Abstract
Although several virologic and immunologic factors associated with an increased risk of perinatal human immunodeficiency virus type 1 (HIV-1) transmission have been described, the mechanism of mother-to-child transmission is still unclear. More specifically, the question of whether selective pressures influence the transmission remains unanswered. The aim of this study was to assess the genetic diversity of the transmitted virus after in utero transmission and after peripartum transmission and to compare the viral heterogeneity in the child with the viral heterogeneity in the mother. To allow a very accurate characterization of the viral heterogeneity in a single sample, limiting-dilution sequencing of a 1,016-bp fragment of the env gene was performed. Thirteen children were tested, including 6 with in utero infections and 7 with peripartum infections. Samples were taken the day after birth and at the ages of 6 and 14 weeks. A homogeneous virus population was seen in six (46.2%) infants, of whom two were infected in utero and four were infected peripartum. A more heterogeneous virus population was detected in seven infants (53.8%), four infected in utero and three infected peripartum. The phylogenetic trees of the mother-child pairs presented a whole range of different tree topologies and showed infection of the child by one or more maternal variants. In conclusion, after HIV-1 transmission from mother to child a heterogeneous virus population was detected in approximately one-half of the children examined. Heterogeneous virus populations were found after peripartum infection as well as after in utero infection. Phylogenetic tree topologies argue against selection processes as the major mechanism driving mother-to-child transmission but support the hypothesis that virus variability is mainly driven by the inoculum level and/or exposure time.
Transmission of human immunodeficiency virus type 1 (HIV-1) from an infected mother to her child can occur in utero, during birth, or postnatally through breast milk and is the main source of pediatric HIV-1 infection. Antiviral therapy during pregnancy and delivery, combined with elective caesarean section and replacement feeding, has been shown to reduce the incidence of HIV-1 transmission to less than 2% (15). Despite this success and although many factors that affect mother-to-child transmission are known, the exact mechanism remains unclear, and a thorough scientific base to further improve both therapeutic and preventive interventions is missing.
Most molecular-variability studies of recently infected individuals, adults as well as children, have shown that the majority of recent infections are characterized by the presence of highly homogeneous virus (26, 31). This contrasts with the extensive genetic diversity seen in chronic infections (4, 23). These observations led to the assumption of selective transmission of HIV-1. Already in 1993 Zhu et al. (32) proposed three possible models for selective transmission: selective penetration, selective amplification, and low inoculum level; but it is still unclear which of these models is applicable. Selective transmission is definitely not the overall rule. Several cases of recent infections with a heterogeneous virus population have been described (20). One recent study of adults suggested gender-related differences in HIV-1 diversity at the time of infection, with women being more often infected with a heterogeneous population than men (14).
The number of infants found to harbor a heterogeneous virus population compared to the number found to be infected with homogeneous virus varies from study to study, with heterogeneous infection/homogeneous infection ratios of 0/7 (1), 3/1 (3), 6/17 (6), 2/1 (12), 1/3 (19), 2/8 (24), 3/2 (28), and 0/3 (29). This results in an overall incidence of a heterogeneous virus population in the children of 28.8%. In most of the studies the number of mother-child pairs is small and information concerning the infection time of the infants is lacking. Moreover the ages of the infants at the time of sample collection vary considerably, and follow-up samples from the infants are seldom studied.
To further study the molecular mechanisms involved in mother-to-child transmission of HIV-1, we compared proviral env sequences (V1 to V4) from 13 mothers and their infected children. We also looked for differences in virus heterogeneity between in utero-infected infants and infants infected at or around birth.
MATERIALS AND METHODS
Study population.
Patients were recruited in Coast Provincial General Hospital, Mombasa, Kenya, in the nonintervention arm of a study performed in 1996 to 1999 aimed at evaluating the effect of vaginal lavage with diluted chlorhexidine on mother-to-child transmission (8). Recruitment procedures have been described (7). After informed consent, a maternal blood sample was obtained the day after delivery. HIV-1 status was determined by enzyme-linked immunosorbent assay. HIV-1 viral load was determined with the Cobas Amplicor HIV-1 Monitor assay, version 1.5 (Roche Diagnostics). Cervicovaginal secretions (CVS) were collected from the women during labor and tested for proviral DNA and viral RNA as described earlier (7). A venous blood sample from each infant was collected the day after birth for HIV-1 testing. Mothers were asked to return for a follow-up visit 6 weeks after delivery, during which their HIV-1 status was disclosed and a venous blood sample from each infant was taken. None of the mothers or infants included in this study received any treatment for their HIV-1 infection. The HIV-1 status of the infants was established by DNA PCR as described earlier (7). An infant with a positive PCR at day 1 was considered infected in utero (group IU). A PCR that was negative on day 1 but positive at 6 weeks was considered indicative of an infection intrapartum or early postpartum. As breast-feeding was the rule in this study population, discrimination between intrapartum and early postpartum was not possible. However, for reasons of clarity this group is referred to below as infected intrapartum (group IP). Of the seven infants with intrapartum or early postpartum infection, two (IP1 and IP5) were never breast-fed and one (IP7) was weaned off at 4 weeks. All other infants were breast-fed at least up to the age of 6 weeks.
