Abstract
The ovine betaretroviruses jaagsiekte sheep retrovirus (JSRV) and enzootic nasal tumor virus (ENTV) cause contagious cancers in the lungs and upper airways of sheep and goats. Oncogenic transformation assays using mouse and rat fibroblasts have localized the transforming activity to the Env proteins encoded by these viruses, which require the putative lung and breast cancer tumor suppressor hyaluronidase 2 (Hyal2) to promote virus entry into cells. These results suggested the hypothesis that the JSRV and ENTV Env proteins cause cancer by inhibiting the tumor suppressor activity of Hyal2. Consistent with this hypothesis, we show that human Hyal2 and other Hyal2 orthologs that can promote virus entry, including rat Hyal2, can suppress transformation by the Env proteins of JSRV and ENTV. Furthermore, we provide direct evidence for binding of the surface (SU) region of JSRV Env to human and rat Hyal2. However, mouse Hyal2 did not mediate entry of virions bearing JSRV or ENTV Env proteins, bound JSRV SU poorly if at all, and did not suppress transformation by the JSRV or ENTV Env proteins, indicating that mouse Hyal2 plays no role in transformation of mouse fibroblasts and that the Env proteins can transform at least some cells by a Hyal2-independent mechanism. Expression of human Hyal2 in mouse cells expressing JSRV Env caused a marked reduction in Env protein levels, indicating that human Hyal2 suppresses Env-mediated transformation in mouse cells by increasing Env degradation rather than by exerting a more general Env-independent tumor suppressor activity.
Jaagsiekte sheep retrovirus (JSRV) and enzootic nasal tumor virus (ENTV) are closely related betaretroviruses that are the causative agents of contagious adenocarcinomas of sheep and goats (7). JSRV induces oncogenic transformation of bronchiolar and alveolar epithelial cells, including type II secretory pneumocytes, while ENTV induces oncogenic transformation of the glandular epithelial cells of the nasal mucosa. Both viruses induce the secretion of copious nasal fluid containing the viruses. JSRV can induce multifocal cancer in newborn sheep in as little as 2 weeks (20), indicating that JSRV carries an acutely active oncogene and that insertional mutagenesis by this retrovirus is not critical for oncogenesis. However, both JSRV and ENTV are simple retroviruses that express retroviral structural and enzymatic proteins without apparent oncogenes derived from the host organism.
Recently we and others have demonstrated that the Env proteins from both JSRV (11, 19) and ENTV (1, 5), but not other viral proteins, can transform mouse and rat fibroblasts in culture and thus are likely to be responsible for oncogenesis in animals. Site-directed mutagenesis has shown that a YXXM motif present in the cytoplasmic tails of the JSRV and ENTV Env proteins, a consensus binding site for phosphatidylinositol 3-kinase (PI3K) after tyrosine phosphorylation of the motif, is critical for transformation of rodent fibroblasts by these proteins (1, 16). Furthermore, Akt, a downstream mediator of PI3K signaling, is activated in JSRV and ENTV Env-transformed cells, and this activation is reversed by treatment with the PI3K inhibitor LY294002, indicating that activation is PI3K dependent (1). These results suggest that JSRV and ENTV Env proteins transform rodent fibroblasts by activating the PI3K pathway. However, a recent study shows that the YXXM motif is not required for JSRV transformation of DF-1 chicken fibroblasts (2), suggesting that more than one mechanism may be involved in Env-mediated transformation.
Given the role of the Env protein in oncogenic transformation, we hypothesized that the cellular receptor that binds Env and mediates retrovirus entry might play a role in virus transformation. To identify the receptor, we constructed retroviral vectors using Moloney murine leukemia virus (MoMLV) gag and pol, a MoMLV-based retroviral vector encoding marker proteins, and the Env protein from JSRV. This JSRV vector was able to transduce human cells but not hamster cells, and by screening hamster cells carrying different assortments of human DNA fragments, Hyal2 was identified as the glycosylphosphatidylinositol-anchored cell surface protein required for JSRV vector entry into cells (19). Subsequently it was shown that Hyal2 is also required for ENTV Env-mediated vector entry into cells (1, 5). Interestingly, Hyal2 is located in a region of human chromosome 3p21.3 that is frequently deleted in human lung and breast cancer and is thought to harbor one or more tumor suppressor genes (10). This association led us to hypothesize that Hyal2 is a tumor suppressor and that Env binding to Hyal2 inhibits this function, resulting in oncogenic transformation. However, neither mouse nor rat cells are transduced by MoMLV-based retroviral vectors bearing JSRV or ENTV Env proteins, indicating that mouse and rat Hyal2 cannot bind the viral Env proteins and thus would be unlikely participants in Env-mediated viral oncogenesis.
Here we show that human Hyal2 can indeed suppress transformation by JSRV and ENTV Env proteins. To see if Env and Hyal2 interact directly, we constructed a hybrid JSRV Env surface (SU) domain-human immunoglobulin G (IgG) Fc fragment fusion protein, and we show that the JSRV SU binds to human Hyal2 expressed on cells. To examine the properties of the Hyal2 proteins expressed by the NIH 3T3 mouse and 208F rat cells used in the assays for Env transformation, we cloned the Hyal2 cDNAs from both cell types. Surprisingly, overexpressed rat Hyal2 can mediate JSRV vector entry, can bind the JSRV SU-IgG fusion protein, and can suppress transformation by JSRV and ENTV Env proteins. However, the mouse protein exhibits none of these activities. Thus, it is unlikely that mouse Hyal2 plays any direct role in transformation by the JSRV and ENTV Env proteins in mouse cells. We show by immunofluorescence analysis that expression of human Hyal2 in mouse cells expressing JSRV Env protein causes a reduction in Env protein levels, and we conclude that this mechanism explains the tumor suppressor activity of human and rat Hyal2 in cells transformed by the JSRV or ENTV Env proteins.
MATERIALS AND METHODS
Cell culture.
Cell lines used here include 208F Fischer rat embryo fibroblasts (17), SSF-123 primary sheep skin fibroblasts (a gift from William Osborne, University of Washington, Seattle), a morphologically flat subclone of NIH 3T3 Swiss mouse embryo fibroblasts suitable for transformation studies (a gift from Maxine Linial, Fred Hutchinson Cancer Research Center, Seattle, Wash.; originally from Doug Lowy, National Cancer Institute, Bethesda, Md.), 293T human embryonic kidney cells (6), and PJ4 JSRV pseudotype (18), PN172 ENTV pseudotype (5), and PT67 10A1-MLV pseudotype (12) retrovirus packaging cells. Unless otherwise stated, cells were grown in Dulbecco's modified Eagle medium (DMEM) with high glucose (4.5 g/liter) and 10% fetal bovine serum at 37°C in a 10% CO2-90% air atmosphere at 100% relative humidity.
Cloning of mouse and rat Hyal2 cDNAs and corresponding genomic regions.
