Abstract
Ebola virus VP30 is an essential activator of viral transcription. In viral particles, VP30 is closely associated with the nucleocapsid complex. A conspicuous structural feature of VP30 is an unconventional zinc-binding Cys3-His motif comprising amino acids 68 to 95. By using a colorimetric zinc-binding assay we found that the VP30-specific Cys3-His motif stoichiometrically binds zinc ions in a one-to-one relationship. Substitution of the conserved cysteines and the histidine within the motif led to a complete loss of the capacity for zinc binding. Functional analyses revealed that none of the tested mutations of the proposed zinc-coordinating residues influenced binding of VP30 to nucleocapsid-like particles but, concerning its role in activating viral transcription, all resulted in a protein that was inactive.
Ebola virus (EBOV) and Marburg virus, the two members of the family Filoviridae, cause a severe hemorrhagic disease with an exceptionally high fatality rate in humans and monkeys (22). Due to their genome organization, filoviruses are grouped in the order Mononegavirales. The EBOV genome consists of a single-stranded nonsegmented negative-sense RNA molecule with a coding capacity for eight proteins. Four of the proteins, NP, VP35, VP30, and L, are associated with the viral genome forming the nucleocapsid complex (6). NP, VP35, and L are sufficient to mediate viral replication in a reconstituted replication and transcription system (21). Transcription, however, is strongly dependent on the presence of the fourth nucleocapsid protein, VP30 (21, 27). Recently it has been shown that VP30 acts as a transcription antitermination factor immediately after transcription initiation while transcription elongation is not affected by the protein. The function of VP30 as transcription activator has been found to be dependent on the formation of an RNA secondary structure at the transcription start site of the first gene (28). Furthermore, VP30 is heavily phosphorylated, and this posttranslational modification has been shown to regulate VP30 activity during transcription (18). However, the molecular mechanism by which VP30 activates viral transcription is presently unknown.
Sequence analysis of filoviral VP30 revealed that it contains a motif similar to that of an unconventional Cys3-His zinc finger that was first characterized for Nup475, a mammalian nuclear protein (5). On the basis of nuclear magnetic resonance data, a structure of the Nup475 metal-binding domain was proposed in which the zinc ion is coordinated by the conserved cysteines and histidine (29). Comparable motifs have been discovered in various proteins from different eukaryotic and viral species (29). The suggestion that the Cys3-His motif within VP30 forms a zinc finger with biological relevance is supported by the fact that it is highly conserved among filoviruses (Fig. 1A). A similar motif was also identified in the M2-1 protein of pneumoviruses, which are closely related to filoviruses (12). Like VP30, the M2-1 protein of human respiratory syncytial virus (hRSV) is functional in the course of viral transcription. In contrast to EBOV VP30, however, M2-1 supports transcriptional elongation and formation of readthrough mRNAs (3, 7, 11). Although binding of zinc has not been demonstrated directly for M2-1, the Cys3-His motif was shown to be of functional significance for the protein and, moreover, for viral growth (12, 25).
FIG. 1.
