Abstract
The formation of nontransmissible virus-like particles (NTVLP) by cells infected with F-deficient Sendai virus (SeV/ΔF) was found to be temperature sensitive. Analysis by hemagglutination assays and Western blotting demonstrated that the formation of NTVLP at 38°C was about 1/100 of that at 32°C, whereas this temperature-sensitive difference was only moderate in the case of F-possessing wild-type SeV. In order to reduce the NTVLP formation with the aim of improving SeV for use as a vector for gene therapy, amino acid substitutions found in temperature-sensitive mutant SeVs were introduced into the M (G69E, T116A, and A183S) and HN (A262T, G264R, and K461G) proteins of SeV/ΔF to generate SeV/MtsHNtsΔF. The use of these mutations allows vector production at low temperature (32°C) and therapeutic use at body temperature (37°C) with diminished NTVLP formation. As expected, the formation of NTVLP by SeV/MtsHNtsΔF at 37°C was decreased to about 1/10 of that by SeV/ΔF, whereas the suppression of NTVLP formation did not cause either enhanced cytotoxicity or reduced gene expression of the vector. The vectors showed differences with respect to the subcellular distribution of M protein in the infected cells. Clear and accumulated immunocytochemical signals of M protein on the cell surface were not observed in cells infected by SeV/ΔF at an incompatible temperature, 38°C, or in those infected by SeV/MtsHNtsΔF at 37 or 38°C. The absence of F protein in SeV/ΔF and the additional mutations in M and HN in SeV/MtsHNtsΔF probably weaken the ability to transport M protein to the plasma membrane, leading to the diminished formation of NTVLP.
Sendai virus (SeV) is an enveloped virus with a nonsegmented negative-strand RNA genome and is a member of the family Paramyxoviridae. The SeV genome contains six major genes arranged in tandem. The three virus-derived proteins nucleoprotein (NP), phosphoprotein (P), and large protein (L) form a ribonucleoprotein complex (RNP) with the SeV genomic RNA, and the RNP acts as a template for transcription and replication. Matrix protein (M) is a lining protein of the viral particle and is involved in the assembly of the particle. Two spike proteins, hemagglutinin-neuraminidase (HN) and fusion protein (F), mediate the attachment of virions and penetration of RNPs into infected cells, respectively (18). SeV replication appears to be independent of nuclear functions, and SeV does not have a DNA phase throughout its life cycle, so the virus is unable to transform cells by integrating its genetic information into the cellular genome (18).
Since SeV infects and replicates in most mammalian cells, including human cells, and directs high-level expression of the genes it carries, vectors derived from SeV are expected to be useful for gene therapy by expressing therapeutic genes and vaccine antigens (16, 21, 29, 36, 38). In particular, we plan the clinical application of an SeV vector carrying human fibroblast growth factor 2 for the treatment of peripheral arterial disease. F-deficient SeV (SeV/ΔF) has thus been produced and demonstrated to be non-self-transmissible due to loss of the F gene in its genomic RNA (19). Recently, we have found that nontransmissible virus-like particles (NTVLP) are formed in cells infected with SeV/ΔF. The NTVLP formation should be reduced for the next-generation SeV vector rather than retained, although so far it has not been proven that NTVLP formation brings about any kind of adverse effect in vivo. In this study, we first characterized the virion formation of SeV/ΔF and showed that loss of the F gene from the SeV genome caused temperature sensitivity of NTVLP formation. We further succeeded in reducing NTVLP formation by introducing amino acid substitutions that had been found in the M and HN proteins of SeV strains Cl.151 (17) and ts271 (33), respectively. Both strains showed temperature sensitivity of virion formation and had been shown to have amino acid substitutions in the M and HN proteins, which are considered to support virus assembly and budding, like the F protein deleted in SeV/ΔF. The use of temperature-sensitive mutations allows vector production to be carried out at low temperature and therapeutic use to be done at body temperature, thereby diminishing NTVLP production. We found that the subcellular localization of M protein was altered by introducing the mutations into M protein itself, and this might be one of the mechanisms of the reduction of NTVLP formation.