Hyal2 cDNAs were obtained by reverse transcription-PCR (RT-PCR) using the SuperScript One-Step RT-PCR System with Platinum Taq (Invitrogen, Carlsbad, Calif.). The mouse Hyal2 cDNA was generated from total RNA isolated from NIH 3T3 mouse cells with forward primer 5′ AGC TGC TAC CAG GCA GGT AAC and reverse primer 5′ TGG GAG CAC TGC CTA CTC CAG. Rat Hyal2 cDNA was generated from total RNA isolated from 208F rat cells with forward primer 5′ TGC GAG TTC CTG AGC TGC TAC and reverse primer 5′ GCC AGC TGG ACT GCT ATC TGC. RT-PCR products were cloned into the mammalian expression vector pCR3.1 (Invitrogen). The genomic regions corresponding to the mouse Hyal2 and rat Hyal2 cDNAs were also amplified from NIH 3T3 mouse cell or 208F rat cell genomic DNA with the cDNA primer sets described above. Genomic PCR products were subcloned into the TOPO TA vector (Invitrogen).
Plasmid expression vectors.
Plasmids used to express JSRV Env (pSX2.Jenv [18]), ENTV Env (pSX2.Eenv [5]), and 10A1-MLV Env (pSX2 [12]) contain the respective Env coding regions cloned into the pSX2 expression vector, which employs a MoMLV promoter and enhancers, splice signals, and the early polyadenylation signal from simian virus 40 to drive transcription. A FLAG-tagged version of the JSRV Env protein was made by adding a sequence encoding a FLAG tag (DYKDDDDK) at the 3′ end of the Env coding region in the JSRV Env expression vector. The pFBJ/R plasmid, which expresses a highly active Fos oncoprotein (14), was used as a positive control for transformation.
A JSRV SU domain-human IgG Fc fragment (JSRV SU-IgG) fusion protein expression construct was generated by cloning the JSRV SU coding region into a vector designed for expression of an amphotropic MLV SU-human IgG Fc fusion protein (8) in place of the amphotropic SU. To avoid possible cleavage by host proteases, the last 7 amino acids of JSRV SU (ALSRPKR) were deleted in this construct. Fusion protein expression in this construct is driven by a human cytomegalovirus immediate-early promoter/enhancer.
Retrovirus vectors.
The LAPSN vector (15) expresses human placental alkaline phosphatase (AP) from the retroviral long terminal repeat (LTR) and neomycin phosphotransferase (Neo) from an internal simian virus 40 early promoter. Retrovirus vectors that express Hyal1 and Hyal2 cDNAs from different species were made by cloning the cDNAs into the LXSN retroviral vector (13). Stable vector-producing cell lines were generated as described previously (13), by transfection of the plasmids into PE501 ecotropic packaging cells, harvest of virus, transduction of PT67 10A1-MLV pseudotype packaging cells (12), and isolation of transduced clones in G418. The clones that produced the highest titer of unrearranged vector were identified and used, and in the case of the mouse and rat Hyal2 vector-producing cells, the Hyal2 inserts were sequenced following amplification by PCR to confirm the absence of mutations. To harvest vectors, the culture medium was incubated with vector-producing packaging cells for 12 to 16 h; then the medium was filtered to remove cells and frozen at −70°C until use.
Transformation assays.
Plasmids were assayed for transforming activity by transfection of NIH 3T3 cells using a standard calcium phosphate transfection procedure (13) or by transfection of 208F cells using a modified calcium phosphate transfection procedure (3). These methods gave the highest transfection efficiencies for the two cell types. For the assay, cells were seeded at 5 × 105 per 6-cm dish and were transfected the next day with 10 μg of the test plasmid plus 1 μg of a plasmid encoding AP (pLAPSN). One day after transfection, the cells were trypsinized, divided 1:5 into 6-cm dishes, and fed every 3 to 4 days thereafter. After reaching confluence, the NIH 3T3 cells were fed with DMEM plus 5% fetal bovine serum (FBS) and the 208F cells were fed with the same medium plus 1 μM dexamethasone. About 2 weeks after transfection, the transformed foci were counted, the cells were fixed and stained for AP, and AP+ foci were counted as a measure of transfection efficiency.
Production and purification of the JSRV SU-IgG fusion protein.
The JSRV SU-IgG construct described above was transfected by calcium phosphate coprecipitation (3) into 293T cells. Cells were fed with low-IgG serum (Life Technologies, Rockville, Md.) 1 day after transfection, and the medium was harvested 48 to 72 h posttransfection and filtered through 0.45-μm-pore-size filters. For binding studies, either this medium was used directly or the JSRV SU-IgG fusion protein was purified by affinity chromatography using protein A columns and was concentrated by using Ultrafree Biomax-50K centrifugal filter devices (Millipore Inc., Bedford, Mass.). JSRV SU-IgG was detected by denaturing polyacrylamide gel separation followed by immunoblot analysis, and the concentration was determined by comparison of the JSRV SU-IgG band with albumin protein standards following Coomassie blue staining.
In vitro binding assay, flow cytometry, and Scatchard analysis.
Binding between Hyal2 orthologs and the JSRV SU-IgG fusion protein was performed by using a protocol similar to that of Kurre et al. (8). Briefly, cells were resuspended by treatment with 5 mM EDTA in phosphate-buffered saline (PBS) (without calcium and magnesium) and were washed three times with PBS (with calcium and magnesium) containing 2% FBS. Next, 5 × 105 cells were incubated with the purified or unpurified JSRV SU-IgG fusion protein on ice for 3.5 h, washed three times, and incubated with a fluorescein isothiocyanate (FITC)-conjugated rabbit antibody against human IgG Fc (DAKO Inc., Glostrup, Denmark) on ice for 45 to 60 min. Cells were washed twice, resuspended in PBS containing 2% FBS and 2 μg of propidium iodide/ml, and analyzed by use of a fluorescence-activated cell sorter (FACS; Becton Dickinson, San Jose, Calif.).
For Scatchard analysis, the free JSRV SU-IgG was estimated to be equal to the total JSRV SU-IgG molecules added, assuming that only a small percentage binds to cells (which we confirmed experimentally; see Results) and assuming that all added protein was active. Bound JSRV SU-IgG was measured by FACS, and a conversion factor relating FACS fluorescence per cell to the number of JSRV SU-IgG molecules bound per cell was determined as follows. The fluorescence of a known amount of an FITC-conjugated secondary antibody was measured in a spectrofluorometer to calculate the fluorescence per molecule of antibody. The fluorescence of a known number of cells incubated with JSRV SU-IgG and the secondary antibody (cells were washed before and after antibody incubation) was measured in the spectrofluorometer to determine the number of JSRV SU-IgG molecules bound per cell, assuming a one-to-one interaction between the JSRV SU-IgG fusion protein and the secondary antibody. The same cells were analyzed by FACS to provide a conversion factor relating FACS fluorescence to molecules bound per cell.
Env immunostaining.