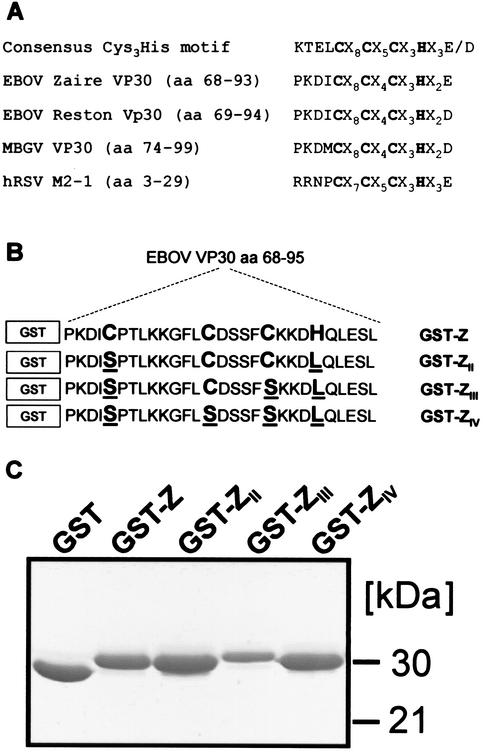
Conserved Cys3-His motifs within viral proteins and expression of GST-VP3068-95 fusion proteins. (A) Alignment of a Cys3-His zinc-binding motif consensus sequence (29) and VP30 sequences of different filoviruses as well as the M2-1 protein sequence of hRSV. Predicted zinc-coordinating residues are printed in bold. MBGV, Marburg virus. (B) Diagram of EBOV VP30-specific peptides expressed as GST fusion proteins. GST (in boxes) represents the amino-acid sequence of GST. The C-terminally fused VP30-specific amino acids are listed. Mutated amino acids are underlined. (C) Bacterial expression and purification of GST fusion proteins. Purified proteins were analyzed by SDS-PAGE in 12% polyacrylamide gels and visualized by staining with Coomassie blue.
In the present study, we investigated the relevance of the zinc-binding motif of EBOV VP30. We examined the region between amino acids 68 and 95 of VP30 to prove that VP30 contains in fact a zinc-binding domain. We further mutated the Cys3-His motif within the full-length protein to check whether a destroyed zinc finger influences the nucleocapsid association of VP30 or the ability of VP30 to mediate activation of EBOV transcription.
Construction and expression of GST-VP30 fusion proteins.
To investigate whether VP30 is able to coordinate zinc ions, first, glutathione S-transferase (GST)-VP30 fusion genes were constructed containing the entire open reading frame of VP30. Since the expressed proteins showed a strong tendency to aggregate, it was not possible to purify them. Therefore, we focused on the VP30-specific Cys3-His motif which was fused to GST. The open reading frame coding for amino acids 68 to 95 of VP30, which included the Cys3-His motif plus 4 and 5 flanking amino acids, was ligated with the GST gene in the fusion protein expression vector pGEX-2X-5X (Amersham-Pharmacia) (Fig. 1B). For construction of pGEX-VP3068-95 expressing the wild-type fusion protein GST-Z (Fig. 1B), a DNA fragment spanning nucleotides 8710 to 8793 of the EBOV genome (GenBank accession number AF086833) was amplified by PCR with primers containing either a BamHI or an EcoRI restriction site. Subsequently, the PCR fragment was inserted in the BamHI-EcoRI restriction sites of pGEX-2X-5X. In vitro mutagenesis of pGEX-VP3068-95 was performed by using the QuikChange site-directed mutagenesis kit (Stratagene), resulting in plasmids expressing the mutants GST-ZII, GST-ZIII, and GST-ZIV (Fig. 1B). The chimeric proteins were expressed in Escherichia coli BL21 cells upon the addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma) at 30°C for 2 h in Luria-Bertani medium. For lysis, cells were collected by centrifugation, resuspended in phosphate-buffered saline containing 2 mg of lysozyme/ml, and incubated for 30 min on ice. Subsequently, cells were frozen and thawed three times. After addition of 1% Triton X-100, cells were incubated for 1 h at 4°C and finally sonicated. The GST fusion proteins were purified using glutathione Sepharose beads (Amersham-Pharmacia) according to the manufacturer's protocol. Protein expression and purification were monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 1C). The GST fusion proteins showed the expected apparent molecular mass of about 30 kDa.
The Cys3-His motif of VP30 binds zinc in vitro.
