyzed by FACS. The percentages of cells expressing NS3 relative to an isotype-matched irrelevant IgG are displayed. Values of <1% were considered negative (−). ND, not determined.
We also examined HCV protein expression by immunoprecipitation and FACS analysis. The levels of 35S-labeled NS3 in Huh-7.5 cells transfected with H/SG-Neo (L+I) and H/SG-5′HE (L+I) were lower than those of the corresponding Con1 subgenomic replicons (Fig. 4, compare lane 4 to 3 and 7 to 1), whereas NS3 was undetectable in cells transfected with replicons carrying the A1226D change or pol− (Fig. 4). NS4B and NS5A were visible above background only in Con1/SG-5′HE (I) and Con1/SG-Neo (I) RNA-transfected cells (Fig. 4, lanes 1 and 3). The frequency of HCV antigen-positive cells quantified by FACS analysis yielded trends similar to those noted in the replicon RNA levels, where 17% of cells stained positive for NS3 after transfection of H/SG-5′HE (L+I) RNA compared to 3% for H/SG-5′HE (D+I) RNA (Fig. 4). As expected, a higher percentage of cells transfected with Con1/SG-5′HE (I) expressed NS3 (74 and 65%) (Fig. 4 and 5B). For full-length RNAs, a lower frequency of NS3-positive cells was evident in H/FL (D+I) than in H/FL (L+I) (2 versus 17%), and 35S-labeled NS3 was detectable only in Huh-7.5 cells transfected with H/FL (L+I) (Fig. 4, lanes 10 and 11). Similar frequencies of NS3 antigen-positive cells were observed after transfection of Huh-7.5 cells with full-length and subgenomic H77 replicons carrying the P1496L and S2204I mutations, and they were comparable to those seen with Con1-derived full-length RNA [Con1/FL (I); ∼14%] (Fig. 4 and 5B). These results are consistent with our earlier observation that subgenomic and full-length H77 RNAs containing P1496L and S2204I have the highest replicative capacities in Huh-7.5 cells. Furthermore, we found that subgenomic and full-length RNAs replicate to similar levels, in contrast to Con1, where subgenomic replicons establish replication in a greater proportion of cells than constructs containing the complete coding sequence (Fig. 4) (3).
FIG. 5.
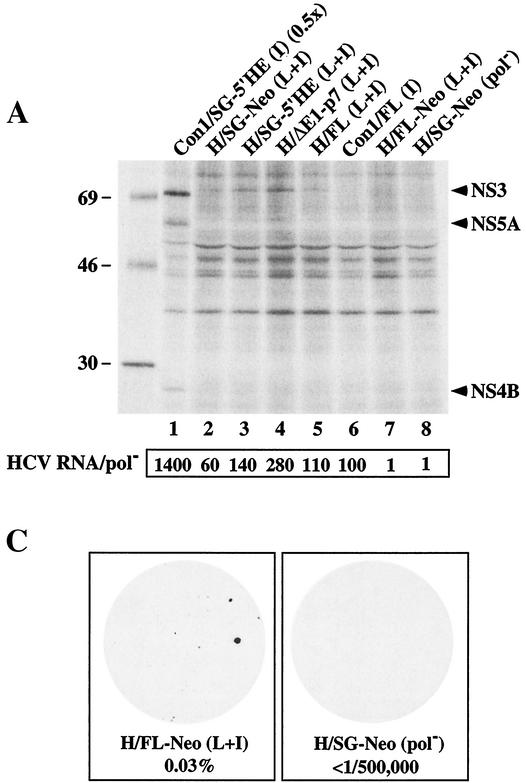
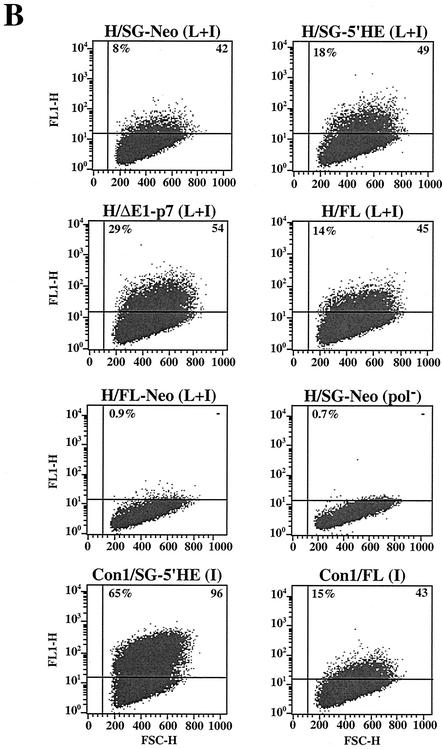
Replication of HCV RNAs with and without heterologous elements. (A) Huh-7.5 cells were transfected with 1 μg (each) of full-length and subgenomic RNAs, and 2 × 105 cells were plated in 35-mm-diameter wells. Ninety-six hours posttransfection, the Huh-7.5 cells were labeled with [35S]methionine and [35S]cysteine for 10 h. The cells were lysed, and HCV proteins were isolated by immunoprecipitation using a patient serum specific for NS3, NS4B, and NS5A. HCV proteins and the positions of protein molecular-mass standards (in kilodaltons) are shown. In lane 1, half the amount of immunoprecipitated sample was loaded (0.5x). The ratio of HCV RNA relative to the pol− negative control (HCV RNA/pol−) is shown below each lane. (B) Transfected Huh-7.5 cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% saponin, and the frequency of cells expressing NS3 antigen was quantified by FACS. The percentage of cells expressing NS3 relative to pol−-transfected