NIH 3T3 cells transformed by pSX2.Jenv-FLAG were transduced by human Hyal2, human Hyal1, rat Hyal2, mouse Hyal2, or the LAPSN vector on day 1 and were trypsinized and seeded at varying ratios onto coverslips in 12-well plates in the presence of G418 (750 μg/ml; active concentration) on day 2. After 7 to 10 days of G418 selection, the expression level of JSRV Env in the cells was examined by immunostaining. Briefly, cells were fixed by 3.7% formaldehyde for 10 min, washed three times with PBS, permeabilized with 0.5% Triton X-100 for 5 min, washed twice, and blocked with 20% normal goat serum for 30 min at room temperature. Cells were then incubated with a mouse monoclonal anti-FLAG antibody (Sigma, St. Louis, Mo.) for 1 h at room temperature, washed three times with PBS containing 1 mM glycine, and then incubated with an FITC-conjugated goat anti-mouse antibody (Upstate Biotechnology, Lake Placid, N.Y.) at room temperature for 1 h. Cells were washed three times, mounted, and examined using a DeltaVision microscope (Applied Precision, Issaquah, Wash.).
Nucleotide sequence accession numbers.
The consensus cDNA sequences of mouse Hyal2 and rat Hyal2 have been deposited in GenBank with accession numbers AF535140 and AF535141, respectively.
RESULTS
JSRV and ENTV Env proteins can transform rodent fibroblasts with similar efficiencies.
Plasmids encoding the JSRV, ENTV, and 10A1-MLV Env proteins, and a plasmid that expresses a highly transforming mutant Fos oncoprotein derived from the FBJ/R retrovirus (14), were tested for oncogenic transformation activity in NIH 3T3 mouse and 208F rat fibroblasts (Table 1). The plasmids encoding the JSRV and ENTV Env proteins and the Fos oncoprotein all induced transformed foci with similar sizes in NIH 3T3 and 208F cells, while no foci were detected following transfection of the 10A1-MLV Env expression plasmid. The Fos plasmid induced fourfold more foci than either the ENTV or the JSRV Env plasmid in NIH 3T3 cells, while the three plasmids induced similar numbers of foci in 208F cells. These results show that both JSRV and ENTV Env proteins can transform rodent fibroblasts with similar efficiencies in the absence of other viral proteins. In contrast, the 10A1 Env protein is derived from an MLV that is believed to induce cancer by insertional mutagenesis and had no transforming activity in this assay.
TABLE 1.
JSRV and ENTV Env proteins transform cultured rodent fibroblastsa
| Expressed protein | No. of transformed foci per μg of DNA for the following cells:
|
|
|---|---|---|
| 208F | NIH 3T3 | |
| JSRV Env | 115 ± 18 | 28 ± 14 |
| ENTV Env | 106 ± 13 | 24 ± 6 |
| Fos | 100 ± 15 | 100 ± 50 |
| 10A1 Env | <0.3 | <0.3 |
Cells were transfected with 10 μg of plasmid DNA encoding JSRV Env (pSX2.Jenv), ENTV Env (pSX2.Eenv), Fos (pFBJ/R), or amphotropic 10A1-MLV hybrid Env (pSX2) plus 1 μg of plasmid pLAPSN by using calcium phosphate coprecipitation. Transformed foci were counted ∼2 weeks posttransfection, and AP staining was performed on the same day. For each cell type the transfection efficiency was similar for all plasmids, as measured by counting AP+ foci. Results are means from two to three experiments.
JSRV and ENTV vector transduction of rodent fibroblasts is low to undetectable, but the cells are rendered susceptible by expression of human Hyal2.
As shown here (Table 2) and reported previously (5, 18, 19), transduction of 208F rat cells by JSRV and ENTV vectors is undetectable (<2 focus-forming units [FFU]/ml) while transduction of NIH 3T3 mouse cells at low levels (<1 to 30 FFU/ml) can be detected in some experiments. Expression of human Hyal2 in these cells confers susceptibility to JSRV and ENTV vector transduction, while expression of human Hyal1 has no effect (Table 2) (5, 19). The same vector with a 10A1 MLV pseudotype transduced all of the NIH 3T3 and 208F cells and their derivatives at high levels, showing that the restriction to JSRV vector entry is due to the JSRV Env protein (data not shown). Thus, transformation of the mouse and rat cells can occur in the absence of functional receptors for JSRV and ENTV Env-mediated virus entry. However, it is still possible that the JSRV and ENTV Env proteins interact with the mouse and rat Hyal2 proteins to cause transformation but that these interactions are not sufficient to allow virus entry.
TABLE 2.
JSRV vector transduction of NIH 3T3 mouse cells and 208F rat cells with and without expression of Hyal2
| Target cellsa | Titer of LAPSN vectorb made by using:
|
|
|---|---|---|
| PJ4 cells | PT67 cells | |
| NIH 3T3/LXSN | 10 | 2 × 107 |
| NIH 3T3/LhHyal2SN | 6 × 105 | 2 × 107 |
| NIH 3T3/LhHyal1SN | 30 | 1 × 107 |
| 208F | <10 | 4 × 106 |
| 208F/LhHyal2SN | 2 × 104 | 4 × 106 |
| 208F/LhHyal1SN | <10 | 3 × 106 |
NIH 3T3 and 208F cells that express human Hyal2 and Neo, human Hyal1 and Neo, or Neo only were generated by transduction of the cells with the retrovirus vector LhHyal2SN, LhHyal1SN, or LXSN, respectively, and selection of the cells in G418.
Results are means of duplicate determinations from a typical experiment.
Expression of human Hyal2 inhibits transformation by JSRV and ENTV Env proteins in NIH 3T3 and 208F cells.
We used NIH 3T3 and 208F cells expressing an empty vector or the vectors encoding human Hyal1 or Hyal2 described above to test whether expression of a functional Hyal2 receptor would have any effect on transformation by JSRV and ENTV Env proteins (Table 3). Prior to manipulation, there were no visible differences in the phenotype of the cells expressing human Hyal1 or Hyal2 either in log-phase growth or at stationary phase, and the cells exhibited similar growth inhibition at confluence. Transformation by the plasmid encoding JSRV Env was similar in NIH 3T3 cells expressing the control vector LXSN or the human Hyal1 vector, while transformation was significantly suppressed in NIH 3T3 cells expressing human Hyal2. The same was true for transformation by the JSRV Env plasmid in 208F cells and for the ENTV Env plasmid in NIH 3T3 cells, although the suppression was not as great. Transformation by the plasmid expressing the Fos oncoprotein was unaffected by expression of human Hyal2 or human Hyal1 in the NIH 3T3 and 208F cells. Thus, human Hyal2 acts to suppress transformation by the JSRV and ENTV Env proteins, consistent with the model that Hyal2 is a tumor suppressor and the JSRV and ENTV Env proteins antagonize this activity.
TABLE 3.