Additional blood samples taken at the age of 14 weeks were analyzed if available. Two infants (IP5 and IP7) died before the age of 14 weeks. Three (IP1, IP4, and IU6) were lost to follow-up, and for one (IP3) the amount of blood collected at 14 weeks was too small to perform the analyses.
Nucleic acid extraction, PCR amplification, and sequencing.
DNA was extracted from whole venous blood with the QIAamp blood kit (Qiagen GmbH, Hilden, Germany). After extraction, all DNA samples were diluted 10-fold. Nested-PCR amplification of an approximately 1,016-bp fragment spanning the V1-to-V4 region of the env gene (positions 6591 to 7607 in the HIV-HXB2 genome) was performed (outer sense primer, 5′-GAGGATATAATCAGTTTATGG-3′; outer antisense primer, 5′-GGTGGGTGCTATTCCTAATGG-3′; inner sense primer, 5′-GATCAAAGCCTAAAGCCATG-3′; inner antisense primer, 5′-ACTTCTCCAATTGTCCCTCATAT-3′). If only two-thirds of replicate PCRs are positive, the likelihood that the PCR products are the result of the amplification of only one molecule is ∼70% (22). Approximately 40 identical PCRs were run for each 10-fold-diluted sample. If less than two-thirds of those reactions were found positive, the PCR products were selected for sequence analysis. Samples with more than two-thirds positive results were diluted further, and the PCR was repeated until no more than two-thirds of the total number of reactions were positive. Both positive and negative controls were included in all PCR assays to assess the sensitivity of the reaction and to detect possible contamination. The lower limit of detection of the PCR assay was equivalent to 1 copy per reaction.
Direct sequencing of both sense and antisense strands was done with the dRhodamine terminator cycle sequencing ready reaction kit (Applied Biosystems, Foster City, Calif.) by using the inner amplification primers and two internal primers: 5′-AATTCCATGTGTACATTGTACTG-3′ and 5′-TATTGTGC(C/T)CC(A/G)GCTGGTTTTGC-3′. Sequencing reaction products were analyzed on an ABI 310 genetic analyzer (Applied Biosystems). All sequencing products for which inspection of the electropherograms revealed nucleotide mixtures at one or more positions were withdrawn from further analysis.
Sequence analysis.
Nucleotide sequences were assembled with the BioEdit package (9). Sequences were aligned by using Clustal W with manual correction (27). Nucleotide gaps were assigned after amino acid conversion to maintain translation integrity. After deletion of all columns with a gap in at least one of the sequences, the intrapatient genetic diversities were calculated for each pairwise comparison between sequences of the same sample by using the generalized two-parameter Kimura algorithm. Phylogenetic analyses and neighbor-joining tree reconstructions were performed by using programs from version 3.6 of the PHYLIP package, with a maximum-likelihood distance matrix and a transition-to-transversion ratio of 2.0. Approximate confidence limits for individual branches were assigned by bootstrap resampling with 1,000 replicates. Tree diagrams were plotted with Treeview, version 1.4.
Nucleotide sequence accession numbers.
All sequences have been submitted to GenBank and assigned accession no. AY174897 to AY175369.
RESULTS
Patient characteristics.
The mean maternal viral load at the time of delivery was 42,481 copies/ml (log, 4.63; log range, 3.38 to 4.96). All children were born after vaginal delivery; nine were males, and four were females. Six children (four males and two females) were considered infected in utero based on a positive DNA PCR result at birth (Table 1, mother-child pairs IU1 to IU6). Seven children (five males and two females) whose HIV-1 DNA PCR result was negative at birth but positive at 6 weeks were considered infected during the intrapartum period (Table 1, mother-child pairs IP1 to IP7). Information on mothers and children is listed in Table 1.
TABLE 1.