cells and an isotype-matched IgG was determined and is shown in the upper left corner of each plot. The median fluorescence intensity of the gated positive cells is shown in the upper right corner of each plot. FSC-H, forward scatter; FL1-H, fluorescence. (C) One microgram (each) of H/FL-Neo (L+I) and H/SG-Neo (pol−) RNA was electroporated into Huh-7.5 cells, and 2 × 106 cells were plated on 100-mm-diameter dishes. G418 selection was applied 48 h posttransfection, and after 3 weeks in culture, G418-resistant foci were fixed and stained with crystal violet. The G418 transduction efficiency, displayed below each dish, was determined as described in the legend to Fig. 3.
Replicative abilities of subgenomic and full-length RNAs.
Both H77- and Con1-derived subgenomic replicons lacking the neo gene appear to initiate replication more efficiently than selectable subgenomic RNAs (Fig. 4) (3). To investigate these observations further, we compared the replication efficiencies of a number of subgenomic and genomic H77 RNAs in the presence or absence of heterologous elements. Besides the selectable bicistronic replicons (SG-Neo [Fig. 1]) and the replicons in which the HCV 5′ NTR was fused to the encephalomyocarditis virus (EMCV) IRES (SG-5′HE [Fig. 1]), a replicon was constructed in which the 5′ NTR was followed by the entire core sequence fused directly to the NS2-NS5B coding region and the 3′ NTR such that cleavage at the core-NS2 junction would be mediated by signal peptidase and translation was under the control of the homologous IRES [H/ΔE1-p7 (L+I)] (Fig. 1). In parallel, we tested the H77 full-length monocistronic RNA [H/FL (L+I)] (Fig. 1) and a bicistronic derivative [H/FL-Neo (L+I)] (Fig. 1), where the HCV 5′ NTR mediates neo gene translation and the EMCV IRES drives core-NS5B expression. Both subgenomic and genomic constructs were engineered to carry P1496L and S2204I. Ninety-six hours after the transfection of Huh-7.5 cells, the relative levels of HCV RNA and protein were measured as described above. A 280-fold increase in H/ΔE1-p7 (L+I) RNA over pol− was observed (Fig. 5A), whereas modest increases in HCV RNA were evident for H/SG-Neo (L+I) and H/SG-5′HE (L+I) (∼60- and ∼140-fold) (Fig. 5A). A higher frequency of Huh-7.5 cells expressed NS3 antigen after electroporation with H/ΔE1-p7 (L+I) (29%) than after electroporation with H/SG-5′HE (L+I) (18%) and H/SG-Neo (L+I) (8%) (Fig. 5B). NS3 antigen levels in the H77 RNA-transfected cells, as determined by the median fluorescence intensity of the gated antigen-positive cells, were similar, suggesting comparable levels of RNA translation and/or protein stability per cell (Fig. 5B). The relative amounts of immunoprecipitated 35S-labeled NS3 paralleled both the frequency of NS3-positive cells and the relative HCV RNA levels (Fig. 5A). After transfection of H/FL (L+I) RNA into Huh-7.5 cells, HCV RNA levels increased 110-fold relative to pol− (Fig. 5A), 14% of cells expressed NS3 (Fig. 5A), and 35S-labeled NS3 was visible (Fig. 5A, lane 5). In contrast, HCV RNA levels for H/FL-Neo (L+I) were no greater than those of the pol− control (Fig. 5A), and NS3 expression was not detectable by FACS (Fig. 5B) or metabolic labeling (Fig. 5A, lane 7), suggesting that this construct was replication defective. In spite of these results, G418-selected colonies were detectable with a relative transduction efficiency of 0.03% (Fig. 5C). Taken together, these findings suggest that H77 RNA replication is more efficient for subgenomic and genomic constructs that lack the neo gene and the EMCV IRES.