Transformation by JSRV or ENTV Env protein of cells with and without human Hyal2 expressiona
| Cells | Vector expressed by cells | Transformation rate for the following plasmidb (% of value for LXSN vector):
|
||
|---|---|---|---|---|
| pSX2.Jenv | pSX2.Eenv | pFBJ/R | ||
| NIH 3T3 | LXSN | 100 | 100 | 100 |
| LhHyal2SN | 8 ± 2* | 22 ± 3* | 110 ± 8 | |
| LhHyal1SN | 112 ± 28 | 98 ± 18 | 104 ± 12 | |
| 208F | LXSN | 100 | 100 | 100 |
| LhHyal2SN | 32 ± 8* | ND | 102 ± 1 | |
| LhHyal1SN | 82 ± 29 | ND | 88 ± 12 | |
Cells transduced with and expressing the indicated vectors were seeded at 5 × 105 per 6-cm dish and were transfected the next day with 10 μg of the test plasmid and 1 μg of plasmid pLAPSN, which encodes AP. The day after transfection, the cells were trypsinized and split 1:5. Twelve days after transfection, transformed foci were counted, and AP+ foci were counted after fixation and staining for AP. The ratios of transformed foci to AP+ foci were calculated for each condition, and the transformation rate was calculated by dividing this ratio by the ratio obtained using cells expressing the control vector LXSN. For reference, the numbers of foci produced per microgram of plasmid DNA for pSX2.Jenv and pSX2.Eenv in NIH 3T3/LXSN and 208F/LXSN cells ranged from 25 to 28, while pFBJ/R induced 100 and 170 foci per μg of plasmid DNA, respectively, for these cells.
Results are means ± standard deviations from two to three experiments, each performed in triplicate. Asterisks indicate values statistically different (P < 0.05) from those obtained by using cells expressing the control vector LXSN. ND, not done.
Transfer of human Hyal2 to cells transformed by JSRV Env can reverse the transformed phenotype.
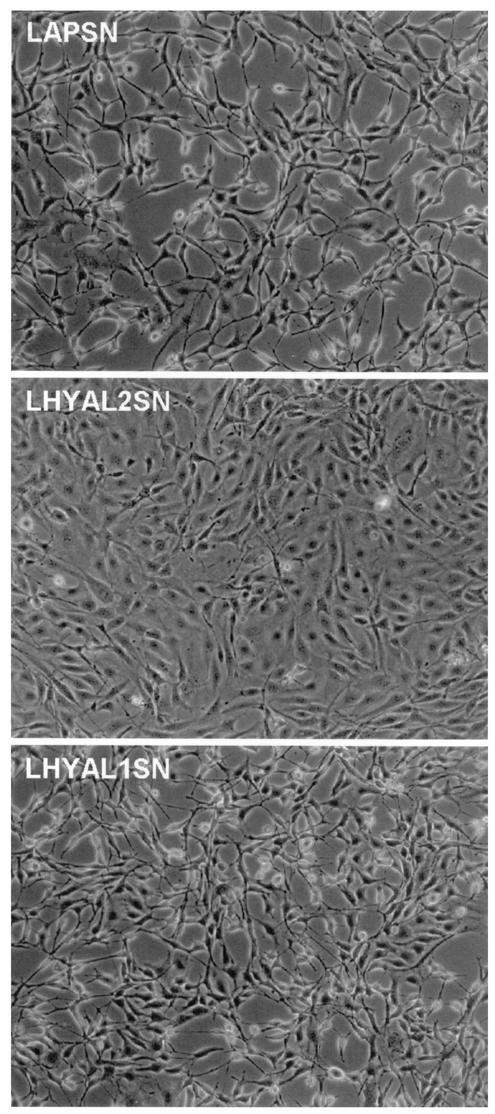
To further explore the tumor suppressor activity of Hyal2, we asked whether the transformed phenotype of cells expressing JSRV Env could be reversed by expression of Hyal2. For this experiment, several transformed foci of cells were isolated from NIH 3T3 cells that had been transfected with the pSX2.Jenv plasmid, and one of the isolates with the most transformed appearance was used for further study. These cells were transduced with vectors encoding AP (LAPSN), Hyal2 (LhHyal2SN), or Hyal1 (LhHyal1SN) and were exposed to G418 to select for cells expressing the vectors. After selection, cells plated at similar densities showed dramatically different phenotypes (Fig. 1): cells expressing Hyal2 showed a flat morphology, indicating a reversal of transformation, while cells expressing the vector encoding Hyal1 or AP had a very transformed appearance, similar to that of the parental cells before vector transduction. The same phenotype was observed when the cells were plated at a low density and allowed to form colonies. Cells expressing Hyal2 formed flat, compact colonies, while cells transduced with the Hyal1 or AP vector formed dispersed colonies consisting of very refractile cells. Quantitation of transformed versus nontransformed colonies in the different populations of cells gave values of 95% transformed colonies for cells transduced with vectors expressing AP or Hyal1 and only 20% transformed colonies for cells transduced with the Hyal2-expressing vector. Thus, transformation by JSRV Env was dramatically suppressed by expression of human Hyal2.
FIG. 1.
Human Hyal2 can reverse the transformed phenotype induced by JSRV Env in NIH 3T3 cells. NIH 3T3 cells transformed by JSRV Env protein were exposed to LXSN-based retroviral vectors encoding human Hyal1, human Hyal2, or alkaline phosphatase, or to no vector, and were selected in a medium containing G418 starting 1 day after vector exposure. Pictures were taken after 5 days of selection, when untransduced control cells were all dead.
Cloning and expression of mouse and rat Hyal2.
The data presented above might be explained by one of two hypotheses: (i) that mouse and rat Hyal2 are indeed tumor suppressors and the JSRV and ENTV Env proteins can bind to and inhibit the activities of these Hyal2 orthologs resulting in transformation, or (ii) that the Env proteins can transform cells independently of Hyal2, but overexpressed Hyal2 protein can bind to and either inactivate or cause the degradation of the Env proteins, resulting in reduced transforming activity. The fact that JSRV and ENTV vectors could not transduce the mouse or rat cells does not rule out the possibility that mouse and/or rat Hyal2 could bind the Env proteins but simply were not capable of mediating virus entry.
To address these possibilities, we isolated Hyal2 cDNA and genomic clones from the NIH 3T3 and 208F cells used in the transformation assays so that we could examine the properties of the Hyal2 proteins made by these cells. Small sequence variations were observed in cDNA and genomic clones from the cells. To determine if the variation was due to PCR error or heterozygosity, we sequenced multiple cDNA and genomic clones. Six mouse Hyal2 cDNA clones were completely sequenced, and the sequences of four clones were identical to the exonic regions of two of five mouse genomic sequences. Four rat Hyal2 cDNA clones were completely sequenced, and the sequences of three clones were identical to the exonic regions of two of four rat genomic sequences. The two mouse Hyal2 cDNA clones that were different from the majority of the clones contained single base changes in different positions (resulting in E195K or S383G amino acid changes). The rat Hyal2 cDNA clone that was different from the majority of the clones also contained a single base change (resulting in a W440-to-stop-codon amino acid change). Genomic clones that were different from the majority of the clones contained single base changes in different positions in all three mouse Hyal2 clones (S105G, H280R, or S392R), and in two rat Hyal2 clones (L431P or D169G). None of the minor cDNA clones was identical to any of the minor genomic clones. Thus, only one consensus sequence was found for either mouse Hyal2 or rat Hyal2, indicating that these genes are homozygous in the NIH 3T3 and 208F cell lines. Minor sequence differences observed are best explained as PCR errors. The sequence of the protein encoded by the NIH 3T3 (Swiss mouse) cDNA (AF535140) exactly matches previously determined sequences for mouse Hyal2 from C3H mice (AF302843 and AF422177) and differs by one conservative amino acid change from that of Czech II mice (AF302844). The sequence of the protein encoded by the 208F (Fischer rat) cDNA (AF535141) differs at four residues from that encoded by a previously sequenced Rattus norvegicus cDNA (AF034218).