To determine the content of Zn2+ bound to the fusion proteins, a colorimetric assay using 4-(2-pyridylazo)resorcinol (PAR) was performed (14, 23). First, the purified protein complexes were washed two times in HSD (50 mM HEPES-KOH [pH 7.5], 200 mM NaCl, 1 mM dithiothreitol [DTT]) and afterwards resuspended in an equal volume of HSD. About 20 μg of protein was incubated in 500 μl of HSD for 15 min at 30°C with zinc acetate and EDTA in succession in differing orders (see also reference 23) as follows. (i) For Z→E incubation, the protein suspensions were first incubated with 0.1 mM zinc acetate, washed four times with 1 ml HSD-5 mM DTT, and finally incubated with 1 mM EDTA. (ii) For E→Z incubation, the suspensions were incubated with 1 mM EDTA, washed four times with 1 ml HSD-5 mM DTT, and then incubated with 0.1 mM zinc acetate. To determine the protein concentration, 5-μl aliquots of each sample were removed and analyzed by SDS-12% PAGE. The gels were stained with Coomassie blue, destained, dried, and scanned. The amount of protein was compared to bovine serum albumin standards and quantified using TINA2.0 software (Raytest, Freiburg, Germany).
For the determination of bound divalent cations, protein samples were digested in a total volume of 50 μl with 40 μg of proteinase K (Ambion) at 60°C for 30 min. Subsequently, an equal volume of HSD containing 0.2 mM PAR was added. The metallochromic indicator PAR forms chelate complexes with divalent metallic cations as Zn2+ that can be quantified by measuring their optical density at 490 nm (23). The amount of Zn ions bound to the GST fusion proteins was determined by comparing it with a Zn2+ standard curve.
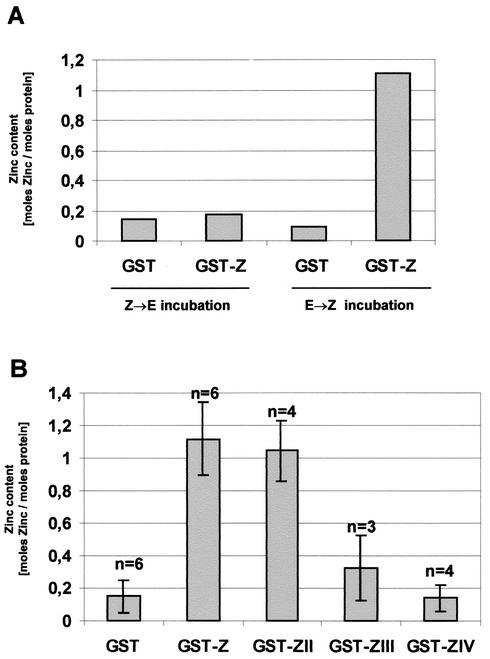
Following the Z→E incubation of GST or GST-Z, only trace amounts of zinc ions were detected (Fig. 2A). However, following E→Z incubation, 1.1 mol of Zn2+ per mol of GST-Z was bound. For GST, only trace amounts of bound metal ions were detected. The subtraction of this background from the value determined for GST-Z after E→Z incubation revealed that the protein contained about 1 mol of zinc per mol of protein (Fig. 2A). The assumption made by Pfister et al. (23) that GST itself binds zinc could not be confirmed under the conditions we used. The lack of zinc-binding activity of GST in our system might have been due to the addition of DTT to the incubation buffer.
FIG. 2.
Amino acids 68 to 95 of VP30 bind zinc in vitro. (A) Binding of zinc ions by GST and GST-Z as determined by measuring optical density at 490 nm in the presence of 0.1 mM PAR following the Z→E or E→Z incubation series. For Z→E incubation, the purified proteins were first incubated with 0.1 mM zinc acetate and subsequently with 1 mM EDTA; for E→Z incubation, purified proteins were first incubated with 1 mM EDTA and afterwards with 0.1 mM zinc acetate. (B) Binding of zinc ions by GST, GST-Z, GST-ZII, GST-ZIII, and GST-ZIV after treatment with 0.1 mM zinc acetate. Standard deviations are indicated by bars. n, number of independent experiments.