DISCUSSION
HCV replicons derived from the genotype 1b isolates Con1 and HCV-N are replication competent in Huh-7 cells (2, 3, 9, 10, 13, 16, 17). Earlier efforts to select stable colonies after transfection of Huh-7 cells with H77-derived subgenomic RNAs were unsuccessful, despite the inclusion of the highly adaptive S2204I change in NS5A that was identified in the Con1 genetic background (2). We reasoned that additional or alternative adaptive mutations might be required for replication in these cells or that Huh-7 cells were not permissive for H77 replication. In this study, we demonstrated that replication of subgenomic and full-length RNAs derived from the H77 infectious clone was dependent upon using the Huh-7 subline Huh-7.5. For reasons that are not yet clear, these cells possess a cellular environment highly permissive for the initiation of Con1 replication (3). The ability of H77-derived replicons to replicate in Huh-7.5, but not in the Huh-7 parental cell line, further emphasizes the importance of cellular factors for HCV replication.
Although a comprehensive study remains to be done, our results suggest that efficient H77 replication in Huh-7.5 cells may require at least two adaptive mutations. Initially, G418-resistant colonies were seen only with transfected subgenomic H77 RNAs containing the S2204I adaptive change in NS5A. In all cell clones analyzed, replicating RNAs had acquired a second amino acid substitution in the helicase domain of NS3 (A1226D or P1496L). Both these mutations, when combined with NS5A S2204I, enhanced the colony-forming ability of subgenomic H77 RNA and allowed the detection of HCV RNA and proteins 96 h after RNA transfection of either subgenomic or genomic replicons. Interestingly, replication was greatest for H77 RNAs carrying P1496L in NS3 and S2204I in NS5A. Although S2204I in NS5A increased the replicative capacity of H77-derived replicons, we were able to select G418-resistant colonies following transfection of subgenomic RNAs carrying only P1496L in NS3. Although the P1496L substitution may be sufficient for replication, the low frequency of G418-resistant colony formation with these transcripts suggests that an additional adaptation(s) may be required. Experiments are in progress to explore this possibility, with the hope of finding more optimal combinations of adaptive mutations for subtype 1a replication. Interestingly, NS3 P1496 is poorly conserved among HCV isolates, with Met and Gly found at this position for genotype 1b isolates Con1 and HCV-N, respectively. Given the apparent flexibility of this residue, it will be interesting to test the effects of other substitutions at this position.