Characteristics of the mother-child pairs and intrapatient genetic diversities
| Pair | Sample sourcea | Sex | Birth wt (g) | Viral load (log copies/ml) | Proviral DNA in CVSd | Viral RNA in CVS | ROMb time (h) | nc | Intrapatient genetic distance (% of nucleotides)
|
||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Min | Max | |||||||||
| IP1 | Mother | 4.92 | Yes | Yes | 6.00 | 16 | 4.80 | 0.35 | 7.78 | ||
| 6 wk | M | 2,550 | 11 | 0.04 | 0.00 | 0.12 | |||||
| IP2 | Mother | 4.96 | Yes | Yes | 0.08 | 12 | 16.75 | 0.80 | 31.35 | ||
| 6 wk | F | 3,700 | 8 | 17.07 | 0.11 | 30.90 | |||||
| 14 wk | 8 | 9.13 | 0.23 | 30.63 | |||||||
| IP3 | Mother | 4.40 | No | No | 9.58 | 4 | 0.59 | 0.32 | 0.86 | ||
| 6 wk | M | 3,000 | 22 | 0.07 | 0.00 | 0.32 | |||||
| IP4 | Mother | 4.62 | Yes | No | 0.83 | 9 | 2.00 | 0.58 | 4.78 | ||
| 6 wk | M | 2,700 | 16 | 0.05 | 0.00 | 0.23 | |||||
| IP5 | Mother | 4.85 | Yes | Yes | 6.17 | 18 | 5.34 | 0.35 | 7.14 | ||
| 6 wk | M | 2,400 | 21 | 0.49 | 0.00 | 1.18 | |||||
| IP6 | Mother | 3.68 | Yes | Yes | 20.25 | 17 | 4.97 | 0.12 | 8.39 | ||
| 6 wk | F | 2,800 | 7 | 1.64 | 0.00 | 4.80 | |||||
| IP7 | Mother | 4.4 | No | No | 5.58 | 5 | 2.25 | 1.12 | 3.26 | ||
| 6 wk | M | 2,300 | 20 | 0.13 | 0.00 | 0.48 | |||||
| IU1 | Mother | 3.38 | Yes | No | 0.08 | 16 | 5.02 | 0.12 | 10.76 | ||
| Neonate | M | 2,300 | 11 | 0.02 | 0.00 | 0.12 | |||||
| 6 wk | 13 | 0.11 | 0.00 | 0.35 | |||||||
| 14 wk | 8 | 0.25 | 0.12 | 0.47 | |||||||
| IU2 | Mother | 4.87 | Yes | No | 0.83 | 15 | 2.70 | 0.00 | 6.75 | ||
| Neonate | F | 2,950 | 6 | 0.08 | 0.00 | 0.23 | |||||
| 6 wk | 6 | 0.38 | 0.23 | 0.46 | |||||||
| 14 wk | 3 | 0.46 | 0.23 | 0.58 | |||||||
| IU3 | Mother | 4.56 | Yes | No | 10.50 | 18 | 0.54 | 0.00 | 1.14 | ||
| Neonate | F | 2,600 | 11 | 0.25 | 0.00 | 0.80 | |||||
| 6 wk | 13 | 0.32 | 0.00 | 0.68 | |||||||
| 14 wk | 9 | 0.36 | 0.11 | 0.80 | |||||||
| IU4 | Mother | 4.85 | Yes | Yes | 1.83 | 27 | 1.36 | 0.00 | 3.31 | ||
| Neonate | M | 3,300 | 12 | 0.21 | 0.00 | 0.44 | |||||
| 6 wk | 7 | 0.58 | 0.00 | 0.67 | |||||||
| 14 wk | 10 | 0.30 | 0.11 | 0.67 | |||||||
| IU5 | Mother | 4.45 | Yes | Yes | 0.45 | 14 | 2.60 | 0.24 | 4.9 | ||
| Neonate | M | 3,500 | 13 | 0.95 | 0.00 | 3.28 | |||||
| 6 wk | 11 | 1.90 | 0.00 | 3.79 | |||||||
| 14 wk | 11 | 2.10 | 0.00 | 3.79 | |||||||
| IU6 | Mother | 4.05 | No | No | 0.17 | 10 | 2.84 | 0.34 | 4.56 | ||
| Neonate | M | 3,100 | 9 | 0.56 | 0.00 | 1.49 | |||||
| 6 wk | 11 | 1.74 | 0.23 | 4.22 | |||||||
Six weeks and 14 weeks, infants at 6 and 14 weeks, respectively.
ROM, rupture of membrane.
n, number of single sequences for each sample.
CVS, cervicovaginal secretions.
Global analysis of env sequences from mother-child pairs.
A total of 38 samples from 13 mother-child pairs were analyzed. A mean of 12.3 independent single-copy sequences were obtained per sample (range, 3 to 27 sequences). A neighbor-joining phylogenetic tree inferred from analysis of all the 468 available env sequences showed that, with the exception of those from mother-child pair IP2, sequences from each mother-child pair clustered together. Inclusion of the subtype reference sequences selected by the Los Alamos National Laboratory HIV sequence database (A-U455, A-92UG.37.1, B-HXB2, B-JRFL, B-OYI, B-RF, C-ETH2220, C-92BR025.8, D-NDK, D-Z2Z6, D-ELI, F-93BR0201, H-90CR056.1, J-SE9208-9, J-SE9173-3, and G-SE6165) revealed that, withthe exception of those from IP2, all sequences clustered in the subtype A branch of the tree. For IP2, 5 maternal and 13 infant sequences clustered in the subtype A branch and 8 maternal and 4 infant sequences clustered in the subtype D branch, indicating a dual infection in both the mother and the infant. All viruses sequenced had neutral or negatively charged amino acids at positions 11 and 25 of the V3 loop and might therefore be considered R5 or NSI (non-syncytium-inducing) viruses (17).