To transfer the Hyal2 cDNAs to cells, we cloned each cDNA into the LXSN retroviral vector and generated vector-producing cells by transfection of ecotropic packaging cells followed by transduction of PT67 10A1-MLV-based packaging cells with the transiently produced virus. Approximately 15 G418-resistant clones of PT67 cells were isolated for each vector, and three of the highest-titer vector producers for each cDNA were identified. All of these were tested in the following experiments, instead of just one clone for each cDNA, to rule out artifacts due to the possibility that mutations might have been introduced into the vectors by reverse transcriptase during the transduction of individual vector-producing packaging cell lines. Given that reverse transcription errors occur at ∼1 in 10,000 bases per retrovirus replication cycle, it is very unlikely that errors that might change the phenotype of the vectors would be present in three independent packaging cell clones.
Properties of mouse and rat Hyal2 in comparison to human Hyal2.
Overexpression of the NIH 3T3 mouse Hyal2 cDNA in NIH 3T3 and 208F cells did not confer susceptibility to JSRV or ENTV vector transduction, while expression of the human Hyal2 cDNA resulted in efficient JSRV vector transduction (Table 4). This result is consistent with the fact that NIH 3T3 cells are poorly transduced with the JSRV vector. In contrast, overexpression of the 208F rat Hyal2 cDNA in NIH 3T3 and 208F cells did confer susceptibility to JSRV vector transduction at levels similar to those of the human Hyal2 cDNA (Table 4). Thus, the 208F rat Hyal2 protein is capable of serving as an efficient receptor for JSRV Env-mediated virus entry. The ENTV vector was able to transduce only sheep cells and NIH 3T3 cells expressing human Hyal2, showing that ENTV does not utilize mouse or rat Hyal2 as a receptor for cell entry. As expected, human Hyal1 showed no receptor activity.
TABLE 4.
Receptor activity of Hyal2 proteins from different species in mouse and rat cellsa
| Cells | Expressed protein | Titer (AP+ FFU/ml) of:
|
|
|---|---|---|---|
| JSRV vector | ENTV vector | ||
| NIH 3T3 mouse | None | <5 | <5 |
| Human Hyal1 | <5 | <5 | |
| Human Hyal2 | 5.9 × 104 | 87 | |
| Rat Hyal2 | 2.8 × 104 | <5 | |
| Mouse Hyal2 | <5 | <5 | |
| 208F rat | None | <5 | <5 |
| Human Hyal1 | <5 | <5 | |
| Human Hyal2 | 2.3 × 104 | <5 | |
| Rat Hyal2 | 1.5 × 104 | <5 | |
| Mouse Hyal2 | <5 | <5 | |
| SSF sheep | None | 3.8 × 105 | 6.0 × 103 |
NIH 3T3 or 208F cells were transduced either by LXSN-based retroviral vectors encoding either rat, mouse, or human Hyal2 or human Hyal1 or by the empty LXSN vector and were selected for the presence of the vectors in G418 (750 μg/ml; active concentration). The transduced cell lines were then exposed to either the LAPSN/PJ4 vector (JSRV vector) or the LAPSN/PN172 vector (ENTV vector). Three days after vector exposure, the cells were fixed and stained for AP, and vector titers were determined. Results are means from at least two experiments that varied by no more than 10%.
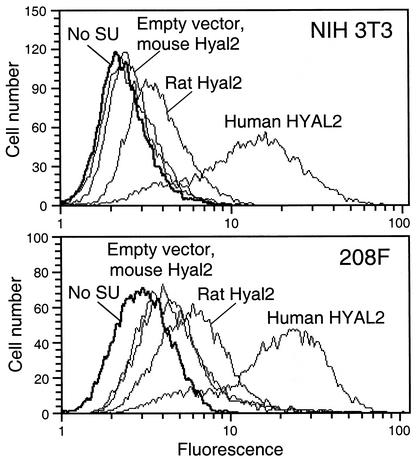
Genetic evidence supports the role of Hyal2 as a receptor for JSRV and ENTV entry, but actual binding of the Env proteins of these viruses to Hyal2 has not been demonstrated. To address this issue and determine if the JSRV Env protein might bind mouse Hyal2 without mediating virus entry, we generated a JSRV SU-human IgG Fc fusion protein for binding studies. Binding of this protein to cells was monitored by using fluorescently labeled secondary antibodies directed against human IgG Fc. The highest level of fusion protein binding was to NIH 3T3 cells expressing human Hyal2, but clear binding to cells expressing rat Hyal2 was observed (Fig. 2). Fusion protein binding to cells expressing mouse Hyal2 was slightly but reproducibly higher than that to cells expressing the empty vector (Fig. 2), consistent with the observation of very low and sporadic transduction by the JSRV vector in mouse cells. The fusion protein bound at similar low levels to NIH 3T3 cells, NIH 3T3 cells transduced with an empty vector, or NIH 3T3 cells transduced with a vector expressing Hyal1 (data not shown). These results are consistent with the hypothesis that Env directly interacts with Hyal2 to mediate virus entry into susceptible cells, and they show that the JSRV Env protein interacts poorly if at all with mouse Hyal2.
FIG. 2.
Flow cytometric analysis of JSRV SU-IgG binding to cells expressing Hyal2 orthologs. Cells transduced by retroviral vectors expressing rat, mouse, or human Hyal2 or human Hyal1 or by the empty retroviral vector LXSN were incubated with the JSRV SU-IgG fusion protein (1 ml of unpurified culture medium containing ∼30 ng of fusion protein for NIH 3T3 cells or 0.1 ml of PBS containing ∼60 ng of purified fusion protein and 2% FBS for 208F cells) on ice for 3.5 h. The cells were washed, incubated with an FITC-conjugated rabbit anti-human Fc antibody, washed again, and analyzed by flow cytometry. Histograms for cells expressing human Hyal1 were virtually identical to those for cells expressing the empty vector LXSN and are not shown in order to simplify the figure. Results shown are representative of multiple experiments.
The pattern of JSRV Env SU-human IgG fusion protein binding to 208F cells expressing the same set of proteins was similar to the pattern for NIH 3T3 cells (Fig. 2). Fusion protein binding increased in cells transduced with the rat Hyal2 vector, and increased most in cells expressing human Hyal2, compared to that of cells expressing the empty vector or a vector encoding Hyal1. Again, a small but reproducible increase in binding was detected in cells expressing mouse Hyal2 in comparison to cells expressing the empty vector or the Hyal1 vector. Unlike the NIH 3T3 cells, 208F cells expressing the empty vector or the Hyal1 vector and incubated with JSRV SU-IgG showed a higher signal than did cells incubated with the fluorescent secondary antibody but without fusion protein, indicating either some endogenous binding activity in the 208F cells, perhaps the endogenous rat Hyal2, or a higher level of nonspecific binding of the fusion protein. Analysis of binding with different concentrations of fusion protein suggested that this binding was nonspecific (data not shown).
