Abstract
Progress toward development of better therapies for the treatment of hepatitis C virus (HCV) infection has been hampered by poor understanding of HCV biology and the lack of biological assays suitable for drug screening. Here we describe a powerful HCV replication system that employs HCV replicons expressing the β-lactamase reporter (bla replicons) and subpopulations of Huh7 cells that are more permissive (or “enhanced”) to HCV replication than naïve Huh7 cells. Enhanced cells represent a small fraction of permissive cells present among naïve Huh7 cells that is enriched during selection with replicons expressing the neomycin phosphotransferase gene (neo replicons). The level of permissiveness of cell lines harboring neo replicons can vary greatly, and the enhanced phenotype is usually revealed upon removal of the neo replicon with inhibitors of HCV replication. Replicon removal is responsible for increased permissiveness, since this effect could be reproduced either with alpha interferon or with an HCV NS5B inhibitor. Moreover, adaptive mutations present in the replicon genome used during selection do not influence the permissiveness of the resulting enhanced-cell population, suggesting that the mechanisms governing the permissiveness of enhanced cells are independent from viral adaptation. Because the β-lactamase reporter allows simultaneous quantitation of replicon-harboring cells and reporter activity, it was possible to investigate the relationship between genome replication activity and the frequency with which transfected genomes can establish persistent replication. Our study demonstrates that differences in the replication potential of the viral genome are manifested primarily in the frequency with which persistent replication is established but modestly affect the number of replicons observed per replicon-harboring cell. Replicon copy number was found to vary over a narrow range that may be defined by a minimal number required for persistent maintenance and a maximum that is limited by the availability of essential host factors.
Hepatitis C virus (HCV) is the major cause of parenterally transmitted non-A, non-B hepatitis, which affects approximately 3% of people worldwide (14). Until recently, the study of HCV replication and the development of cell-based assays to screen for inhibitors of HCV replication have been hampered by the inability to culture the virus ex vivo. The first robust replication system for studying HCV replication in cell culture, described by Lohmann et al. (7), utilizes bicistronic subgenomic viral RNAs, or replicons, that encode the nonstructural proteins and have the cis RNA elements required for autonomous replication (4, 11-13). These replicons were engineered to express neomycin phosphotransferase (Neor) so that cells harboring replication-competent replicons could be isolated under suitable selective pressure. Subsequent studies have demonstrated that efficient replication of HCV-con1 replicons requires cell culture-adaptive mutations that are not observed in vivo (1, 6). Although HCV-con1 adaptive mutations have been found in several nonstructural genes, many are clustered within a small region of NS5A (1, 3, 6). More recently, replicons derived from the HCV-N isolate were also shown to be replication competent in cell culture (2, 3). Replication of the HCV-N genome does not require the cell culture-adaptive mutations found in con1 replicons but instead requires a natural four-amino-acid insertion that maps within the interferon sensitivity-determining region of NS5A (3). The finding that diverse mutations in NS5A act to enhance replication in cell culture suggests that NS5A plays a critical role in cell culture adaptation. The mechanisms of NS5A-dependent cell culture adaptation are not known, but it is reasonable to speculate that they involve direct interactions with viral and/or cellular factors. An intriguing possibility is that adaptive mutations in NS5A may prevent the action of negative factors involved in the innate intracellular antiviral response (9).
Better understanding of HCV biology will require more efficient methods for measurement of viral replication. Viral replication can be measured by using a variety of techniques that either measure the number of cells in which replication is established (neo selection experiments) or directly measure viral RNA (Northern blotting or RNase protection assay) or proteins (Western blotting or enzyme-linked immunosorbent assay) or by using reporters such as luciferase expressed by replicons (1-3, 5-7, 10). Colony selection is both time consuming and assumes that the frequency of Neor colonies observed with a given genome is directly proportional to its intrinsic replication activity. In addition, colony selection does not provide a measure of the number of replicon copies per cell. As with colony selection, direct quantitation of viral RNA or protein products is time consuming and not practical for high-throughput screening. Alternatively, reporters such as luciferase have been used to detect and quantitate replication and are compatible with high-throughput screening for identification of inhibitors of replication. Reporters such as luciferase provide a measure of the amount of replicon RNA in a sample but do not provide a direct measure of the number of cells supporting replication. Here, we constructed subgenomic replicons in which the Neor gene is replaced by a β-lactamase reporter (bla). A replication assay with bla replicons has the advantages of both neo selection experiments and reporter assays in that it provides a means to measure both the number of cells harboring replicon and the number of replicons per cell. In addition, bla reporter assays are compatible with live-cell formats such that viable cells can be recovered. We used an RNA transfection assay and the bla reporter to identify cells susceptible to persistent replication in the absence of drug selection. Here we describe the characterization of several cell lines that are more permissive or “enhanced” to persistent HCV replication and present evidence that the interplay of viral and cellular factors govern HCV replication efficiency in cell culture.
MATERIALS AND METHODS
Replicon constructs.