Subsequently, we were interested in determining whether substitution of the predicted zinc-coordinating amino acids influences the zinc-binding activity of GST-Z. To address this issue, the GST-Z mutants GST-ZII, GST-ZIII, and GST-ZIV, in which two, three, or four of the putative zinc-coordinating residues, respectively, were exchanged (Fig. 1B), were constructed. Cysteine residues were replaced with serines, and the histidine residue was replaced with leucine. The mutant proteins were expressed, purified as described above, and analyzed in the colorimetric zinc-binding assay. Surprisingly, GST-ZII showed almost the same zinc-binding capacity as GST-Z, indicating that mutations C72S and H90L did not impair the zinc-binding activity (Fig. 2B). However, when all of the four putative zinc-coordinating amino acids were exchanged, specific zinc binding was abolished (Fig. 2B, lane GST-ZIV). Zinc-binding activity of the GST-ZIII construct with three predicted zinc-coordinating residues exchanged (C72,86S and H90L) was considerably reduced. Taken together, these data indicate that the VP30-specific Cys3-His motif is an autonomous zinc-binding domain which specifically mediates the binding of one ion of zinc.
Mutations within the zinc-binding motif of VP30 do not influence binding to NP-derived inclusion bodies.
It has been shown recently that VP30 interacts with homoaggregates formed by NP, the major structural component of the nucleocapsid (18). When NP is expressed in the absence of other viral proteins, it forms characteristic inclusion bodies that contain highly ordered tubular structures resembling the nucleocapsids in virus-infected cells (15). Coexpression of VP30 and NP in a recombinant system leads to relocalization of VP30 into the NP-derived inclusion bodies, indicating the nucleocapsid association of VP30. For this interaction, the phosphorylation state of VP30 plays an important role: only the phosphorylated form of VP30 binds to the NP inclusions, whereas nonphosphorylated VP30 is unable to interact (18). Since classical zinc fingers have been found to mediate DNA/RNA binding and protein-protein interactions (8, 10, 16, 19), it was of interest to investigate whether the VP30-specific zinc-binding domain might be important for the nucleocapsid association of the protein.
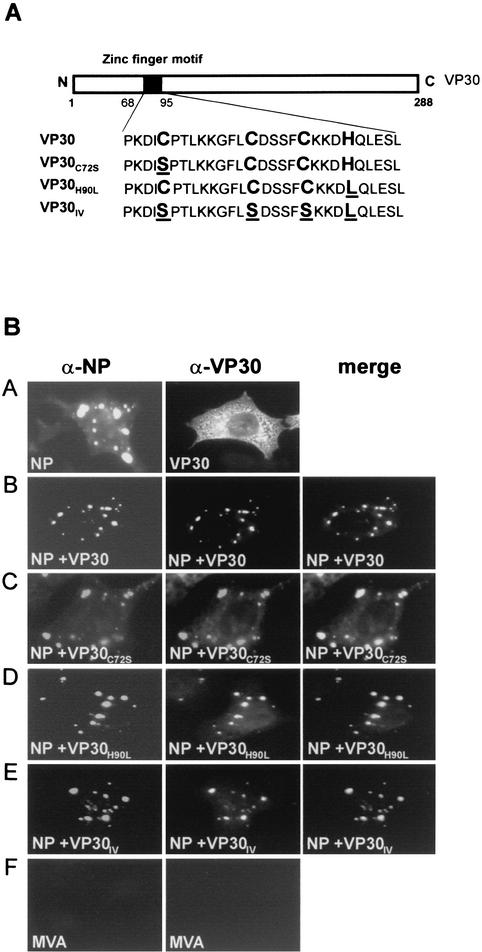
To analyze binding of VP30 to NP-derived inclusions, VP30 and NP were expressed either solely or in combination and protein interaction was determined by immunofluorescence analyses. Subconfluent HeLa cells (50%) were infected with the recombinant vaccinia virus MVA-T7 containing the T7 RNA polymerase gene (24) with a multiplicity of infection of 3 to 5 PFU per cell. At 1 h postinfection, cells were transfected, using the Lipofectin (GIBCO-BRL) precipitation method (20), with the respective plasmids encoding NP, VP30, or VP30 mutants. Construction of plasmids pT-NPEBO and pT-VP30EBO, coding for NP and VP30, respectively, has been described previously (21). The plasmids pT-VP30H90L, pT-VP30C72S, and pT-VP30IV, coding for the VP30 mutants VP30H90L, VP30C72S, and VP30IV, respectively, were constructed using the QuikChange site-directed mutagenesis kit with pT-VP30EBO as the template (Fig. 3A). At 8 h posttransfection, cells were fixed and prepared for immunofluorescence analysis as described by Modrof et al. (17). When VP30 was expressed in the absence of NP, it was homogenously distributed in the cytoplasm (Fig. 3B, panel A). Upon coexpression with NP, VP30 colocalized with the NP-induced inclusion bodies, indicating the interaction of both proteins (Fig. 3B, panel B). The same distribution pattern was observed when the different VP30 mutants were coexpressed with NP, suggesting that mutations within the zinc-binding domain did not influence the interaction between VP30 and NP inclusions (Fig. 3B, panels C to E). Since at least mutant VP30IV had lost the capacity to coordinate zinc, it can be concluded that binding of zinc ions is dispensable for the nucleocapsid association of VP30.