MATERIALS AND METHODS
Cells and viruses.
Two monkey kidney cell lines, LLC-MK2 and CV-1, were cultured in Eagle's minimal essential medium (MEM) (Gibco-BRL, Rockville, Md.) supplemented with 10% heat-inactivated fetal bovine serum (FBS). A human lung epithelial cell line, BEAS-2B, was cultured in a 1:1 mixture of RPMI 1640 (Gibco-BRL) and Dulbecco's modified Eagle medium (DMEM) (Gibco-BRL) supplemented with 10% FBS. A rat myoblast cell line, A-10, was cultured in DMEM supplemented with 10% FBS. All cells were cultured in a humidified atmosphere of 5% CO2-95% air at 37°C. The genome of the attenuated SeV Z strain was used as the basis for genome modifications in this study. Wild-type SeV carrying the enhanced green fluorescent protein (GFP) gene inserted before the NP open reading frame (ORF) (SeV+18GFP) (26) was grown in 10-day-old embryonated chicken eggs (15). A series of F-deficient SeV vectors were prepared by using recombinant LLC-MK2 cells carrying F genes (LLC-MK2/F7) (19). An adenovirus vector, AxCANCre (14), expressing Cre recombinase was used for the induction of F protein in LLC-MK2/F7 cells (referred to as LLC-MK2/F7/A). Recombinant vaccinia virus vTF7-3 (6) carrying T7 RNA polymerase was inactivated with psoralen and long-wave UV irradiation and then used for RNP recovery experiments (35).
Antibodies.
The rabbit polyclonal anti-M antibody N-39F was raised against three mixed synthetic peptides of SeV M protein, i.e., MADIYRFPKFSYE+Cys, LRTGPDKKAIPH+Cys, and Cys+NVVAKNIGRIRKL, corresponding to amino acids 1 to 13, 23 to 35, and 336 to 348 of SeV M, respectively. Anti-HN antibody HN-2 is a mouse monoclonal antibody (23). Goat anti-rabbit immunoglobulin G (IgG) conjugated with horseradish peroxidase was purchased from ICN (Aurora, Ohio). Goat anti-rabbit IgG conjugated with Alexa Fluor 488 or Alexa Fluor 568 and goat anti-mouse IgG conjugated with Alexa Fluor 488 were purchased from Molecular Probes (Eugene, Oreg.).
Introduction of temperature-sensitive mutations in the M and HN genes of F-deficient SeV.
To introduce mutations into the M gene of F-deficient SeV carrying the reporter GFP gene (SeV18+/ΔF-GFP), the 4.9-kb NaeI fragment containing the M gene of pSeV18+/ΔF-GFP (19) was cloned into pBluescript II (Stratagene, La Jolla, Calif.) to generate pBlueNaeIfrg-ΔFGFP. Site-directed mutagenesis was conducted by using the QuikChange site-directed mutagenesis kit (Stratagene) and the M gene of pBlueNaeIfrg-ΔFGFP as a template. For mutagenesis of the M gene, the primer pairs 5′-GAAACAAACAACCAATCTAGAGAGCGTATCTGACTTGAC-3′ (sense) and 5′-GTCAAGTCAGATACGCTCTCTAGATTGGTTGTTTGTTTC-3′ (antisense) for G69A, 5′-ATTACGGTGAGGAGGGCTGTTCGAGCAGGAG-3′ (sense) and 5′-CTCCTGCTCGAACAGCCCTCCTCACCGTAAT-3′ (antisense) for T116A, and 5′-GGGGCAATCACCATATCCAAGATCCCAAAGACC-3′ (sense) and 5′-GGTCTTTGGGATCTTGGATATGGTGATTGCCCC-3′ (antisense) for A183S were used. The 2.6-kb SalI-digested and partially ApaLI-digested fragment containing the mutagenized M gene was cloned into LITMUS 38 (New England Biolabs, Beverly, Mass.) with the 6.3-kb ApaLI and NheI fragment of pSeV18+/ΔF-GFP, which contained the HN gene, to generate LitmusSalINheIfrg-MtsΔFGFP. Site-directed mutagenesis of the HN gene was conducted by using LitmusSalINheIfrg-MtsΔFGFP or LitmusSalINheIfrg-ΔFGFP, which has no mutation in the M gene, as a template to generate LitmusSalINheIfrg-MtsHNtsΔFGFP or LitmusSalINheIfrg-HNtsΔFGFP, respectively. For mutagenesis of the HN gene, the primer pairs 5′-CATGCTCTGTGGTGACAACCCGGACTAGGGGTTATCA-3′ (sense) and 5′-TGATAACCCCTAGTCCGGGTTGTCACCACAGAGCATG-3′ (antisense) for A262T/G264R and 5′-CTTGTCTAGACCAGGAAATGAAGAGTGCAATTGGTACAATA-3′ (sense) and 5′-TATTGTACCAATTGCACTCTTCATTTCCTGGTCTAGACAAG-3′ (antisense) for K461G were used. The 8.9-kb SalI- and NheI-digested fragment containing both mutagenized M and HN genes was substituted for the corresponding fragment of pSeV18+/ΔF-GFP to generate pSeV18+/MtsHNtsΔF-GFP. pSeV18+/MtsΔF-GFP or pSeV18+/HNtsΔF-GFP, containing mutagenized M (and nonmutagenized HN) or mutagenized HN (and nonmutagenized M), respectively, was constructed by replacing the 8.9-kb SalI- and NheI-digested fragment of LitmusSalINheIfrg-MtsΔF-GFP or LitmusSalINheIfrg-HNtsΔF-GFP, respectively, with that of pSeV18+/ΔF-GFP.
Insertion of the SEAP gene upstream of the NP ORF.
To quantify the expression level of a foreign gene carried in the SeV genome, the gene for the secreted form of human placental alkaline phosphatase (SEAP) was inserted upstream of the NP ORF. The site for insertion was previously created and consists of 18 nucleotides (GAGGGCCCGCGGCCGCGA) containing a unique NotI restriction site (9). The SEAP gene with the SeV transcriptional termination (E) and restart (S) signals connected with a trinucleotide intergenic sequence at its 3′ end was amplified by PCR with pSEAP-Basic (Clontech, Palo Alto, Calif.) (34). The amplified 1.7-kb fragment was introduced into the NotI sites of pSeV18+/ΔF-GFP, pSeV18+/MtsHNtsΔF-GFP, pSeV18+/MtsΔF-GFP, and pSeV18+/HNtsΔF-GFP to generate pSeV18+SEAP/ΔF-GFP, pSeV18+SEAP/MtsHNtsΔF-GFP, pSeV18+SEAP/MtsΔF-GFP, and pSeV18+SEAP/HNtsΔF-GFP, respectively. In order to introduce the single mutation (G69E, T116A, or A183S) in M of pSeV18+SEAP/ΔF-GFP, the 8.9-kb SalI- and NheI-digested fragment of LitmusSalINheIfrg-ΔFGFP containing the respective mutagenized M gene was substituted for the corresponding fragment of pSeV18+SEAP/ΔF-GFP, thus generating pSeV18+SEAP/M(G69E)ΔF-GFP, pSeV18+SEAP/M(T116A)ΔF-GFP, and pSeV18+SEAP/M(A183S)ΔF-GFP.
Recovery of F-deficient SeV vectors from cDNA.