Standard protocols were used for all manipulation of nucleic acids. bla replicon constructs were derived from pHCVneo17.wt (8), a template for T7 transcription of RNA identical in sequence to the replicon I377neo/NS3-3′/wt (7). The bla coding region was PCR amplified by using pcDNA3-blaM (Aurora Biosciences) as the template with primers that introduced AscI and PmeI sites at the 5′ and 3′ ends, respectively, and subcloned into the corresponding sites of pHCVneo17.wt. A silent mutation was subsequently introduced to eliminate a ScaI restriction site from the bla coding region.
The cell culture-adaptive mutations S2204I (1), A2199T (1), and R2884G (6) were introduced by PCR mutagenesis using a QuikChange PCR mutagenesis kit (Stratagene) and a template generated by subcloning the 2,694-bp SalI fragment from pHCVneo17.wt into Litmus38 (New England Biolabs).
Lipotransfections of Huh7 cell lines.
Complete medium for maintenance of Huh7 cells consisted of Dulbecco's modified Eagle's medium (Cellgro) supplemented with 10% fetal calf serum, nonessential amino acids, 2 mM GlutaMAX (Invitrogen Life Technologies), 100 U of penicillin/ml, and 100 μg of streptomycin/ml. Cells were seeded in six-well plates at a density of 300,000 cells per well and incubated for 16 to 20 h. The lipotransfection medium was prepared by vigorously vortexing 2 ml of OptiMEM I (Invitrogen Life Technologies), 12 μl of DMRIE-C (Invitrogen Life Technologies), and 5 μg of replicon RNA. Cells were washed once with OptiMEM I and then incubated with the lipotransfection medium for 7 to 8 h at 37°C in a humidified incubator containing 5% CO2. The lipotransfection mixture was removed by aspiration and replaced with complete medium, followed by incubation overnight at 37°C. Transfection efficiency was determined by quantitation of bla-positive cells 24 h after transfection, and then cells were split into T75 flasks. The cells were kept subconfluent by splitting 1:4 to 1:6 as needed throughout the experiment.
Assays for bla reporter activity.
Medium was removed, and cells were stained for 90 min with CCF4-AM (Aurora Biosciences Corp.) in Dulbecco's modified Eagle's medium supplemented with 25 mM HEPES, pH 8.0. For quantitation of the fraction of cells harboring bla replicons, cells were photographed by using a digital charge-coupled device color camera and green and blue cells were counted by DIP with Image-Pro Plus software. Alternatively, fluorescence was measured by using a CytoFluor 4000 fluorescence plate reader.
Colony formation assays.
For colony formation experiments, 2 × 105 Huh7 cells were lipotransfected with 100 ng of the indicated neo replicon in T75 flasks. Immediately following transfection, the medium was supplemented with 250 μg of G418/ml. Colonies were counted at 4 weeks.
Generation of cured cells.
For curing with alpha interferon (IFN-α), cells harboring neo replicons were seeded at 80% confluence and then cultured at high cell density in the presence of 100 U of IFN-α/ml for 11 days. Cells treated with a nucleoside NS5B inhibitor were seeded at 10% confluence, allowed to settle overnight, and then incubated with a 5 μM concentration of compound 1 (95% inhibitory concentration) for 7 days.
RNase protection assays.
A template construct for transcription of probes to NS3 was generated by amplifying a portion of NS3 with primers 5′-CGGAATTCGCACTCATTTTCTGCCATTCCAAG and 5′-CGGGATCCCGTTGTGGCACGGTCGTCGTCTCAAT and then subcloning the resulting PCR product into the EcoRI and BamHI sites of pSP72 (Promega). 32P-labeled NS3 probes were transcribed with T7 RNA polymerase by using a Maxiscript kit (Ambion) and an XhoI-digested template. Probes were purified by using a Qiagen RNeasy kit and were diluted to 10,000 cpm/μl. Total RNA was purified from cells by Trizol extraction. RNase protection assays were done by using an RPA III kit (Ambion) with 5 μg of total RNA, 50,000 cpm of probe, and a hybridization temperature of 44°C. Following RNase treatment, samples were analyzed by electrophoresis using 10% acrylamide Tris-borate-EDTA-urea gels (Invitrogen). The products were quantitated by phosphor autoradiography with a Storm PhosphorImager and ImageQuant software (Molecular Dynamics). HCV replicon RNA signal was normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase). The number of bla replicons per cell was normalized to GAPDH and then divided by the fraction of blue cells determined by DIP.
RESULTS
Quantitation of HCV replication by using the bla reporter.
To facilitate quantitation of viral replication, we replaced the Neor gene with the bla reporter to generate replicon bla-NS3-3′/wt, which is otherwise identical to the replicon I377/NS3-3′ described by Lohmann et al. (7). bla activity was measured in live cells by using the membrane-permeant fluorescent substrate CCF4-AM. Cells that lack bla emit green fluorescence, but cells expressing bla cleave the CCF4 substrate and emit blue fluorescence (15). Reporter activity can be measured either by quantitation of blue and green light emission with a plate reader or by quantitation of blue and green cells by DIP of images acquired with a charge-coupled device camera mounted on a fluorescence microscope (see Materials and Methods). The latter readout represents an important advantage over other reporter systems because it allows direct quantitation of the fraction of cells in a population, expressed as the percentage of blue cells (%BC), that express bla. Below, we refer to assays that use bla to quantitate HCV replication as Bla-Rep and the units (light emission or %BC) indicate the assay readout.