FIG. 3.
Mutations within the zinc-binding motif do not influence binding to NP-induced inclusions. (A) Diagram of the mutations inserted in the Cys3-His motif of the full-length VP30. Mutated amino acids are underlined. (B) HeLa cells grown on glass coverslips were infected with MVA-T7 and cotransfected with 200 ng of the DNA plasmids encoding the wild-type VP30 or the VP30 substitution mutants together with 1 μg of pT-NPEBO (encoding EBOV NP). At 12 h postinfection, cells were fixed, permeabilized using 0.2% Triton X-100, and subjected to immunofluorescence analysis using a monoclonal anti-NP immunoglobulin G (IgG) antibody (1:20) and a monoclonal anti-VP30 IgM antibody (1:10). As secondary antibodies, a μ chain-specific fluorescein isothiocyanate-conjugated F(ab′)2 fragment of goat anti-mouse IgM (1:100; Dianova) and a rhodamine-conjugated goat anti-mouse IgG (1:100; Dianova) were used. Panel A, NP and VP30 expressed alone; panel B, coexpression of VP30 and NP; panels C to E, coexpression of VP30 mutants with NP; panel F, MVA-T7-infected cells.
Transcription activation mediated by VP30 requires an intact zinc-binding motif.
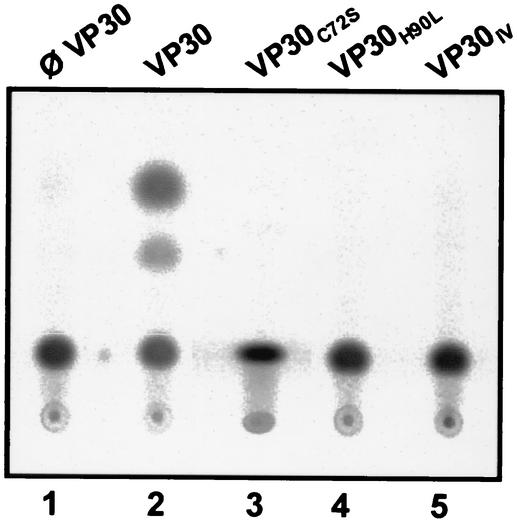
It has been shown previously that replication of minigenomic RNA is unaffected by VP30 whereas transcription is strongly dependent on the presence of VP30 (21). To observe the relevance of the zinc-binding motif for the role of VP30 as transcription activator, VP30 substitution mutants were tested in a reconstituted EBOV-specific replication and transcription system (21). A total of 5 × 105 BSR T7/5 cells (1) seated in a 7-cm2 well were transfected, using FuGENE 6 (Roche) as described previously (18), with plasmids encoding the EBOV nucleocapsid proteins NP, VP35, L, and either VP30 (21) or VP30 mutants. Simultaneously, a plasmid encoding the EBOV-specific minigenome 3E-5E, which contained the leader and trailer regions of the EBOV genome flanking the chloramphenicol acetyltransferase (CAT) reporter gene (21), was transfected. After an incubation period of 8 h, cells were washed two times with Dulbecco's modified Eagle medium and further incubated for 36 h at 37°C. Subsequently, cells were harvested and, as a readout for transcription activity, CAT activity was determined using a standard protocol. The radioactive signals were detected with a BAS-1000 Bio-Imaging Analyzer (Fujifilm) and TINA 2.0 software (Raytest). In lanes 1 and 2 of Fig. 4, it is shown that the presence of VP30 in the transcription system is necessary for EBOV-specific transcription. All VP30 mutants with a destroyed zinc-binding motif failed to mediate viral transcription (Fig. 4, lanes 3 to 5). The loss of VP30 activity was observed even when only one amino acid involved in zinc binding was replaced.