A series of F-deficient SeV vectors were recovered from cDNAs according to previously described procedures (19) with slight modifications. Briefly, approximately 107 LLC-MK2 cells seeded in a 10-cm-diameter dish were infected with psoralen- and long-wave-UV-treated (11) vTF7-3 at a multiplicity of infection (MOI) of 2. After a 1-h incubation at room temperature, the cells were washed three times with MEM and transfected with the plasmid mixture described below. For each transfection, plasmids pGEM-NP (4 μg) (15), pGEM-P (2 μg) (15), pGEM-L (4 μg) (15), and pGEM-FHN (4 μg) (20) and the indicated SeV cDNA plasmid (12 μg) were suspended in 200 μl of OptiMEM (Gibco-BRL) with 110 μl of Superfect transfection reagent (Qiagen, Tokyo, Japan). After standing at room temperature for 15 min, the mixture was added to the cells in 3 ml of OptiMEM containing 3% FBS and cultured further for 3 h. The cells were washed twice with MEM and then cultured for 24 h in MEM containing 1-β-d-arabinofuranosylcytosine (AraC) (40 μg/ml) and trypsin (7.5 μg/ml). Approximately 107 LLC-MK2/F7/A cells, suspended in MEM containing AraC (40 μg/ml) and trypsin (7.5 μg/ml), were layered on the transfected cells (11) and cultured at 37°C for an additional 48 h. The cells were harvested, and the pellet was resuspended in OptiMEM (107 cells/ml). After three cycles of freezing and thawing, the cell lysate was transfected into LLC-MK2/F7/A cells. They were cultured at 32°C in MEM containing AraC (40 μg/ml) and trypsin (7.5 μg/ml) for 3 to 12 days. The viruses recovered in the culture supernatants could be further amplified by a few serial infections in LLC-MK2/F7/A cells at 32°C. The spread of GFP expression from cells to neighboring cells was observed by fluorescence microscopy. The titers of recovered viral vectors were expressed as cell infectious units estimated from GFP-expressing cells (GFP-CIU per milliliter) (20). As the seed virus for all experiments, the third or fourth passage of culture supernatants, stored at −80°C after addition of bovine serum albumin solution (Gibco-BRL) to a final concentration of 1% (wt/vol), was used.
Kinetic analysis of NTVLP formation.
LLC-MK2 cells (106) grown in six-well plates were infected at an MOI of 3 with SeV18+GFP, SeV18+/ΔF-GFP, or SeV18+/MtsHNtsΔF-GFP and incubated at 32, 37, or 38°C in serum-free MEM. The culture supernatants were collected every 24 h, with immediate addition of MEM to the remaining cells. The NTVLP were quantified by the hemagglutination (HA) assay according to a method described previously (15). One HA unit was estimated to be equivalent to 106 SeV virions, and the HA activity was also accordingly expressed as the number of virus-like particles.
Semiquantitative Western blotting.
LLC-MK2 cells (106) grown in six-well plates were infected at an MOI of 3 with SeV18+GFP, SeV18+/ΔF-GFP, or SeV18+/MtsHNtsΔF-GFP and incubated in serum-free MEM at 37°C. Two days after the infection, the culture supernatants were passed through a 0.45-μm-pore-size filter and centrifuged at 48,000 × g for 45 min to collect NTVLP. Cells recovered from one well of a six-well plate were frozen at −80°C and then thawed in 100 μl of 1× sample buffer for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Red Loading Buffer Pack; New England Biolabs, Beverly, Mass.) and heated at 98°C for 10 min. After centrifugation, a 10-μl aliquot of the supernatant was loaded onto a sodium dodecyl sulfate-polyacrylamide gel (Multigel 10/20; Daiichi Pure Chemicals Co., Ltd., Tokyo, Japan). After electrophoresis at 15 mA for 2.5 h, proteins were transferred to a polyvinylidene difluoride membrane (Immobilon PVDF transfer membrane; Millipore, Bedford, Mass.) by the semidry method at 100 mA for 1 h. The membrane was immersed in blocking solution (Block Ace; Snow Brand Milk Products Co., Ltd., Sapporo, Japan) at 4°C for 1 h or more, soaked in a primary antibody solution containing 10% Block Ace supplemented with anti-M antibody diluted 1:1,000, and allowed to stand at 4°C overnight. After being washed with Tris-buffered saline (TBS) containing 0.05% Tween 20 and with TBS (three times each), the membrane was immersed in a secondary antibody solution containing 10% Block Ace and supplemented with a 1:5,000 dilution of anti-rabbit IgG conjugated with horseradish peroxidase and then agitated at room temperature for 1 h. After the membrane was washed three times each with TBS-0.05% Tween 20 and with TBS, the proteins on the membrane were detected by the chemiluminescence method (ECL Western blotting detection reagents; Amersham Pharmacia Biotech, Uppsala, Sweden). The ratio of the M protein level in NTVLP and in cells was estimated from the relative densities detected with chemical fluorescent Lumi-Phos Plus and an LAS 1000 image analyzer (Fuji Film, Tokyo, Japan).