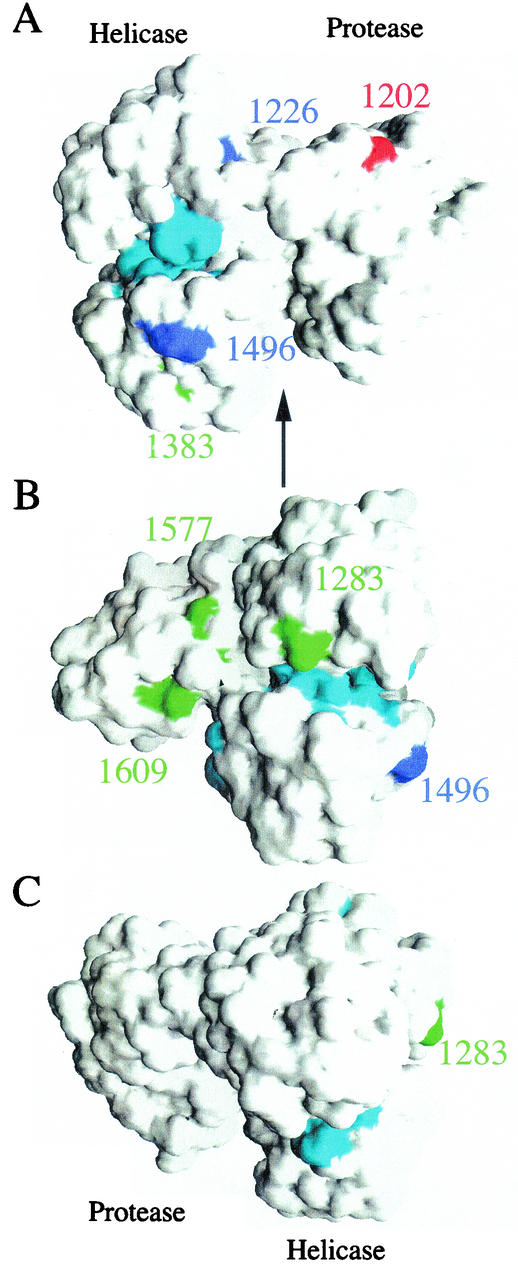
Adaptive mutations in NS3 have been reported in two other studies (13, 16). Lohmann et al. and Kreiger et al. identified and analyzed several spontaneous NS3 mutations individually and together for effects on replication. The mutations R1283G, E1383A, and K1609E had 7-, 2-, and 3.5-fold increases in RNA replication, respectively, compared to the wild-type Con1 subgenomic replicon, while K1577R had a modest decrease of ∼2-fold. However a replicon that harbored all four mutations displayed a 10-fold increase in RNA levels over the wild-type sequence. Kreiger and coworkers isolated two other mutations, E1202G and T1280I, which displayed 14- and 7-fold increases, respectively, compared to the parental wild-type sequence. A replicon containing both mutations (E1202G and T1280I) displayed an additive effect, with a 25-fold increase in RNA levels. Interestingly, the three studies (including this one) failed to find a mutation at the same position within the protein; however, seven of the eight mutations are located within the helicase domain of NS3. These NS3 mutations generally act synergistically with mutations in NS4B or NS5A to enhance replication. In an attempt to determine if these adaptive mutations cluster within the three-dimensional structure, the location of each position was mapped onto the structure of NS3-4A (23) (Fig. 6). As noted earlier (13), mutant residues were located on the solvent-accessible surface, with the exception of 1280, which is buried. Interestingly, residue 1280 makes several van der Waals contacts with the solvent-exposed residue 1283, also identified as a position for an adaptive mutation. The seven helicase mutations appear to be confined to one surface of the helicase domain (compare Fig. 6A and B with C). Although these mutations are not located within the seven conserved sequence motifs of RNA helicases (Fig. 6), they do appear to reside on adjacent surfaces of the active site.
FIG. 6.
Locations of NS3 adaptive mutations. Solvent-accessible surface of the NS3/4A crystal structure (23) highlighting the locations of several adaptive mutations. Adaptive mutations described in this paper are colored blue, while published mutations from references 13 and 16 are colored red and green, respectively. The seven conservedmotifs of the RNA helicase are colored cyan. The numbering corresponds to the genotype 1 sequences. (A) The NS4A peptide and protease domain are on the right, with the helicase domain on the left. (B and C) Rotations (90 and 180°, respectively) about a vertical axis (represented by the arrow in panel A).