FIG. 4.
Transcription activation mediated by VP30 requires an intact zinc-binding motif. Approximately 5 × 105 BSR T7/5 cells were transfected with DNA plasmids encoding EBOV nucleocapsid proteins NP, VP35, L, and either the VP30 or the VP30 mutant, together with a DNA plasmid encoding the EBOV-specific artificial minigenome 3E-5E. At 2 days posttransfection, cells were lysed and CAT activity was determined. Lane 1, control without VP30; lane 2, wild-type VP30; lanes 3 to 5, VP30 substitution mutants (the names of the respective mutants are given at the top of the panel).
When the VP30 zinc-binding motif was fused to GST, it was necessary to replace at least three proposed zinc-coordinating amino acids to obtain a peptide incapable of binding zinc. This might have been due to a very strong zinc-binding capacity of the VP30 peptide (amino acids 68 to 95) when fused to GST under the conditions used. It has been reported that single-amino-acid substitutions within the zinc-binding domain of Sendai virus V protein led to the reduction of zinc-binding activity to between 22% and 68% of that of the wild type but not to a complete loss of activity (13). However, the function of the protein in viral pathogenicity was strongly impaired by single-amino-acid substitutions. A similar phenomenon was observed with the metallo-β-lactamase CcrA from Bacteroides fragilis. Individual substitutions of zinc-ligating residues resulted in a significant reduction in β-lactam hydrolysis, while the metal binding capacity of the respective enzymes was only partly reduced (30). This also seemed to be the case for VP30. A single-amino-acid substitution within the zinc-binding domain of the full-length protein resulted in a complete loss of the transactivating function of VP30 during transcription, pointing to the importance of the integrity of this domain for the role of VP30 as transcription activator. The observed discrepancy between the number of mutations necessary to impair either the VP30 function or the binding of zinc raises the question of whether the bound zinc ion is involved directly in the function of VP30 as transcription activator. Thus, it is possible that the structural integrity of the zinc-binding domain is a prerequisite for VP30 function and that the bound zinc ion is not involved in formation but is involved in stabilizing this structure.
The exact function of the zinc-binding motif within VP30 is not yet clear. Other proteins possessing a comparable Cys3-His domain have been shown to be involved in RNA metabolism, suggesting that the Cys3-His finger mediates an RNA interaction (29). The Drosophila protein SU(S) contains two Cys3-His motifs. However, RNA binding has been assigned to two arginine-rich motifs, and the zinc-binding motifs are proposed to influence the stability of the SU(S)-RNA interaction (26). So far it is not known whether VP30 is an RNA-binding protein. The M2-1 protein of hRSV, which shows striking structural similarities to VP30 and also contains the Cys3-His motif, has been reported to bind RNA (2, 4). However, the mapped RNA binding domain does not include the predicted zinc finger motif (4). Nevertheless, the Cys3-His motif of M2-1 has not only been found to be essential for the transcription antitermination function of the protein but also for virus growth (12, 25). Like VP30, M2-1 is associated with the nucleocapsid via binding to the nucleoprotein (9). In contrast to VP30, however, interaction with the nucleoprotein was prevented when the predicted zinc-coordinating amino acids within the Cys3-His motif of M2-1 were mutated (12).