Plaque-forming assay.
LLC-MK2/F7/A cells (106/well) grown in six-well plates were infected at an MOI of 0.1 with SeV18+/ΔF-GFP or SeV18+/MtsHNtsΔF-GFP and overlaid with a 1:1 mixture of 2× MEM (Gibco-BRL) and 2% agarose solution (SeaPlaque GTG agarose; BMA, Rockland, Maine) containing final concentrations of 7.5 μg of trypsin per ml and 0.1% bovine serum albumin. After the agar was solidified, the infected cells were cultured for 6 days at 32 or 37°C. GFP-expressing cells were counted under a DM IRB-SLR fluorescence microscope (Leica, Wetzlar, Germany).
Quantitative analysis of cytotoxicity.
LLC-MK2, BEAS-2B, and CV-1 cells (4 × 104/well) grown in 96-well plates were infected at an MOI of 0.001, 0.01, 0.03, 0.1, 0.3, 1, 3, or 10 with SeV18+/ΔF-GFP or SeV18+/MtsHNtsΔF-GFP and incubated in their respective serum-free medium at 37°C. The culture supernatants were recovered 3 days after the infection and subjected to a cytotoxicity test with a cytotoxicity detection kit (Roche, Basel, Switzerland) to measure lactate dehydrogenase (LDH) activity released from damaged cells.
SEAP assay.
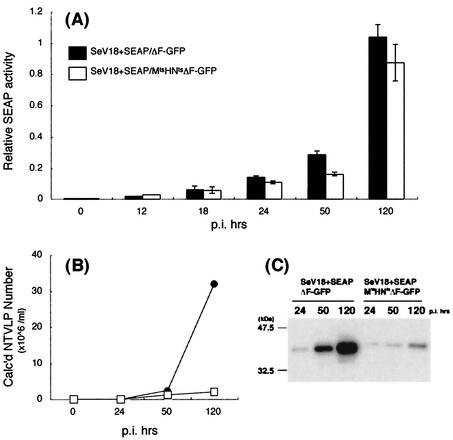
LLC-MK2 cells (106/well) grown in six-well plates were infected at an MOI of 3 with SeV18+SEAP/ΔF-GFP or SeV18+SEAP/MtsHNtsΔF-GFP and incubated in serum-free MEM at 37°C. The culture supernatants were recovered at 12, 18, 24, 50, and 120 h after the infection, and their SEAP activities were measured with an SEAP Reporter Assay Kit (Toyobo, Osaka, Japan) with chemical fluorescent Lumi-Phos Plus and an LAS 1000 image analyzer (Fuji Film).
Immunofluorescence microscopy.
Monolayer cultures of LLC-MK2 cells in eight-well slide chambers (Nulgenunc, Rochester, N.Y.) were infected at an MOI of 1 with SeV18+GFP, SeV18+/ΔF-GFP, or SeV18+/MtsHNtsΔF-GFP and incubated in serum-free MEM for 2 days. After being washed with PBS, cells were fixed with cold methanol at 4°C for 15 min and then washed three times with PBS. The fixed cells were permeabilized with 0.1% Triton X-100 with 2% goat serum (Gibco-BRL) in PBS. SeV M was detected by incubating the cells with a polyclonal anti-M antibody (N-39F) in PBS with 2% goat serum at 37°C for 15 min followed by labeling with Alexa Fluor 488-conjugated anti-rabbit IgG (Molecular Probes) at 37°C for 30 min. The cells were washed three times with PBS after each of these incubations, mounted with an antifade reagent in glycerol buffer (Vector Laboratories, Inc., Burlingame, Calif.), and observed under a DM IRB-SLR fluorescence microscope (Leica).