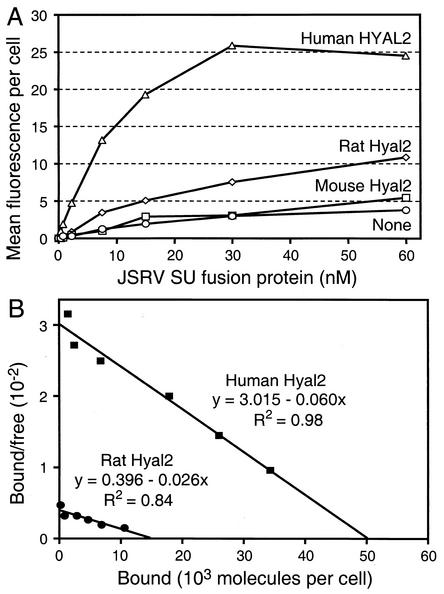
Further analysis of binding with variable amounts of fusion protein (Fig. 3) showed specific binding of the fusion protein to human Hyal2 with a Kd of 9 ± 6 nM and to rat Hyal2 with a Kd of 23 ± 12 nM when the proteins were expressed on 208F rat cells (means of two experiments). The numbers of Hyal2 receptors per cells were estimated to be 46,000 ± 8,000 for human Hyal2 and 14,000 ± 2,000 for rat Hyal2 (means of two experiments), assuming that SU-IgG molecules bind to receptors in a 1:1 ratio.
FIG. 3.
Scatchard analysis of JSRV SU-IgG fusion protein binding to Hyal2 orthologs. (A) 208F cells transduced by vectors encoding rat Hyal2, mouse Hyal2, or human Hyal2 or by the empty retroviral vector LXSN were incubated with increasing amounts of purified JSRV SU-IgG fusion protein, washed, incubated with an FITC-conjugated rabbit anti-human antibody, washed again, and analyzed by flow cytometry. The geometric means (log10) of fluorescence (y axis) were plotted against the concentration of fusion protein used (x axis). The experiment was repeated once with similar results. (B) Scatchard analysis of JSRV SU-IgG binding to 208F cells expressing rat or human Hyal2. The bound/free ratio is plotted against the geometric means of bound fluorescence measured by flow cytometric analysis and expressed as the number of bound JSRV SU-IgG molecules per cell. Results are from a representative experiment.
Human Hyal2 has been reported to exhibit hyaluronidase activity (9), but it is very low compared to other hyaluronidases such as the serum hyaluronidase Hyal1 and the sperm hyaluronidase Spam1 (19). It is possible that during evolution Hyal2 orthologs variably lost the ability to degrade hyaluronan, and this might help explain their different activities as binding partners for Env. To address this question, we assayed the hyaluronidase activities of mouse cells expressing the different Hyal2 orthologs or human Hyal1. While hyaluronidase activity was high in cells expressing Hyal1, it was undetectable in cells expressing mouse, rat, or human Hyal2 (<2% of the activity of cells expressing Hyal1). Thus, differences in the hyaluronidase activities of the Hyal2 orthologs cannot explain the different properties of these proteins.
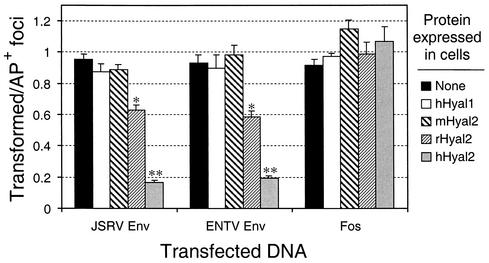
Mouse Hyal2 is unable to suppress transformation by JSRV or ENTV Env proteins.
The results above show that mouse Hyal2 from NIH 3T3 cells does not mediate JSRV or ENTV vector entry and does not appreciably bind the SU domain of JSRV; thus, a model for Env transformation involving Env binding to and inhibition of the tumor suppressor activity of mouse Hyal2 seems unlikely. As a final test of this model, we asked whether mouse Hyal2 could suppress transformation by Env proteins. 208F cells expressing mouse, rat, or human Hyal2, human Hyal1, or the empty vector LXSN were transfected with the JSRV or ENTV Env or Fos expression plasmids, and transformed foci were quantitated (Fig. 4). Expression of human and, to a lesser extent, rat Hyal2 significantly suppressed transformation by the JSRV and ENTV Env proteins but had no effect on Fos transformation. Mouse Hyal2 expression had no effect on transformation by any of the proteins.
FIG. 4.
208F cell transformation by JSRV and ENTV Env proteins is suppressed by human Hyal2 and rat Hyal2 but not by mouse Hyal2. 208F cells were transduced by LXSN-based retroviral vectors encoding rat Hyal2 (rHyal2), mouse Hyal2 (mHyal2), human Hyal2 (hHyal2), or human Hyal1 (hHyal1) or by an empty LXSN vector (none) and were selected for the presence of the vectors in G418. The transduced cells were then cotransfected with 10 μg of plasmid DNA encoding JSRV Env (pSX2.Jenv), ENTV Env (pSX2.Eenv), a Fos oncoprotein (pFBJ/R), or 10A1 MLV Env (pSX2) plus 1 μg of plasmid pLAPSN, which expresses AP. Transformed foci were counted approximately 2 weeks posttransfection, and the cells were fixed and stained for AP. Results are presented as the ratios of transformed foci to AP+ foci and are means from two independent experiments. In this experiment, the numbers of foci induced by pSX2.Jenv, pSX2.Eenv, and pFBJ/R in 208F/LXSN cells were similar and the means ranged from 120 to 160 foci per μg of plasmid DNA. Asterisks indicate values statistically different from those obtained by using cells expressing the control vector LXSN (*, P < 0.05; **, P < 0.01).
In a variation of this experiment, we asked whether the Hyal2 orthologs could reverse the transformed phenotype of NIH 3T3 cells transformed by the JSRV and ENTV Env proteins. NIH 3T3 cells transformed by transfection of plasmids encoding JSRV Env, a FLAG-tagged JSRV Env, or ENTV Env were transduced with vectors expressing mouse, rat, or human Hyal2, human Hyal1, or AP. All of these vectors also express Neo, and the cells were grown in G418 to select for cells expressing the vectors. While human and rat Hyal2 could reverse the transformed phenotype of cells expressing the Env proteins, mouse Hyal2, human Hyal1, and AP had no activity (Table 5). Together these data indicate that the JSRV and ENTV Env proteins can transform NIH 3T3 mouse cells in the absence of any significant binding to or genetic interaction with mouse Hyal2.
TABLE 5.
Reversion of JSRV/ENTV Env transformation following expression of Hyal2 proteins from different speciesa
| Plasmid used to transform NIH 3T3 cells | Nontransformed colonies (% of total) following expression of:
|
||||
|---|---|---|---|---|---|
| Human Hyal2 | Rat Hyal2 | Mouse Hyal2 | Human Hyal1 | Human AP | |
| pSX2-Jenv | 85, 60 | 79, 75 | 2, 1 | 2, 1 | 1, 0 |
| pSX2-Jenv-FLAG | 88, 80 | 85, 74 | 4, 5 | 3, 4 | 1, 4 |
| pSX2-Eenv | 52, 40 | 5, 2 | 2, 4 | 3, 6 | 2, 5 |
NIH 3T3 cells were transfected with the indicated Env expression plasmids, and a transformed focus of cells was isolated by micromanipulation and expanded for each plasmid. The transformed cells were transduced with LXSN-based vectors expressing the indicated proteins and were grown in G418 to select for the presence of the vectors, and the numbers of G418-resistant colonies with flat and contact-inhibited (nontransformed) or refractile and diffuse (transformed) phenotypes (see Fig. 1) were counted. At least 100 colonies were scored for each condition, and the results of two experiments are shown.