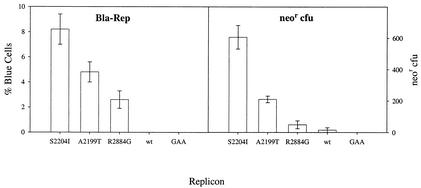
The %BC readout is a direct measure of the fraction of cells in a population that harbor replicon and should be equivalent to assays that count G418-resistant (Neor) colonies. We compared the Bla-Rep and colony formation activities of replicons harboring con1 adaptive mutations S2204I (position 232 in NS5A), A2199T (position 227 in NS5A), and R2884G (position 465 in NS5B) (1, 6) by using replicons expressing Neor or bla. The results summarized in Fig. 1 show that replicon activity rank order correlated well in both assays but also indicate that differences in replication activity were magnified in the Neor CFU assay (Fig. 1). The fluorescent dye used in Bla-Rep is capable of identifying cells that harbor as few as 100 copies of the bla reporter (15). Despite the high sensitivity of the assay format used, to ensure that the %BC truly reflected the fraction of cells harboring replicon and that it did not miss cells that could potentially harbor replicons at levels below the limits of detection, blue and green fluorescing cells were segregated by fluorescence-activated cell sorter and then assayed for the presence of replicon RNA. Neither blue cells nor bla replicon RNA was detected in the sorted population of green cells (data not shown). The colony selection data were obtained under modest selective pressure by using 250 μg of G418/ml to maximize colony formation efficiency. The differences in colony formation efficiency were more exaggerated at greater G418 concentrations (data not shown). The relative activities of replicons harboring adaptive mutations A2199T and R2884G normalized with respect to the activity of replicons harboring adaptive mutation S2204I (the most active replicon in both assays) were approximately 0.5 and 0.25, respectively, in Bla-Rep but only 0.3 and 0.10, respectively, in the Neor CFU assay (Fig. 1). Importantly, replicons without adaptive mutations (wild type) have low but measurable activity in the Neor CFU assay but are at background levels in Bla-Rep. The dynamic range of Bla-Rep is best after 72 h (data not shown), when the signal obtained with the replication-deficient replicon bla-NS3-3′/S2204I-gaa (containing Asp-to-Ala substitutions in the catalytic triad of NS5B) has completely decayed. In our hands, bla-NS3-3′/S2204I replicons had the highest replication efficiency and were used for subsequent assay optimization studies.
FIG. 1.
The %BC readout correlates with the number of Neor CFU. Left panel, Huh7 cells were lipotransfected with 5 μg of the indicated bla-NS3-3′ replicon and stained for bla expression at day 6. The fraction of cells harboring replicon was determined by DIP of stained cells (>1,000 cells/data point). Right panel, 2 × 105 Huh7 cells were lipotransfected with 100 ng of the indicated neo-NS3-3′ replicon in T75 flasks. Cells were cultured in complete medium supplemented with 250 μg of G418/ml for 4 weeks, at which time colonies were counted.
Huh7 cells selected with neo replicons support persistent replication of bla replicons.
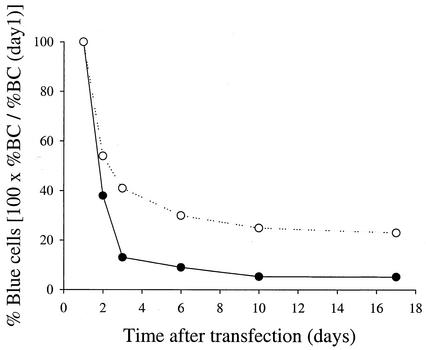
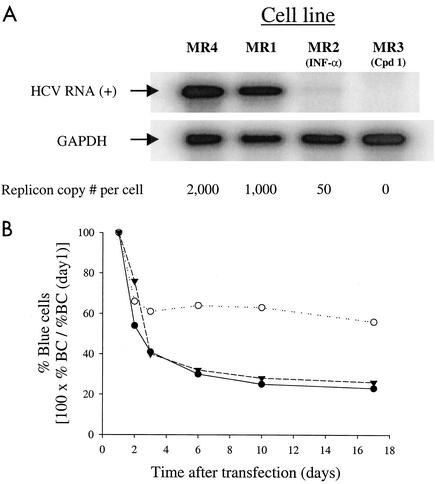
Although replication of bla replicons in Huh7 cells was easily detected, replication was transient and only a small fraction of cells harbored replicons (Fig. 1). We reasoned that drug-selected cells that support persistent replication of neo replicons may also support persistent replication of bla replicons. To test this hypothesis, several Huh7 cell lines harboring neo replicons were generated and tested for their ability to support replication of bla replicons. Replicons harboring various adaptive mutations expressing the complete HCV nonstructural region (neo-NS2-3′) and replicons that lacked NS2 (neo-NS3-3′) were used to select the cell lines described below (Table 1). Remarkably, a cell line selected following transfection of Huh7 cells with a neo-NS2-3′/S2204A replicon (MR1 [Table 1]) was more permissive to replication of bla replicons than were