Immunofluorescent confocal laser scanning microscopy.
A-10 cells cultured on glass-bottom dishes (Asahi Technoglass Corp., Tokyo, Japan) were infected at an MOI of 1 with SeV18+SEAP/ΔF-GFP or SeV18+SEAP/MtsHNtsΔF-GFP and incubated in serum-free DMEM for 1 day at 32 or 37°C. After being washed with PBS, the cells were fixed with cold methanol at 4°C for 15 min and then washed three times with PBS. The fixed cells were then treated with 0.1% Triton X-100 with 2% goat serum in PBS. For double staining, the cells were incubated with a mixture of rabbit polyclonal anti-M antibody and mouse monoclonal anti-HN antibody in PBS with 2% goat serum at 37°C for 15 min. Next, they were stained with Alexa Fluor 568-conjugated anti-rabbit IgG and Alexa Fluor 488-conjugated anti-mouse IgG (Molecular Probes) at 37°C for 30 min. TO_PRO3 (Molecular Probes) was added to the cells for staining the nuclei, and the cells were allowed to stand at room temperature for 15 min. Finally, to prevent quenching, Slow Fade antifade solution (Molecular Probes) was substituted for the staining solution, and then the cells were observed under an MRC-1024 confocal laser microscope (Bio-Rad, Hercules, Calif.).
RESULTS
Construction of F-deficient SeV cDNA carrying temperature-sensitive mutations.
Temperature-sensitive mutations in M (G69E, T116A, and A183S) and HN (A262T, G264R, and K461G) were introduced into SeV/ΔF (19), since SeV/ΔF, a nontransmissible type of SeV, is being considered for development as a first-generation SeV vector for gene therapy and should be improved. At first, all of the above-described mutations were introduced by site-directed mutagenesis into SeV/ΔF genomic cDNA carrying the GFP gene at the site of the F deletion (pSeV18+/ΔF-GFP) (19) to generate pSeV18+/MtsHNtsΔF-GFP. The tactical idea of introduction of the mutations into the M and HN genes came from the sequences of SeV temperature-sensitive strains Cl.151 (17) and ts271 (33), respectively. Further, to quantify the expression level of a foreign gene carried in the SeV genome, the SEAP gene was inserted upstream of the NP ORF, and thus pSeV18+SEAP/ΔF-GFP and pSeV18+SEAP/MtsHNtsΔF-GFP were generated. In addition, for analyzing the roles of individual mutations, some mutations were introduced singly or in combination, thus generating pSeV18+SEAP/MtsΔF-GFP, pSeV18+SEAP/HNtsΔF-GFP, pSeV18+SEAP/M(G69E)ΔF-GFP, pSeV18+SEAP/M(T116A)ΔF-GFP, and pSeV18+SEAP/M(A183S)ΔF-GFP, the last three of which have just one mutation in M protein.
Recovery of F-deficient SeV vectors carrying temperature-sensitive mutations.
Viruses were recovered as previously described (19, 20). Since the F gene has been deleted in the cDNA, all of the F-deficient SeV vectors were recovered by utilizing a packaging cell line that expresses SeV-F protein (LLC-MK2/F7/A) (19). For the recovery of SeV vectors carrying the temperature-sensitive mutations, the culture at the initial passage (P1) and the subsequent cultures were carried out at 32°C because the original temperature-sensitive viruses had been reported to propagate well at 32°C (17, 33). The titers of recovered SeV vectors were about (or more than, in most cases) 108 GFP-CIU/ml; that is, all of the vectors could be recovered in high titers at 32°C.