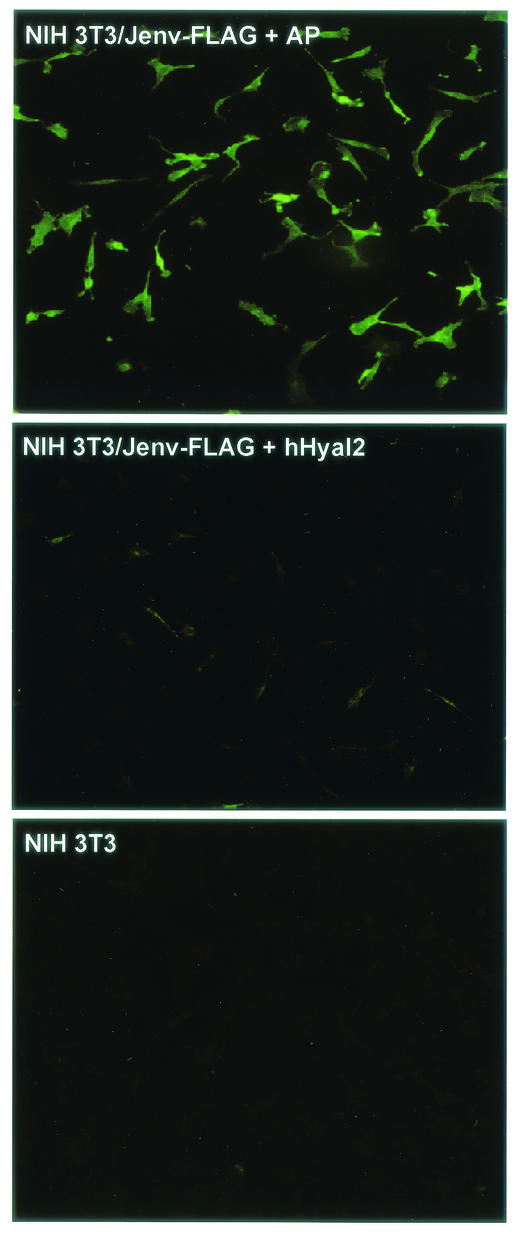
Expression of human Hyal2 reduces Env protein levels in cells.
To explore whether Hyal2 binding to Env could inhibit transformation by reducing the level of Env in cells, possibly by increasing Env degradation, we examined expression of FLAG-tagged JSRV Env protein in NIH 3T3 cells transformed by this protein after transduction with vectors encoding human Hyal2, human Hyal1, rat Hyal2, or mouse Hyal2, or with the LAPSN vector. Cells expressing human Hyal2 showed a clear decrease in Env levels relative to levels in cells expressing the control vector that encodes AP (Fig. 5). Little background staining was observed in NIH 3T3 cells that did not express the FLAG-tagged JSRV Env (Fig. 5). Expression of rat Hyal2 caused a similar decrease in Env levels, whereas expression of mouse Hyal2 or human Hyal1 had no effect on Env levels relative to those in LAPSN-transduced or untransduced cells (data not shown). Thus, the ability of Hyal2 proteins to suppress transformation by Env is likely due to the reduction in Env levels resulting from Hyal2 binding. This reduction in Env levels could result from increased degradation of Env before transport to the cell surface or increased internalization and degradation following cell surface transport.
FIG. 5.
Hyal2 expression reduces JSRV Env protein levels. NIH 3T3 cells transformed by pSX2-Jenv-FLAG were transduced with retroviral vectors encoding human Hyal2 or AP. After 10 days of selection in G418, these cells and untreated NIH 3T3 cells were immunostained using anti-FLAG antibodies. The same conditions of illumination and photography were used for all panels.
DISCUSSION
Previous work showed that cells which are not susceptible to JSRV vector transduction can be rendered susceptible by expression of human, ovine, or bovine Hyal2 proteins in the cells (5, 19), suggesting that Hyal2 is the binding receptor that mediates JSRV infection. Hyal2 is a glycosylphosphatidylinositol-anchored protein, and it was shown that removal of Hyal2 from the surfaces of susceptible cells with phosphatidylinositol-specific phospholipase C resulted in a large decrease in the susceptibility of the cells to JSRV vector transduction, further supporting the conclusion that Hyal2 is a binding receptor for JSRV (19). Here we provide direct evidence that human Hyal2 binds the SU region of Env, indicating that Hyal2 is the binding receptor for JSRV, and likely also for ENTV. It is not clear yet whether Hyal2 mediates virus fusion as well or whether other cellular proteins are required.
The findings that (i) JSRV and ENTV cause cancer in the lung and upper respiratory tract, (ii) the Env proteins of JSRV and ENTV can transform cells, (iii) the receptor for JSRV and ENTV entry is Hyal2, and (iv) Hyal2 is located in the p21.3 region of human chromosome 3, which is commonly deleted in lung and breast cancer, led to the attractive hypothesis that Hyal2 is a tumor suppressor and oncogenesis is the result of inhibition or reversal of this function by Env. However, the facts that NIH 3T3 mouse and 208F rat fibroblasts could be transformed by Env but could not be transduced by JSRV- or ENTV-pseudotype retroviral vectors presented a paradox, since lack of vector entry suggested that the Env proteins could not bind Hyal2 orthologs in these cells and thus could not directly inhibit the putative Hyal2 tumor suppressor activity. Here we have cloned the mouse and rat Hyal2 orthologs from NIH 3T3 and 208F cells, and we find that while rat Hyal2 can bind the JSRV Env SU domain, and thus might participate in transformation by Env, mouse Hyal2 has very low if any binding activity. Furthermore, while rat and human Hyal2 can suppress transformation by JSRV and ENTV Env proteins, mouse Hyal2 did not inhibit transformation. Thus, in NIH 3T3 mouse fibroblasts it is unlikely that the mouse Hyal2 plays any role in transformation by Env.
Introduction of human or rat Hyal2 into NIH 3T3 cells transformed by the FLAG-tagged JSRV Env construct resulted in a marked reduction in Env levels in the cells, indicating that the mechanism of Env transformation suppression by Hyal2 orthologs that can bind Env involves Env degradation. We conclude that the apparent tumor suppressor activity of Hyal2 in cells transformed by Env is due to Hyal2-mediated reduction of Env levels and not to a more general Env-independent tumor suppressor activity of Hyal2.
Although rat Hyal2 can act as a receptor for JSRV vectors when it is expressed in rat or mouse cells, we have found that JSRV vectors do not transduce 208F rat cells. Hyal2 is widely expressed in different tissues of mice and humans (4, 9, 21), and we used RNA from 208F cells to clone the rat Hyal2 cDNA, so Hyal2 is expressed in these cells. We have not explored whether there is a threshold of Hyal2 expression below which transduction does not occur. Alternatively, other proteins in these cells might bind Hyal2 and block Env interaction with Hyal2, resulting in a requirement for high-level Hyal2 expression in order for transduction to occur.