Intrapatient variation for each mother-child pair.
Sequences from each mother-child pair were aligned, and after removal of all columns with gaps the alignment was used to calculate the intrapatient genetic distance for each specimen. Results are presented in Table 1. The mean genetic distance was higher in the mothers (mean, 3.98%; range, 0.5 to 16.8%) than in the first samples from the children (mean, 1.6%; range, 0.04 to 17.1%). Only in pair IP2 did the infant show a higher genetic variability (17.1%) than the mother (16.5%).
For the six children infected in utero there were increases in genetic diversity between the sample obtained at birth and the sample taken at 6 weeks (mean increase of 0.5%) and between the samples taken at 6 and 14 weeks (mean increase, 0.2%; no 14-week sample was available for infant IU6).
A low genetic diversity (less than 0.2%), highly indicative of a homogeneous infection, was observed in six children, two infected in utero (IU1 and IU2) and four infected intrapartum (IP1, IP3, IP4, and IP7). A high intrapatient diversity (1% or more) was observed in three children, one with an in utero infection (IU5) and two with intrapartum infections (IP2 and IP6). The remaining four children (IU3, IU4, IP5, and IP7) had mean genetic diversities between 0.2 and 1%.
Phylogenetic analysis of mother-child pairs.
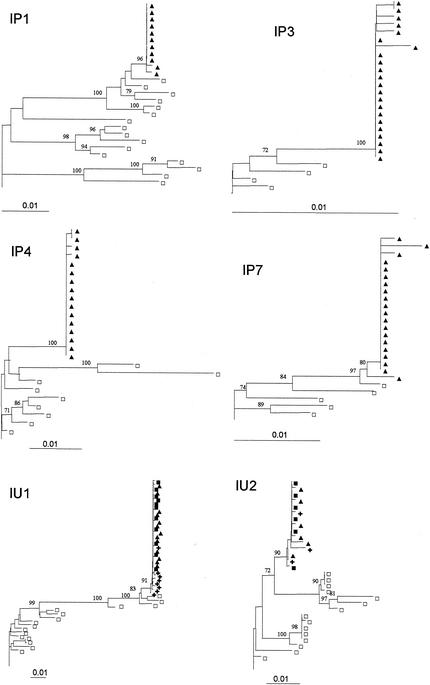
Figure 1 presents the neighbor-joining phylogenetic trees for the six mother-child pairs for whom a homogeneous infection of the infant was presumed based on the level of genetic diversity. All trees revealed high similarity between the sequences from the children and the mothers, with a tight clustering of the infants' sequences in a single branch of the tree. Branch separation was supported by high bootstrap values (mean, 92.7%).
FIG. 1.
Rooted neighbor-joining trees of HIV-1 env (V1 to V4) DNA nucleotide sequences for the six mother-child pairs with a low intrapatient genetic distance in the children. Bootstrap values are expressed as percentages per 1,000 replicates. Only bootstrap proportions higher than 70% are indicated. □, one maternal sequence; ▪, one child sequence isolated at birth; ▴, one child sequence isolated at 6 weeks; +, one child sequence isolated at 14 weeks.
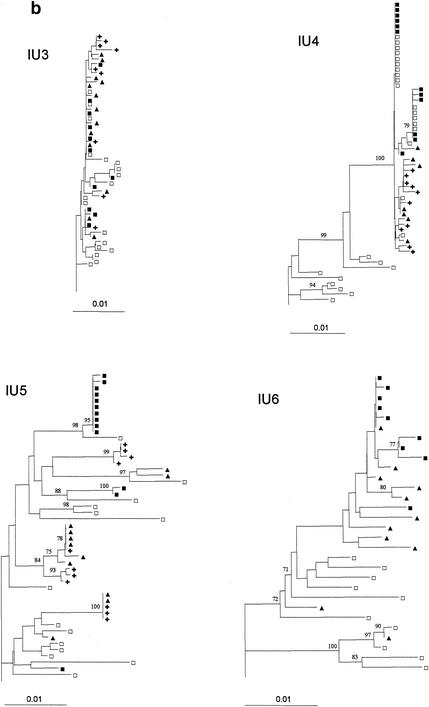
Figure 2 presents the neighbor-joining phylogenetic trees for the seven remaining mother-child pairs. In contrast to what was found for the child sequences presented in Fig. 1, substantial genetic heterogeneity was detected in these infants' sequences. The sequences of the infants are intermingled with the sequences obtained from the mother. This results in very similar distributions of the infants' and the mothers' sequences and supports the transmission of multiple maternal variants in IU3, IU5, and IP2. In IU3 the sequences from both the mother and the child had only limited heterogeneity, possibly indicating a relatively recent infection in the mother.