Huh7 cells (Fig. 2). The fraction of transfected cells that supports HCV replication was approximately three times greater with MR1 than naïve Huh7 cells. Moreover, bla replicons were stably maintained for several weeks in MR1 but not in naïve Huh7 cells (Fig. 2). The increased permissiveness of MR1 cells suggested that cells permissive to HCV replication were indeed enriched during drug selection. Because MR1 cells contain approximately 1,000 copies of neo replicon RNA per cell (Fig. 3A), it was important to investigate whether curing them of the neo replicon could influence their permissiveness. MR1 cells were treated with IFN-α to generate MR2 cells or with a specific nucleoside NS5B inhibitor to generate MR3 cells. The identity and properties of this compound will be published elsewhere (S. S. Carroll, J. Tomassini, M. Bosserman, K. Getty, M. W. Stahlhut, A. B. Eldrup, B. Bhat, D. Hall, A. L. Simcoe, R. LaFemina, C. A. Rutkowski, B. Wolanski, Z. Yang, G. Migliaccio, R. De Francesco, L. C. Kuo, M. MacCoss, and D. B. Olsen, submitted for publication). The neo replicon was completely lost after treatment with the NS5B inhibitor, but small amounts of neo replicon remained in cells treated with IFN-α (Fig. 3A). neo selection experiments indicated that 1 to 4% of the cells in the MR2 population harbored replicon which persisted for a minimum of several months. Since only a small fraction of the cells harbored replicon, the following results are reflective primarily of the overwhelming majority of MR2 cells which have been cured of replicon. Interestingly, the permissiveness of nucleoside-cured cells (MR3) is similar to that of parental MR1 cells but cells partially cured with IFN-α (MR2) became considerably more permissive than untreated cells (Fig. 3B). The increased permissiveness of MR2 cells (IFN treated) is unlikely to be a direct consequence of lower neo replicon copy number since cells that were completely cured with the NS5B inhibitor (MR3) did not become more permissive (Fig. 3B). Together, these findings suggested that neo replicons present in MR1 cells do not interfere with transfected bla replicons. neo replicon-selected cells that are more permissive to persistent replication of bla replicons than are naïve Huh7 cells are referred to as enhanced cells in this paper.
TABLE 1.
Summary of cell lines
| Cell line | Parental cell line | Replicon used during selection | Drug treatment | Enhanced phenotypea |
|---|---|---|---|---|
| MR1 | Huh7 | neo-NS2-3′/S2204A | None | ++ |
| MR2 | MR1 | neo-NS2-3′/S2204A | IFN-α | ++++ |
| MR3 | MR1 | neo-NS2-3′/S2204A | Compound 1 | ++ |
| MR4 | Huh7 | neo-NS3-3′/S2204I | None | − |
| MR5 | MR4 | neo-NS3-3′/S2204I | IFN-α | ++ |
| MR6 | MR4 | neo-NS3-3′/S2204I | Compound 1 | ++ |
| MR7 | Huh7 | neo-NS3-3′/A2199T | None | − |
| MR8 | MR7 | neo-NS3-3′/A2199T | Compound 1 | ++ |
| MR9 | Huh7 | neo-NS3-3′/R2884G | None | − |
| MR10 | MR9 | neo-NS3-3′/R2884G | Compound 1 | ++ |
+ symbols, relative levels of enhancement; −, no observed enhancement of permissiveness.
FIG. 2.
Persistent replication of bla-NS3-3′/S2204I in MR1 cells. Naïve Huh7 cells and MR1 cells (G418-selected population of Huh7 cells harboring neo-NS2-3′/S2204A) were transfected with bla-NS2-3′/S2204I replicon. The fraction of cells harboring replicon was determined for each population by DIP at the indicated times. •, Huh7 cells; ○, Huh7 cells harboring neo-NS2-3′/S2204A.
FIG. 3.
Replicon-enhanced cells. (A) Total RNA was isolated from Huh7 cells harboring either neo-NS3-3′/S2204I (MR4) or neo-NS2-3′/S2204A (MR1) and from MR1 cells treated with IFN-α (MR2 cells) or with a specific NS5B inhibitor (MR3 cells). Replicon RNA levels were estimated by RNase protection assays by comparison with HBI10A cells that have 800 to 1,000 copies/cell (8). (B) MR1 (•), MR2 (○), and MR3 (▾) cells were transfected with bla-NS3-3′/S2204I replicon RNA. The fraction of cells harboring replicon was determined by DIP at the indicated times following transfection.