We have attempted to knock out the Hyal2 gene in mice to see if the animals might get cancer at a higher rate as a result, in support of a general tumor suppressor role for Hyal2, and to see if fibroblasts derived from such animals would still be transformed following expression of JSRV and ENTV Env proteins, but so far we have been unsuccessful in generating viable knockouts. Alternatively, it would be useful to generate Hyal2 knockouts in cell culture either by homologous recombination or by RNA interference technology to provide definitive proof that Hyal2 is not involved in Env transformation.
Although Hyal2 appears to play no role in the transformation of mouse fibroblasts by the JSRV or ENTV Env proteins, these viruses transform epithelial cells in animals, and the mechanism of transformation in these cells may be different. Indeed, it appears that in human epithelial cells, Hyal2 can interact with a cell surface tyrosine kinase(s) to inhibit growth signaling, and that in this case Env binding to Hyal2 stimulates growth signal transduction leading to cell transformation (A. Danilkovitch-Miagkova, F.-M. Duh, I. Kuzamin, D. Angeloni, S.-L. Liu, A. D. Miller, and M. I. Lerman, submitted for publication). Therefore, it appears that the JSRV and ENTV Env proteins can transform cells by Hyal2-dependent and -independent pathways depending on cell type. The mechanism of transformation of DF-1 chicken fibroblasts is unknown but appears different from that in rodent fibroblasts (2), and it will be interesting to see if Hyal2 is involved in the transformation of these cells or whether a third mechanism of transformation is operative.
Acknowledgments
This work was supported by grants DK47754 and HL54881 (to A.D.M.), contracts CO12400 (to F.-M.D.) and CO56000 (to M.I.L.), and training grant T32-CA09437 (to S.-L.L.) from the National Institutes of Health.
REFERENCES
- 1.Alberti, A., C. Murgia, S.-L. Liu, M. Mura, C. Cousens, M. Sharp, A. D. Miller, and M. Palmarini. 2002. Envelope-induced cell transformation by ovine betaretroviruses. J. Virol. 76:5387-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, T. E., K. J. Sherrill, S. M. Crispell, M. R. Perrott, J. O. Carlson, and J. C. DeMartini. 2002. The jaagsiekte sheep retrovirus envelope gene induces transformation of the avian fibroblast cell line DF-1 but does not require a conserved SH2 binding domain. J. Gen. Virol. 83:2733-2742. [DOI] [PubMed] [Google Scholar]
- 3.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Csoka, A. B., S. W. Scherer, and R. Stern. 1999. Expression analysis of six paralogous human hyaluronidase genes clustered on chromosomes 3p21 and 7q31. Genomics 60:356-361. [DOI] [PubMed] [Google Scholar]
- 5.Dirks, C., F. M. Duh, S. K. Rai, M. I. Lerman, and A. D. Miller. 2002. Mechanism of cell entry and transformation by enzootic nasal tumor virus. J. Virol. 76:2141-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DuBridge, R. B., P. Tang, H. C. Hsia, P. M. Leong, J. H. Miller, and M. P. Calos. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan, H. (ed.). 2003. Current topics in microbiology, vol. 275. Jaagsiekte sheep retrovirus and lung cancer. Springer-Verlag, Berlin, Germany.
- 8.Kurre, P., H. P. Kiem, J. Morris, S. Heyward, J.-L. Battini, and A. D. Miller. 1999. Efficient transduction by an amphotropic retrovirus vector is dependent on high-level expression of the cell surface virus receptor. J. Virol. 73:495-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lepperdinger, G., B. Strobl, and G. Kreil. 1998. HYAL2, a human gene expressed in many cells, encodes a lysosomal hyaluronidase with a novel type of specificity. J. Biol. Chem. 273:22466-22470. [DOI] [PubMed] [Google Scholar]
- 10.Lerman, M. I., and J. D. Minna, for The International Lung Cancer Chromosome 3p21.3 Tumor Suppressor Gene Consortium. 2000. The 630-kb lung cancer homozygous deletion region on human chromosome 3p21.3: identification and evaluation of the resident candidate tumor suppressor genes. Cancer Res. 60:6116-6133. [PubMed] [Google Scholar]
- 11.Maeda, N., M. Palmarini, C. Murgia, and H. Fan. 2001. Direct transformation of rodent fibroblasts by jaagsiekte sheep retrovirus DNA. Proc. Natl. Acad. Sci. USA 98:4449-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller, A. D., and F. Chen. 1996. Retrovirus packaging cells based on 10A1 murine leukemia virus for production of vectors that use multiple receptors for entry. J. Virol. 70:5564-5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller, A. D., and G. J. Rosman. 1989. Improved retroviral vectors for gene transfer and expression. BioTechniques 7:980-982. [PMC free article] [PubMed] [Google Scholar]
- 14.Miller, A. D., I. M. Verma, and T. Curran. 1985. Deletion of the gag region from FBR murine osteosarcoma virus does not affect its enhanced transforming activity. J. Virol. 55:521-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller, D. G., R. H. Edwards, and A. D. Miller 1994. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc. Natl. Acad. Sci. USA 91:78-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmarini, M., N. Maeda, C. Murgia, C. De-Fraja, A. Hofacre, and H. Fan. 2001. A phosphatidylinositol-3-kinase (PI-3K) docking site in the cytoplasmic tail of the Jaagsiekte sheep retrovirus transmembrane protein is essential for envelope-induced transformation of NIH 3T3 cells. J. Virol. 75:11002-11009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quade, K. 1979. Transformation of mammalian cells by avian myelocytomatosis virus and avian erythroblastosis virus. Virology 98:461-465. [DOI] [PubMed] [Google Scholar]
- 18.Rai, S. K., J. C. DeMartini, and A. D. Miller. 2000. Retrovirus vectors bearing the jaagsiekte sheep retrovirus Env transduce human cells by using a new receptor localized to chromosome 3p21.3. J. Virol. 74:4698-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rai, S. K., F.-M. Duh, V. Vigdorovich, A. Danilkovitch-Miagkova, M. I. Lerman, and A. D. Miller. 2001. Candidate tumor suppressor HYAL2 is a glycosylphosphatidylinositol (GPI)-anchored cell-surface receptor for jaagsiekte sheep retrovirus, the envelope protein of which mediates oncogenic transformation. Proc. Natl. Acad. Sci. USA 98:4443-4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharp, J. M., K. W. Angus, E. W. Gray, and F. M. M. Scott. 1983. Rapid transmission of sheep pulmonary adenomatosis (jaagsiekte) in young lambs. Arch. Virol. 78:89-95. [DOI] [PubMed] [Google Scholar]
- 21.Strobl, B., C. Wechselberger, D. R. Beier, and G. Lepperdinger. 1998. Structural organization and chromosomal localization of Hyal2, a gene encoding a lysosomal hyaluronidase. Genomics 53:214-219. [DOI] [PubMed] [Google Scholar]