FIG. 2.
Rooted neighbor-joining trees of HIV-1 env (V1 to V4) DNA nucleotide sequences for the seven mother-child pairs with substantial intrapatient genetic distances in the children. Bootstrap values are expressed as percentages per 1,000 replicates. Only bootstrap proportions higher than 70% are indicated. □, one maternal sequence; ▪, one child sequence isolated at birth; ▴, one child sequence isolated at 6 weeks; +, one child sequence isolated at 14 weeks.
For IU4 and IP5 the infants' sequences are clustered in one branch. This branch also contains part of the mothers' sequences but is separated from the other maternal sequences (with 100% bootstrap support). Although the genetic diversity between the infant sequences from IU4 and IP5 is relatively low, the tree topology strongly supports the transmission of a few closely related maternal variants.
For IP6 and IU6 the infant sequences are found in, respectively, two and three branches of the tree. The topology of the tree from IP6 reflects the transmission of two distinct maternal variants. For IU6, 95% of the sequences obtained from the infant, including all sequences from the sample taken at birth, cluster in one branch, though the confidence level for clustering is low. Two of the sequences obtained at 6 weeks show a higher homology to maternal variants than to the other infants' variants.
Evolution of viral sequences after in utero infection.
For the two children with an in utero infection and a highly homogeneous virus population at birth (IU1 and IU2) the virus population remained relatively homogeneous at 6 and 14 weeks. The maximum genetic distances between the sequences obtained at birth and those obtained at 14 weeks were 0.5% for IU1 and 0.7% for IU2. All infants' samples remained clustered in the same branch of the phylogenetic tree.
For the four children with in utero infections and heterogeneous virus populations at birth the mean genetic distance remained constant for IU3 and IU4 but increased over time for IU5 and in IU6.
Factors influencing the transmission of multiple variants.
A comparison of factors known to be associated with mother-to-child transmission, such as maternal viral load, presence of proviral DNA and viral RNA in the CVS, and duration of membrane rupture, between the mother-child pairs characterized by transmission of a single virus variant and the other pairs was performed. There was a tendency for higher viral load, higher frequency of detection of HIV-1 DNA and RNA in the CVS, and longer duration of membrane rupture in the seven mothers who transmitted more variants than in the six mothers who transmitted only one variant (maternal mean log viral loads, 4.48 versus 4.38; detectable proviral DNA in the CVS in six of seven mothers versus four of six mothers; detectable viral RNA in the CVS in five of seven mothers versus one of six mothers; mean durations of membrane rupture, 5.64 versus 3.82 h). The total number of samples was too low to perform reliable statistical analysis.
The influence of viral and proviral load in breast milk could not be assessed because breast milk samples were not available.
DISCUSSION
This study attempted to assess viral heterogeneity in 13 infected children and to compare the viral heterogeneity in infants infected in utero with that in infants infected at or shortly after birth. Samples were taken the day after birth and at the ages of 6 and 14 weeks. Due to the early time points for sampling, heterogeneity resulting from viral evolution in the children after transmission is supposed to be low. Shankarappa et al. showed that in adults substitutions in a partial env fragment accumulated at a rate of approximately 1% per year (25). Although we are unable to estimate the infection time in an in utero infection, we can assume that infants with an “early” in utero infection have a good chance of having clinical symptoms at birth. With the exception of one infant (IU3) which was born with slightly enlarged lymph nodes, none of the infants studied showed any sign of infection at clinical examination the day after birth. The mean birth weight in the in utero-infected infants was higher than that of the infants infected at or after birth (2,958 versus 2,779 g).
Homogeneous virus populations were detectable in 6 (46.2%) of the 13 children studied and were seen after intrapartum or early postpartum infection (in 4 children) as well as after in utero infection (in 2 children), confirming results reported recently by Dickover et al. (6). In seven (53.8%) of the children (four with in utero infections and three with peripartum infections) high genetic diversity indicated infection with more than one maternal variant. Phylogenetic analysis for these seven mother-child pairs showed a spectrum of different tree topologies compatible with either infection with multiple genetically diverse maternal variants as in IU5, infection with a few genetically diverse variants as in IP6, infection with several closely linked variants as in IU4 and IP5, or a combination of the last two as in IU6.
The number of infants found to be infected with a homogeneous virus population is lower than expected from the results of other studies (1, 19, 24, 30). Technical differences can contribute to this finding since in most of the studies the information was obtained through clonal sequencing or heteroduplex mobility assay (HMA). HMA is known to reveal only genetic diversities of about 1% and more (5). Clonal sequencing on the other hand might select for and overestimate certain variants in the population (13). Becker-Pergola et al. recently showed that mixtures of HIV-1 variants are more readily detected by replicate amplification than by cloning techniques (2). We used limiting-dilution PCR followed by sequencing of the single-copy PCR products. Although labor intensive, this method allows a very accurate profile of the HIV-1 heterogeneity in a single sample to be obtained (20). Moreover, we sequenced a relatively large fragment (about 1,000 nucleotides, comprising V1 to V4) of the env gene, while most studies restricted their analysis to the V3 region.