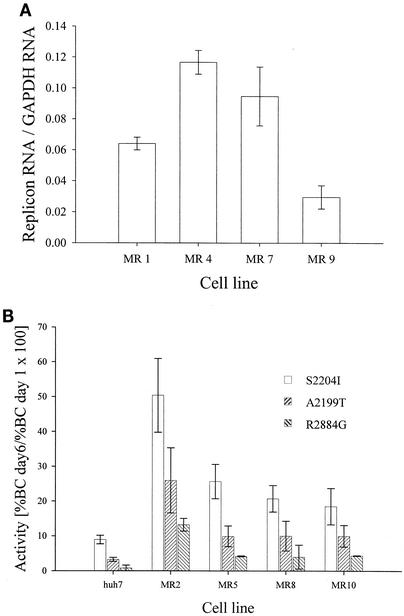
To better understand the factors that influence cellular permissiveness, similar experiments were performed using cell lines selected with different neo replicons. Cell lines MR4, MR7, and MR9 were selected with neo-NS3-3′ replicons harboring adaptive mutations S2204I, A2199T, and R2884G, respectively (Table 1). Bla-Rep assays revealed that none of these cell lines was more permissive than naïve Huh7 cells (Fig. 4 and data not shown). Because all of the cell lines described here were expanded from pools of Neor colonies, it is unlikely that clonal variations within each cell line account for their phenotypic differences. To evaluate whether neo-NS3-3′ replicons could interfere with replication of bla replicons, MR4 cells were treated with IFN-α and a nucleoside NS5B inhibitor to generate cell lines MR5 and MR6, respectively (Table 1). Interestingly, both MR5 and MR6 cells became approximately three times more permissive than naïve Huh7 and parental MR4 cells upon removal of the neo replicon (Fig. 4). The finding that treatments with IFN-α and the NS5B inhibitor led to the same level of increased permissiveness indicated that neo replicon elimination rather than nonspecific drug effects are responsible for the enhanced phenotype of MR5 and MR6 cells. Furthermore, the observation that the restriction of MR4 cells to HCV replication can be reversed by two different HCV inhibitors strongly suggests that neo replicons present in these cells inhibit replication of transfected bla replicons (Fig. 4). Although both MR5 and MR2 cells were treated with IFN-α, MR2 cells are significantly more permissive than MR5 cells, suggesting that the replicon used during selection might contribute to the enhanced phenotype of selected cells.
FIG. 4.
Replicon-cured MR4 cells are enhanced to replication of bla replicons. MR5 and MR6 cells were derived by treating MR4 cells with IFN-α and an NS5B specific inhibitor, respectively. Persistent replication was assayed in these cells as described in the text. •, Huh7 cells; ○, MR4 cells; ▾, MR5 cells; ▿, MR6 cells.
con1 adaptive mutations present in neo replicons do not influence the enhanced phenotype of selected cells.
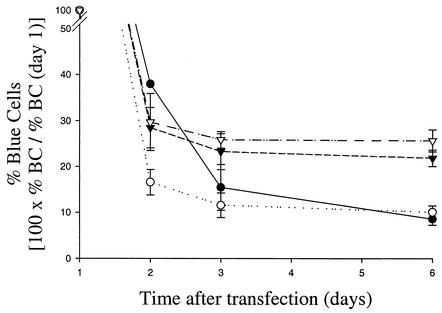
As discussed above, MR4, MR7, and MR9 cell lines (selected with neo-NS3-3′ replicons harboring adaptive mutations S2204I, A2199T, and R2884G, respectively) did not display a highly permissive phenotype (Fig. 4 and data not shown). Quantitation of replicon copy number indicated that neo replicon RNA levels do not appear to correlate with cell line permissiveness to transfected bla replicons (Fig. 5A). To investigate whether restriction to persistent replication of bla replicons in MR7 and MR9 cells could also be reversed by elimination of their neo replicon, both cell lines were treated with the NS5B inhibitor, and the permissiveness of the resulting MR8 and MR10 cell lines was evaluated in Bla-Rep assays. RNase protection analyses indicated that drug treatment completely cured MR8 and MR10 cells of the neo replicon used to isolate them (data not shown). Consistent with the enhancement observed with MR5 cells enriched using the S2204I replicon and cured with the NS5B inhibitor, MR8 and MR10 cells were also enhanced for replication of bla replicons (Fig. 5B). Remarkably, the rank order of the activities of bla replicons harboring the S2204I, A2199T, or R2884G adaptive mutation was the same in all cell lines tested (Fig. 5B). Thus, the permissiveness of enhanced cells is not specific to the adaptive mutation present in the replicon used to isolate them. In addition, the finding that MR5, MR8, and MR10 cells are equally susceptible to HCV replication suggests that there is no obvious correlation between the intrinsic replication activity of the replicon used to isolate enhanced cells and the permissiveness of the resulting cell line. Together, the results described above demonstrate that permissive Huh7 cells are enriched during drug selection and the enhanced permissiveness of resulting cell lines is revealed upon removal of the neo replicon used during selection.
FIG. 5.
Level of cellular permissiveness is not dependent on the con1 adaptive mutation present in the neo replicon used to isolate Neor cells. (A) Replicon RNA per cell in MR1, MR4, MR7, and MR9 cells was measured by RNase protection assays. Data were normalized with respect to GAPDH. (B) Huh7 and replicon-cured MR2, MR5, MR8, and MR10 cells were transfected with bla-NS3-3′ replicons containing the indicated adaptive mutations. The fraction of cells harboring replicon 6 days after transfection was determined by DIP.