In conclusion, our data revealed that EBOV VP30 is a zinc-binding protein. Furthermore, we have demonstrated that the structural integrity of the zinc-binding domain is a prerequisite for protein function during transcription but is dispensable in the interaction with nucleocapsid-like structures. The exact role of the zinc-binding domain for VP30 function remains to be elucidated.
Acknowledgments
We thank Angelika Lander for excellent technical assistance.
This work was supported by the FAZIT Stiftung (to J. Modrof) and by the Deutsche Forschungsgemeinschaft (SFB 535 and 286).
REFERENCES
- 1.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cartee, T. L., and G. W. Wertz. 2001. Respiratory syncytial virus M2-1 protein requires phosphorylation for efficient function and binds viral RNA during infection. J. Virol. 75:12188-12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins, P. L., M. G. Hill, J. Cristina, and H. Grosfeld. 1996. Transcription elongation factor of respiratory syncytial virus, a nonsegmented negative-strand RNA virus. Proc. Natl. Acad. Sci. USA 93:81-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cuesta, I., X. Geng, A. Asenjo, and N. Villanueva. 2000. Structural phosphoprotein M2-1 of the human respiratory syncytial virus is an RNA binding protein. J. Virol. 74:9858-9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DuBois, R. N., M. W. McLane, K. Ryder, L. F. Lau, and D. Nathans. 1990. A growth factor-inducible nuclear protein with a novel cysteine/histidine repetitive sequence. J. Biol. Chem. 265:19185-19191. [PubMed] [Google Scholar]
- 6.Elliott, L. H., M. P. Kiley, and J. B. McCormick. 1985. Descriptive analysis of Ebola virus proteins. Virology 147:169-176. [DOI] [PubMed] [Google Scholar]
- 7.Fearns, R., and P. L. Collins. 1999. Role of the M2-1 transcription antitermination protein of respiratory syncytial virus in sequential transcription. J. Virol. 73:5852-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friesen, W. J., and M. K. Darby. 2001. Specific RNA binding by a single C2H2 zinc finger. J. Biol. Chem. 276:1968-1973. [DOI] [PubMed] [Google Scholar]
- 9.Garcia, J., B. Garcia-Barreno, A. Vivo, and J. A. Melero. 1993. Cytoplasmic inclusions of respiratory syncytial virus-infected cells: formation of inclusion bodies in transfected cells that coexpress the nucleoprotein, the phosphoprotein, and the 22K protein. Virology 195:243-247. [DOI] [PubMed] [Google Scholar]
- 10.Hanas, J. S., D. J. Hazuda, D. F. Bogenhagen, F. Y. Wu, and C. W. Wu. 1983. Xenopus transcription factor A requires zinc for binding to the 5 S RNA gene. J. Biol. Chem. 258:14120-14125. [PubMed] [Google Scholar]
- 11.Hardy, R. W., S. B. Harmon, and G. W. Wertz. 1999. Diverse gene junctions of respiratory syncytial virus modulate the efficiency of transcription termination and respond differently to M2-mediated antitermination. J. Virol. 73:170-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardy, R. W., and G. W. Wertz. 2000. The Cys3-His1 motif of the respiratory syncytial virus M2-1 protein is essential for protein function. J. Virol. 74:5880-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang, C., K. Kiyotani, Y. Fujii, N. Fukuhara, A. Kato, Y. Nagai, T. Yoshida, and T. Sakaguchi. 2000. Involvement of the zinc-binding capacity of Sendai virus V protein in viral pathogenesis. J. Virol. 74:7834-7841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunt, J. B., S. H. Neece, and A. Ginsburg. 1985. The use of 4-(2-pyridylazo)resorcinol in studies of zinc release from Escherichia coli aspartate transcarbamoylase. Anal. Biochem. 146:150-157. [DOI] [PubMed] [Google Scholar]