It has to be taken into account that, of the 13 mother-child pairs studied here, 12 were infected with an HIV-1 strain classified subtype A. Most of the studies reported in the literature were performed on mother-infant pairs of non-African origin. Mechanisms of mother to child virus transmission might differ among subtypes, although so far there are no data to support this possibility. In one mother-child pair a dual infection with subtype A and D viruses was detected. Dual transmission of A and D has been described; one case was reported by Mellquist et al. in 1999 (16), and more recently Murray et al. (18) described three cases in which HIV-1-positive mothers with mixed HIV-1 infections transmitted both subtypes to their children.
The low heterogeneity of HIV-1 env sequences in perinatally infected infants has led some researchers to hypothesize that a selection mechanism by which viruses with specific phenotypic properties are preferentially transmitted is operating (11, 29). However, up to now studies have failed to reveal distinct phenotypic or genotypic characteristics of transmitted viruses (1, 10). The observation reported here that, for several of the infants considered to be infected with multiple variants, the infants' variants clustered in one branch of the phylogenetic tree might indicate some kind of selection mechanism whereby only viruses with specific characteristics are able to pass the mucosa or are able to multiply in the infants’ tissues surrounding the contact site. However the variants transmitted to the children might also have been part of a subpopulation of actively replicating viruses in the mother at the time of infection, or children might have been infected with closely linked viral variants that were the most abundant at the site of contact. Indeed, the latter hypothesis cannot be excluded since independent evolution of different “quasispecies” in blood and CVS has been described previously (21, 32).
Selection might also act posttransmission, i.e., there would be transmission of multiple variants and a quick outgrowth of the strain with the highest fitness. Since we analyzed proviral DNA in the early phase of the infection, this process would have resulted in phylogenetic trees in which different variants are present but one is dominant. Such a tree topology was not observed. Another hypothesis proposes subsequent infections at different times as a mechanism to explain sequence heterogeneity in infected infants (3). Newborns infected in utero are expected to have the highest probability of being infected at different time points. Two of the six children with an in utero infection that were studied here had highly homogeneous virus populations at birth, and both populations remained homogeneous at 6 and at 14 weeks. Both children were breast-fed. No evidence for reinfection was found. Populations in the four infants with in utero infections and with heterogeneous virus populations at 14 weeks already showed a marked degree of heterogeneity at birth, making it impossible to discriminate between simultaneous infection and infection at a later time point. In IU3 the overall genetic diversity is too low to allow conclusions. In IU4, IU5, and IU6 there is only moderate intermingling between the sequences from the first sample and those obtained later. This might indicate reinfection but might also reflect evolution and selection in the child.
Of the viral factors known to be associated with mother-to-child transmission the difference in frequency of detectable viral RNA in the CVS was the most pronounced, with detectable RNA in the CVS of five of the seven women who transmitted multiple variants but in only one of the six women who transmitted a single variant. Presence of viral RNA in the CVS was found to be highly predictive of HIV-1 transmission in the study from which we have extracted the cases described here (7).
In conclusion, the results of this study, performed on a relatively large number of mother-child pairs infected with HIV-1 subtype A virus and using limiting-dilution PCR sequencing on a long fragment of the HIV-1 env gene, showed that, after transmission of the virus from mother to child, a homogeneous virus population, indicative of transmission of a single maternal variant or of early selection, was established in approximately one-half of the children. Homogeneous virus populations were detectable after intrapartum or early postpartum infection as well as after in utero infection. More-heterogeneous infections were found in 7 of the 13 children studied. In six of these, transmission resulted in a loss of sequence heterogeneity, i.e., less heterogeneity in sequences from the infants than in those from the mothers. This finding and the observation that the phylogenetic trees of the mother-child pairs present a whole range of different tree topologies reveal that mother-to-child HIV-1 transmission can result in a monoclonal or oligoclonal infection as well as a polyclonal infection. Inoculum level, exposure time, and host-specific factors such as the condition of the skin and the mucosal surfaces might be important factors determining viral heterogeneity in the infected child. The fact that, although there are not enough data for reliable statistical analysis, the maternal factors known to be related to transmission (viral load, presence of HIV-1 DNA in the CVS, presence of HIV-1 RNA in the CVS, and duration of membrane rupture) were all slightly higher or more frequently present in the group of women who transmitted more than one variant to the infant is an additional argument to support the importance of the effect of inoculum level and duration of exposure on virus heterogeneity after transmission.