Establishment of persistent replication is RNA dose dependent.
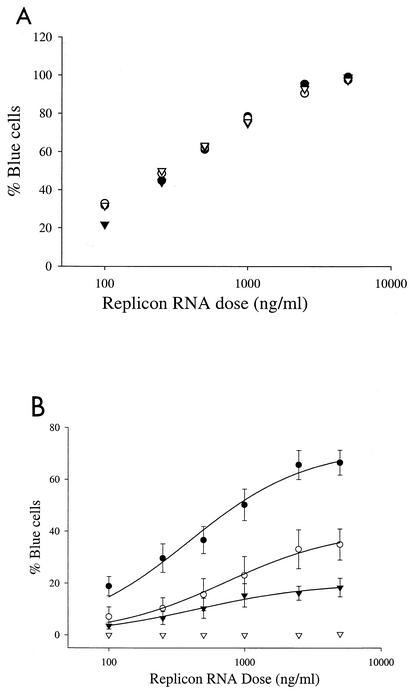
A logical assumption is that the establishment of persistent replication is dependent on the replication competence of transfected genomes. A corollary to this assumption is the prediction that the amount of replicon RNA delivered per cell directly correlates with the efficiency with which persistent replication is established. To study the impact of RNA delivery on persistent replication, MR2 cells were transfected with titrations of replicon RNAs containing different cell culture-adaptive mutations that ranged in their abilities to establish replication. Transfection efficiencies were identical for all RNAs and were linear over a wide range of replicon RNA concentrations (Fig. 6A). Six days after transfection, the number of replicon-harboring cells was proportional to the number of replicon genomes delivered per cell (Fig. 6B). Remarkably, the number of blue cells observed on day 6 at saturating RNA concentrations was dependent on the adaptive mutation present in the replicon, demonstrating that persistent replication is influenced by both RNA delivery and the replication competence of the viral genome.
FIG. 6.
Persistent replication is RNA dose dependent. MR2 cells were transfected with various doses of bla-NS3-3′ replicons containing the indicated adaptive mutation. The fraction of cells harboring replicon was determined by DIP at days 1 (A) and 6 (B). The bla-NS3-3′ replicons used had the following mutations: S2204I (•), A2199T (○), R2884G (▾), and S2204I and the NS5B GAA inactivating mutation (▿).
Correlation between persistent replication and genome replication activity.
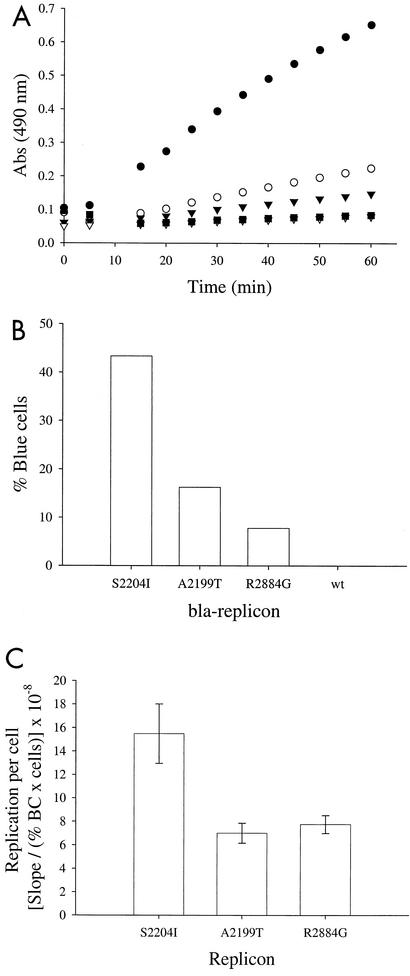
Our data indicate that establishment of persistent replication is a function of several factors, including the host cell, cell culture-adaptive mutations in the replicon, and RNA dose. On the other hand, the relationship between replicon activity in assays that measure the frequency with which viral genomes can establish persistent replication (such as Neor and %BC) and replicon copy number per cell has not been addressed. Replicon copy number in cells supporting persistent replication could be constrained by a minimum required to prevent replicon dilution in rapidly dividing cell lines (or to express sufficient amounts of a marker required for selection) and a maximum limited by the availability of cellular factors essential for replication or by toxic effects. Using Bla-Rep and replicons with various adaptive mutations, the relative number of replicon RNA molecules per replicon-harboring cell (Fig. 7C) was indirectly determined by normalizing the total bla reporter activity (Fig. 7A) by the number of replicon-harboring cells determined by DIP (Fig. 7B). This technique has the advantages that the amount of replicon RNA per cell or intrinsic replication activity is determined in the absence of selective pressure and that data for both the fraction of replicon-harboring cells and the relative number of replicons per cell can be determined from a single population at one time. As shown in Fig. 7C, the intrinsic replication activities of all three replicons are similar, with that of bla-NS3-3′/R2884G being nearly the same as that of bla-NS3-3′/A2199T and approximately one-half of that of replicon bla-NS3-3′/S2204I. In contrast, the activity of bla-NS3-3′/R2884G in unnormalized reporter assays is significantly lower than those of the bla-NS3-3′/S2204I and bla-NS3-3′/A2199T replicons. These results are in agreement with measurements of replicon RNA copy number in stable cell lines selected with neo replicons harboring the same adaptive mutations analyzed above (Fig. 5A), which reveals a strikingly similar pattern to that obtained with %BC. Together, these analyses indicate that levels of viral replication vary over a narrow range in cells supporting persistent replication.