- 15.Kolesnikova, L., E. Mühlberger, E. Ryabchikova, and S. Becker. 2000. Ultrastructural organization of recombinant Marburg virus nucleoprotein: comparison with Marburg virus inclusions. J. Virol. 74:3899-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller, J., A. D. McLachlan, and A. Klug. 1985. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 4:1609-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Modrof, J., C. Möritz, L. Kolesnikova, T. Konakova, B. Hartlieb, A. Randolf, E. Mühlberger, and S. Becker. 2001. Phosphorylation of Marburg virus VP30 at serines 40 and 42 is critical for its interaction with NP inclusions. Virology 287:171-182. [DOI] [PubMed] [Google Scholar]
- 18.Modrof, J., E. Mühlberger, H. D. Klenk, and S. Becker. 2002. Phosphorylation of VP30 impairs Ebola virus transcription. J. Biol. Chem. 277:33099-33104. [DOI] [PubMed] [Google Scholar]
- 19.Morgan, B., L. Sun, N. Avitahl, K. Andrikopoulos, T. Ikeda, E. Gonzales, P. Wu, S. Neben, and K. Georgopoulos. 1997. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J. 16:2004-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mühlberger, E., B. Lötfering, H.-D. Klenk, and S. Becker. 1998. Three of the four nucleocapsid proteins of Marburg virus, NP, VP35, and L, are sufficient to mediate replication and transcription of Marburg virus-specific monocistronic minigenomes. J. Virol. 72:8756-8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mühlberger, E., M. Weik, V. E. Volchkov, H.-D. Klenk, and S. Becker. 1999. Comparison of the transcription and replication strategies of Marburg virus and Ebola virus by using artificial replication systems. J. Virol. 73:2333-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peters, C. J., and A. S. Khan. 1999. Filovirus diseases. Curr. Top. Microbiol. Immunol. 235:85-95. [DOI] [PubMed] [Google Scholar]
- 23.Pfister, T., K. W. Jones, and E. Wimmer. 2000. A cysteine-rich motif in poliovirus protein 2CATPase is involved in RNA replication and binds zinc in vitro. J. Virol. 74:334-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sutter, G., M. Ohlmann, and V. Erfle. 1995. Non-replicating vaccinia vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett. 371:9-12. [DOI] [PubMed] [Google Scholar]
- 25.Tang, R. S., N. Nguyen, X. Cheng, and H. Jin. 2001. Requirement of cysteines and length of the human respiratory syncytial virus M2-1 protein for protein function and virus viability. J. Virol. 75:11328-11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turnage, M. A., P. Brewer-Jensen, W. L. Bai, and L. L. Searles. 2000. Arginine-rich regions mediate the RNA binding and regulatory activities of the protein encoded by the Drosophila melanogaster suppressor of sable gene. Mol. Cell. Biol. 20:8198-8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Volchkov, V. E., V. A. Volchkova, E. Mühlberger, L. V. Kolesnikova, M. Weik, O. Dolnik, and H. D. Klenk. 2001. Recovery of infectious Ebola virus from complementary DNA: RNA editing of the GP gene and viral cytotoxicity. Science 291:1965-1969. [DOI] [PubMed] [Google Scholar]
- 28.Weik, M., J. Modrof, H.-D. Klenk, S. Becker, and E. Mühlberger. 2002. Ebola virus VP30-mediated transcription is regulated by RNA secondary structure formation. J. Virol. 76:8532-8539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Worthington, M. T., B. T. Amann, D. Nathans, and J. M. Berg. 1996. Metal binding properties and secondary structure of the zinc-binding domain of Nup475. Proc. Natl. Acad. Sci. USA 93:13754-13759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang, Y., D. Keeney, X. Tang, N. Canfield, and B. A. Rasmussen. 1999. Kinetic properties and metal content of the metallo-beta-lactamase CcrA harboring selective amino acid substitutions. J. Biol. Chem. 274:15706-15711. [DOI] [PubMed] [Google Scholar]