Understanding the underlying mechanisms of selection of sequence variability and, if possible, their relationship to HIV-1 pathogenesis and clinical manifestation may be of assistance in the design of preventive vaccines or immunotherapies.
REFERENCES
- 1.Ahmad, N., B. M. Baroudy, R. C. Baker, and C. Chappey. 1995. Genetic analysis of human immunodeficiency virus type 1 envelope V3 region isolates from mothers and infants after perinatal transmission. J. Virol. 69:1001-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker-Pergola, G., J. L. Mellquist, L. Guay, F. Mmiro, C. Ndugwa, P. Kataaha, J. B. Jackson, and S. H. Eshleman. 2000. Identification of diverse HIV type 1 subtypes and dual HIV type 1 infection in pregnant Ugandan women. AIDS Res. Hum. Retroviruses 16:1099-1104. [DOI] [PubMed] [Google Scholar]
- 3.Briant, L., C. M. Wade, J. Puel, A. J. Brown, and M. Guyader. 1995. Analysis of envelope sequence variants suggests multiple mechanisms of mother-to-child transmission of human immunodeficiency virus type 1. J. Virol. 69:3778-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burger, H., B. Weiser, K. Flaherty, J. Gulla, P. N. Nguyen, and R. A. Gibbs. 1991. Evolution of human immunodeficiency virus type 1 nucleotide sequence diversity among close contacts. Proc. Natl. Acad. Sci. USA 88:11236-11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delwart, E. L., E. G. Shpaer, J. Louwagie, F. E. McCutchan, M. Grez, H. Rubsamen-Waigmann, and J. I. Mullins. 1993. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science 262:1257-1261. [DOI] [PubMed] [Google Scholar]
- 6.Dickover, R. E., E. M. Garratty, S. Plaeger, and Y. J. Bryson. 2001. Perinatal transmission of major, minor, and multiple maternal human immunodeficiency virus type 1 variants in utero and intrapartum. J. Virol. 75:2194-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaillard, P., C. Verhofstede, F. Mwanyumba, P. Claeys, V. Chohan, K. Mandaliya, J. Bwayo, J. Plum, and M. Temmerman. 2000. Exposure to HIV-1 during delivery and mother-to-child transmission. AIDS 14:2341-2348. [DOI] [PubMed] [Google Scholar]
- 8.Gaillard, P., F. Mwanyumba, C. Verhofstede, P. Claeys, V. Chohan, E. Goetghebeur, K. Mandaliya, J. Ndinya-Achola, and M. Temmerman. 2001. Vaginal lavage with chlorhexidine during labour to reduce mother-to-child HIV transmission: clinical trial in Mombasa, Kenya. AIDS 15:389-396. [DOI] [PubMed] [Google Scholar]
- 9.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 98/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 10.Kampinga, G. A., A. Simonon, P. Van de Perre, E. Karita, P. Msellati, and J. Goudsmit. 1997. Primary infections with HIV-1 of women and their offspring in Rwanda: findings of heterogeneity at seroconversion, coinfection, and recombinants of HIV-1 subtypes A and C. Virology 227:63-76. [DOI] [PubMed] [Google Scholar]
- 11.Kliks, S., C. H. Contag, H. Corliss, G. Learn, A. Rodrigo, D. Wara, J. I. Mullins, and J. A. Levy. 2000. Genetic analysis of viral variants selected in transmission of human immunodeficiency viruses to newborns. AIDS Res. Hum. Retroviruses 16:1223-1233. [DOI] [PubMed] [Google Scholar]
- 12.Lamers, S. L., J. W. Sleasman, J. X. She, K. A. Barrie, S. M. Pomeroy, D. J. Barrett, and M. M. Goodenow. 1994. Persistence of multiple maternal genotypes of human immunodeficiency virus type I in infants infected by vertical transmission. J. Clin. Investig. 93:380-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu, S. L., A. G. Rodrigo, R. Shankarappa, G. H. Learn, L. Hsu, O. Davidov, L. P. Zhao, and J. I. Mullins. 1996. HIV quasispecies and resampling. Science 273:415-416. [DOI] [PubMed] [Google Scholar]
- 14.Long, E. M., H. L. Martin, Jr., J. K. Kreiss, S. M. Rainwater, L. Lavreys, D. J. Jackson, J. Rakwar, K. Mandaliya, and J. Overbaugh. 2000. Gender differences in HIV-1 diversity at time of infection. Nat. Med. 6:71-75. [DOI] [PubMed] [Google Scholar]
- 15.McGowan, J. P., and S. S. Shah. 2000. Prevention of perinatal HIV transmission during pregnancy. J. Antimicrob. Chemother. 46:657-658. [DOI] [PubMed] [Google Scholar]
- 16.Mellquist, J. L., G. Becker-Pergola, J. Gu, L. Guay, L. Himes, P. Kataaha, F. Mmiro, C. Ndugwa, J. B. Jackson, and S. H. Eshleman. 1999. Dual transmission of subtype A and D HIV type 1 viruses from a Ugandan woman to her infant. AIDS Res. Hum. Retroviruses 15:217-221. [DOI] [PubMed] [Google Scholar]