FIG. 7.
Relative amounts of replicon per replicon-harboring cell. Cells were lipotransfected with the indicated bla-NS3-3′ replicon and then assayed at day 3. (A) The relative amounts of bla in each sample are proportional to the rate at which nitrocefin is cleaved to generate a product that absorbs at 490 nm. Cells were lysed by three cycles of freezing and thawing. Reactions were initiated by the addition of nitrocefin, and absorbance (Abs) at 490 nm was measured at 5-min intervals. •, S2204I; ○, A2199T; ▾, R2884G; ▿, wild type; ▪, GAA. (B) The %BC in the same population was determined by DIP. (C) Reporter activity per replicon-harboring cell (or intrinsic genome replication activity) was calculated by normalizing the reporter activity presented in panel A by the number of cells expressing bla at the time of the assay (total number of cells assayed multiplied by the fraction of cells that contain replicon [panel B]).
DISCUSSION
We took advantage of the versatility of the β-lactamase reporter to investigate the relationship between replication activity and the frequency with which transfected genomes can establish persistent replication. We asked whether replication activity per replicon-harboring cell was proportional to the frequency with which a replicon genome is able to establish persistent replication. To this end, we compared the activities of replicons harboring adaptive mutation S2204I, A2199T, or R2884G by using Neor colony formation assays and two assays based on the bla reporter, one that measures blue cell counts (%BC) and another that measures reporter activity (blue light emission). Our analyses indicate that the readout of all three assays is proportional to the number of cells in which persistent replication is established following RNA transfection and that none of these assays provides a direct measure of intrinsic genome replication activity, defined as replication activity per replicon-harboring cell. The intrinsic replication activity of genomes harboring adaptive mutations S2204I, A2199T, and R2884G was determined by normalizing bla activity (Fig. 7A) by the number of replicon-harboring cells at the time of the assay (Fig. 7B). These analyses revealed that the intrinsic replication activity of HCV replicons is not directly proportional to their ability to establish persistent replication (Fig. 7C). Importantly, similar results were obtained from analyses that used viral RNA instead of reporter activity as the readout for viral replication (data not shown). These results suggest that variations in genome replication potential are manifested primarily in the efficiency (or frequency) with which persistent replication is established, but once persistent replication is established, the levels of viral replication vary over a narrow range and are not significantly affected by the replicon genome. Thus, it appears that small differences in replicon activity are amplified in assay readouts that measure the frequency with which persistent replication is established (such as Neor and %BC). In this regard, we find that the %BC readout reflects genome replication activity more accurately than the Neor CFU assay, as drug selection pressure appears to lead to an overestimation of the activity of robust replicons and an underestimation of the activity of weaker ones.
Recently, Krieger et al. (5) reported a direct correlation between the efficiency of Neor colony formation and viral RNA production as well as luciferase reporter activity in transient replication assays. Krieger and colleagues did not normalize their transient assays by the number of replicon-harboring cells. Therefore, their study did not address directly whether the efficiency of the Neor colony assay correlated with genome replication activity defined as the amount of viral RNA produced per replicon-harboring cell. Their results, however, are in agreement with ours and indicate that con1 adaptive mutations stimulate genome replication competence.
Based on the work described here, we propose a “threshold” model that may explain the mechanisms governing the efficiency by which persistent replication is established in RNA transfection assays. According to this model, the rate at which replicons form functional complexes following transfection is a major determinant of persistence. The rate at which HCV replication complexes are formed following RNA transfection is likely to be dependent on the number of genomes delivered per cell as well as on their replication competence. A putative antiviral cellular response and RNA decay may negatively influence viral replication. Because HCV replication complexes are diluted at each cell division, a requirement for persistent replication is that sufficient replicon RNA is present following cell division for translation of viral proteins at rates that support generation of replication complexes. Replicon RNA in persistently replicating cells is present within a narrow copy number range, with the minimum being set by the requirement to replenish the number of functional complexes between cell divisions. The maximum replicon copy number per cell might be capped by mechanisms that restrict replicon RNA copy number per cell. Two potential reasons for this limit may be that host cell factors required for replication may become limiting or elevated expression of viral proteins may be toxic to cells.