The mechanism by which the changes in NS3 and NS5A act individually and often synergistically to enhance HCV replication is unclear. Since the function of NS5A in RNA replication is unknown, possibilities range from direct alterations in replicase activity (via interaction with other HCV nonstructural proteins or cellular factors) to modulation of the cellular environment, including host antiviral defense pathways (22). For NS3, although the role of the HCV RNA helicase activity in replication is not known, adaptive mutations could affect RNA binding, enzymatic activity of the helicase, or higher-order interactions in the replicase that enhance the efficiency of productive replication. In future studies, it will be interesting to examine the effects of different adaptive mutations on the relative syntheses of positive- and negative-strand RNAs. However, to understand the mechanism(s) of cell culture adaptation, it is clear that more work is needed to define the components and the higher-order structure of an active HCV replicase.
For H77-derived subgenomic replicons, the inclusion of the neo gene or the EMCV IRES generally decreased the ability of RNAs to initiate replication. For instance, transient analyses demonstrated the hierarchy of replication efficiencies H/ΔE1-p7 (L+I) > H/SG-5′HE (L+I) > H/SG-Neo (L+I), demonstrating that the EMCV IRES is not a requirement for efficient expression of the HCV replicase region and replication in Huh-7 cells. A similar trend has also been seen for Con1-derived replicons, where Con1/SG-5′HE (I) initiates replication more efficiently than Con1/SG-Neo (I) (this report and reference 3). In the case of replicons containing the full-length polyprotein, addition of the neo-EMCV IRES module also decreases replication efficiency, although G418-resistant colonies can be selected for both H77 and Con1 bicistronic derivatives (this work and references 3 and 20). Hence, both subgenomic and full-length HCV RNA replication can now be studied in the absence of heterologous sequences. Furthermore, inclusion of these or other adaptive mutations, as well as minimizing or eliminating non-HCV sequences, may help to enhance replication of other, as yet nonreplicating HCV genotypes or strains.
Full-length Con1 RNA replicated in ∼15% of Huh-7.5 cells compared to ∼65% for the subgenome [Con1/SG-5′HE (I)], whereas the replication efficiencies of H77 subgenomic [H77/SG-5′HE (L+I)] and genomic HCV RNAs were comparable (15 to 18%) (Fig. 5B). Moreover, the levels of NS3 per cell (mean fluorescence intensity [Fig. 5B]) were similar for all RNAs except Con1/SG-5′HE (I), where it was twice that of the other RNAs. This surprising difference in replicative capacity between H77 and Con1 subgenomic and full-length RNAs underscores the importance of studying more than one isolate.
Thus far, there is no evidence for HCV particle assembly and release from Huh-7 cells supporting replication of Con1 (3, 20) or HCV-N (10) full-length RNAs. Although Huh-7 cells may be nonpermissive for one or more of these steps, it is not known whether this will be generally true for all HCV genotypes. The well characterized H77 strain is highly infectious in chimpanzees and replicates to high titers, suggesting that it may be a good candidate for establishing a complete replication cycle in cell culture. Since the E1E2 glycoproteins are retained in the endoplasmic reticulum (ER) via retention signals in their C-terminal hydrophobic tails (6, 8), assembly (presumably in the ER or an ER-Golgi intermediate compartment) and release of HCV particles should be associated with movement of viral particles and their surface glycoproteins through the secretory pathway. This should result in transit through the Golgi and the acquisition of complex N-linked glycans. Although endoglycosidase-H sensitivity of the HCV H77 glycoproteins has not been examined, the E2 staining pattern is consistent with ER retention in cells transfected with full-length cell culture-adaptive RNAs (data not shown). Whether a low level of particle release from these cells is occurring is under study.