- 17.Milich, L., B. H. Margolin, and R. Swanstrom. 1997. Patterns of amino acid variability in NSI-like and SI-like V3 sequences and a linked change in the CD4-binding domain of the HIV-1 Env protein. Virology 239:108-118. [DOI] [PubMed] [Google Scholar]
- 18.Murray, M. C., J. E. Embree, S. G. Ramdahin, A. O. Anzala, S. Njenga, and F. A. Plummer. 2000. Effect of human immunodeficiency virus (HIV) type 1 viral genotype on mother-to-child transmission of HIV-1. J. Infect. Dis. 181:746-749. [DOI] [PubMed] [Google Scholar]
- 19.Pasquier, C., C. Cayrou, A. Blancher, C. Tourne-Petheil, A. Berrebi, J. Tricoire, J. Puel, and J. Izopet. 1998. Molecular evidence for mother-to-child transmission of multiple variants by analysis of RNA and DNA sequences of human immunodeficiency virus type 1. J. Virol. 72:8493-8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poss, M., H. L. Martin, J. K. Kreiss, L. Granville, B. Chohan, P. Nyange, K. Mandaliya, and J. Overbaugh. 1995. Diversity in virus populations from genital secretions and peripheral blood from women recently infected with human immunodeficiency virus type 1. J. Virol. 69:8118-8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poss, M., A. G. Rodrigo, J. J. Gosink, G. H. Learn, D. D. V. Panteleeff, H. L. Martin, Jr., J. Bwayo, J. K. Kreiss, and J. Overbaugh. 1998. Evolution of envelope sequences from the genital tract and peripheral blood of women infected with clade A human immunodeficiency virus type 1. J. Virol. 72:8240-8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodrigo, A. G., P. C. Goracke, K. Rowhanian, and J. I. Mullins. 1997. Quantitation of target molecules from polymerase chain reaction-based limiting dilution assays. AIDS Res. Hum. Retroviruses 13:737-742. [DOI] [PubMed] [Google Scholar]
- 23.Saag, M. S., B. H. Hahn, J. Gibbons, Y. Li, E. S. Parks, W. P. Parks, and G. M. Shaw. 1988. Extensive variation of human immunodeficiency virus type-1 in vivo. Nature 334:440-444. [DOI] [PubMed] [Google Scholar]
- 24.Scarlatti, G., T. Leitner, E. Halapi, J. Wahlberg, P. Marchisio, M. A. Clerici-Schoeller, H. Wigzell, E. M. Fenyo, J. Albert, M. Uhlen, et al. 1993. Comparison of variable region 3 sequences of human immunodeficiency virus type 1 from infected children with the RNA and DNA sequences of the virus populations of their mothers. Proc. Natl. Acad. Sci. USA 90:1721-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simmonds, P., L. Q. Zhang, F. McOmish, P. Balfe, C. A. Ludlam, and A. J. Brown. 1991. Discontinuous sequence change of human immunodeficiency virus (HIV) type 1 env sequences in plasma viral and lymphocyte-associated proviral populations in vivo: implications for models of HIV pathogenesis. J. Virol. 65:6266-6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van't Wout, A. B., N. A. Kootstra, G. A. Mulder-Kampinga, N. Albrecht-van Lent, H. J. Scherpbier, J. Veenstra, K. Boer, R. A. Coutinho, F. Miedema, and H. Schuitemaker. 1994. Macrophage-tropic variants initiate human immunodeficiency virus type1 infection after sexual, parenteral, and vertical transmission. J. Clin. Investig. 94:2060-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wike, C. M., B. T. Korber, M. R. Daniels, C. Hutto, J. Munoz, M. Furtado, W. Parks, A. Saah, M. Bulterys, J. B. Kurawige, et al. 1992. HIV-1 sequence variation between isolates from mother-infant transmission pairs. AIDS Res. Hum. Retroviruses 8:1297-1300. [DOI] [PubMed] [Google Scholar]
- 30.Wolinsky, S. M., C. M. Wike, B. T. Korber, C. Hutto, W. P. Parks, L. L. Rosenblum, K. J. Kunstman, M. R. Furtado, and J. L. Munoz. 1992. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science 255:1134-1137. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, L. Q., P. MacKenzie, A. Cleland, E. C. Holmes, A. J. Brown, and P. Simmonds. 1993. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J. Virol. 67:3345-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu, T., H. Mo, N. Wang, D. S. Nam, Y. Cao, R. A. Koup, and D. D. Ho. 1993. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science 261:1179-1181. [DOI] [PubMed] [Google Scholar]