The replicon-enhanced cells in combination with the bla-expressing replicons described here represent a significant advance in our ability to quantitate HCV replication in cell culture by providing a means to perform high-volume structure-function and drug-screening assays. A major practical advantage of replicon-enhanced cells is that transfected replicons expressing reporter proteins are stably maintained in these cells, thereby allowing generation of stable reporter cell lines, effectively bypassing the need for drug selection. The data presented here indicate that only a fraction of permissive cells are present in a population of naïve Huh7 cells and demonstrate that permissive cells are isolated during drug selection of cells that stably support replication of neo replicons. Although all drug-selected cell lines should theoretically be more permissive to HCV replication than parental Huh7 cells, their capacity for replication of transfected replicons can vary greatly. Three different cell lines selected with neo-NS3-3′ replicons harboring adaptive mutations S2204I, A2199T, and R2884G (MR4, MR7, and MR9, respectively) were not permissive to replication of transfected bla replicons but became permissive after curing of the neo replicon with a specific inhibitor of HCV replication (Fig. 4 and 5B). In contrast, the enhanced phenotype of MR1 cells (selected with neo-NS2-3′/S2204A replicon) does not require curing the neo replicon, and replicon removal with an HCV NS5B inhibitor does not increase their permissiveness. The replicon copy number of MR1 cells is lower than that of MR4 cells but higher than that of MR9 cells (Fig. 5), indicating that their phenotype does not correlate with neo replicon copy number. Because MR1 cells but not MR4, MR7, and MR9 cells were selected with a neo replicon expressing NS2, it is possible that the permissiveness of MR1 cells may be related to the presence of NS2. Evaluation of this possibility will require the generation and characterization of additional cell lines selected with neo replicons expressing HCV NS2. Alternatively, the permissiveness of MR1 cells prior to being cured may reflect the possibility that the supertransfected bla-NS3 replicon outcompetes the neo-NS2 replicon. Our most permissive cell line, MR2, was derived by treatment of MR1 cells with IFN-α. The effect of IFN-α in MR1 cells is not reproducible, since subsequent experiments in which MR1 cells were treated with IFN-α have failed to yield cell lines with the MR2 phenotype. The increased permissiveness of MR2 cells compared with MR1 cells is not due to replicon removal, since MR1 cells cured with an HCV NS5B inhibitor (MR3 [Table 1]) are not more permissive than untreated MR1 cells. Moreover, IFN-α treatment produced identical results as treatment with an HCV NS5B inhibitor (Fig. 4 and data not shown) in all subsequent experiments, suggesting that the principal effect of the cytokine on the enhanced phenotype is due to removal of neo replicons. The hallmark of replicon-enhanced cells is that both the number of transfected cells that can establish persistent replication and the stability of those cell populations harboring the replicon (i.e., replicon maintenance) are increased compared with parental Huh7 cells. We did not notice much variability in terms of replicon maintenance among different replicon-enhanced cells, but significant variability with respect to the percentage of transfected cells that can establish persistent replication was observed (Fig. 5B). Cellular changes present in replicon-enhanced cells that are responsible for persistent HCV replication are not yet known, but proteomic and genomic approaches promise to be useful tools in addressing this interesting question.
Acknowledgments
E.M.M. and J.A.G. contributed equally to this work.
We thank Giovanni Migliaccio, Steve Ludmerer, Robert LaFemina, and Joanne Tomassini for critical reading of the manuscript and Charles Rice for providing HCV-con1 replicon constructs.
REFERENCES
- 1.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 2.Guo, J.-T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ikeda, M., M. Yi., K. Li., and S. M. Lemon. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 76:2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kolykhalov, A. A., S. M. Feinstone, and C. M. Rice. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 70:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lohmann, V., F. Körner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lohmann, V., F. Körner, J.-O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 8.Mottola, G., G. Cardinali, A. Ceccacci, C. Trozzi, L. Bartholomew, M. R. Torrisi, E. Pedrazzini, S. Bonatti, and G. Migliaccio. 2002. Hepatitis C virus nonstructural proteins are localized in a modified endoplasmic reticulum of cells expressing viral subgenomic replicons. Virology 293:31-43. [DOI] [PubMed] [Google Scholar]
- 9.Pflugheber, J., B. Fredericksen, R. Sumpter, Jr., C. Wang, R. Ware, D. L. Sodora, and M. Gale, Jr. 2002. Regulation of PKR and IRF-1 during hepatitis C virus RNA replication. Proc. Natl. Acad. Sci. USA 99:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pietschmann, T., V. Lohmann, G. Rutter, K. Kurpanek, and R. Bartenschlager. 2001. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J. Virol. 75:1252-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka, T., N. Kato, M.-J. Cho, and K. Shimotohno. 1995. A novel sequence found at the 3′ terminus of hepatitis C virus genome. Biochem. Biophys. Res. Commun. 215:744-749. [DOI] [PubMed] [Google Scholar]
- 12.Tsukiyama-Kohara, K., N. Iizuka, M. Kohara, and A. Nomoto. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Want, C., P. Sarnow, and A. Siddiqui. 1993. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J. Virol. 67:3338-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wasley, A., and M. J. Alter. 2000. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin. Liver Dis. 20:1-16. [DOI] [PubMed] [Google Scholar]
- 15.Zlokarnik, G., P. A. Negulescu, T. E. Knapp, L. Mere, N. Burres, L. Feng, M. Whitney, K. Roemer, and R. Y. Tsien. 1998. Quantitation of transcription and clonal selection of single living cells with β-lactamase as reporter. Science 279:84-88. [DOI] [PubMed] [Google Scholar]