In conclusion, we have established a cell culture system for replication of HCV RNAs derived from the infectious H77 isolate. These genotype 1a subgenomic and full-length replicons should be useful for basic studies of HCV RNA replication and HCV-host cell interactions and for antiviral drug screening and evaluation.
Acknowledgments
We thank Arash Grakoui for helpful discussion and Brett Lindenbach for critical reading of the manuscript. We are also grateful to Raffaele De Francesco and Jean Dubuisson for providing HCV-specific monoclonal antibodies.
This work was supported in part by grants from the Public Health Service (CA57973 and AI40034) and the Greenberg Medical Research Institute.
REFERENCES
- 1.Blight, K. J., A. Grakoui, H. L. Hanson, and C. M. Rice. 2002. The molecular biology of hepatitis C virus, p. 81-108. In J.-H. J. Ou (ed.), Hepatitis viruses. Kluwer Academic Publishers, Boston, Mass.
- 2.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 3.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for hepatitis C virus genomic and subgenomic RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornberg, M., H. Wedemeyer, and M. P. Manns. 2001. Hepatitis C: therapeutic perspectives. Forum 11:154-162. [PubMed] [Google Scholar]
- 5.Cornberg, M., H. Wedemeyer, and M. P. Manns. 2002. Treatment of chronic hepatitis C with PEGylated interferon and ribavirin. Curr. Gastroenterol. Rep. 4:23-30. [DOI] [PubMed] [Google Scholar]
- 6.Dubuisson, J. 1999. Folding, assembly, and subcellular localization of the hepatitis C virus glycoproteins. Curr. Top. Microbiol. Immunol. 242:135-148. [DOI] [PubMed] [Google Scholar]
- 7.Fishman, J. A., R. H. Rubin, M. J. Koziel, and B. J. Periera. 1996. Hepatitis C virus and organ transplantation. Transplantation 62:147-154. [DOI] [PubMed] [Google Scholar]
- 8.Flint, M., E. R. Quinn, and S. Levy. 2001. In search of hepatitis C virus receptor(s). Clin. Liver Dis. 5:873-893. [DOI] [PubMed] [Google Scholar]
- 9.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikeda, M., M. Yi, K. Li, and S. M. Lemon. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 76:2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolykhalov, A. A., E. V. Agapov, K. J. Blight, K. Mihalik, S. M. Feinstone, and C. M. Rice. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570-574. [DOI] [PubMed] [Google Scholar]
- 12.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lanford, R. E., and C. Bigger. 2002. Advances in model systems for hepatitis C virus research. Virology 293:1-9. [DOI] [PubMed] [Google Scholar]
- 15.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott-Raven Publishers, Philadelphia, Pa. [Google Scholar]
- 16.Lohmann, V., F. Korner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lohmann, V., F. Korner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 18.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 19.Mottola, G., G. Cardinali, A. Ceccacci, C. Trozzi, L. Bartholomew, M. R. Torrisi, E. Pedrazzini, S. Bonatti, and G. Migliaccio. 2002. Hepatitis C virus nonstructural proteins are localized in a modified endoplasmic reticulum of cells expressing viral subgenomic replicons. Virology 293:31-43. [DOI] [PubMed] [Google Scholar]
- 20.Pietschmann, T., V. Lohmann, A. Kaul, N. Krieger, G. Rinck, G. Rutter, D. Strand, and R. Bartenschlager. 2002. Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J. Virol. 76:4008-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenberg, S. 2001. Recent advances in the molecular biology of hepatitis C virus. J. Mol. Biol. 313:451-464. [DOI] [PubMed] [Google Scholar]
- 22.Tan, S. L., and M. G. Katze. 2001. How hepatitis C virus counteracts the interferon response: the jury is still out on NS5A. Virology 284:1-12. [DOI] [PubMed] [Google Scholar]
- 23.Yao, N., P. Reichert, S. S. Taremi, W. W. Prosise, and P. C. Weber. 1999. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase. Structure Fold. Des. 7:1353-1363. [DOI] [PubMed] [Google Scholar]