NTVLP formation by F-deficient SeV is temperature sensitive and reduced successfully by mutations in M and HN.
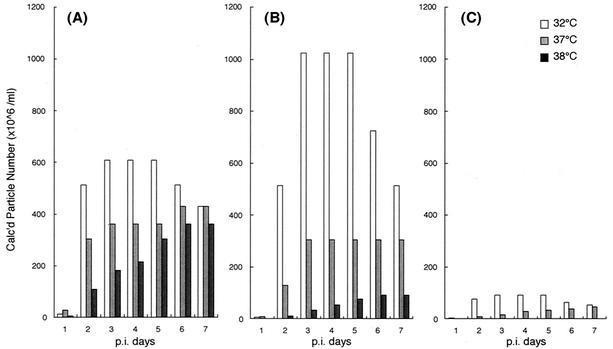
Virus-like particles with no transmission function (NTVLP) are produced by cells infected with F-deficient strain SeV18+/ΔF-GFP (20). In order to characterize the NTVLP formation by SeV18+/ΔF-GFP and to examine the effect of the temperature-sensitive mutations in SeV18+/MtsHNtsΔF-GFP, the kinetics of particle formation were quantified for these two types of F-deficient SeV as well as wild-type SeV carrying the GFP gene (SeV18+GFP). After the infection of LLC-MK2 cells at an MOI of 3, the cells were cultured at 32, 37, or 38°C. Observation of GFP expression 3 days after the infection under a fluorescence microscope revealed almost equal infection efficiency and expression of GFP for all three of these types of virus vectors at all of these temperatures (data not shown). The appearance of NTVLP in the medium was quantified by the HA assay. In the case of SeV18+/ΔF-GFP, the NTVLP formation at 38°C was about 1/100 of that at 32°C at 2 and 3 days after infection, whereas the reduction was moderate in the case of SeV18+GFP (Fig. 1). This indicates that the deletion of the F gene caused temperature sensitivity of NTVLP formation. The NTVLP formation by SeV18+/MtsHNtsΔF-GFP was remarkably small at 37°C and was about 1/10 of that of SeV18+/ΔF-GFP at all times postinfection (Fig. 1).
FIG. 1.
Quantification of kinetics of NTVLP production by HA assay. The appearance of virus-like particles was quantified by assaying HA activity in the culture supernatant of LLC-MK2 cells cultured at 32, 37, or 38°C over time (at the time of sampling, the medium was replaced with fresh medium) after infection with SeV18+GFP (A), SeV18+/ΔF-GFP (B), or SeV18+/MtsHNtsΔF-GFP (C) at an MOI of 3.
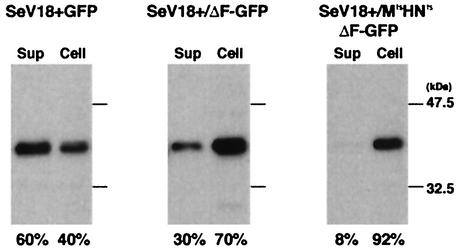
To quantify the NTVLP formation by using another indicator, semiquantitative Western blotting was performed. SeV-M proteins from culture supernatants and cells, both prepared 2 days after the infection, were detected by using the anti-M antibody (N-39F). In the cases of all three vectors, M proteins were detected at similar levels in the cells, but the levels in the supernatants were quite different. Whereas 60% of the total M protein was in the supernatant for SeV18+GFP, 30% and only 8% was in the supernatant for SeV18+/ΔF-GFP and SeV18+/MtsHNtsΔF-GFP, respectively (Fig. 2). Thus, the reduction of NTVLP formation was also confirmed by Western blotting.
FIG. 2.
Quantification of NTVLP by Western blotting. The ratio of the M protein level in virus-like particles and in the cells was determined by Western blotting with an anti-M antibody. The culture supernatant (Sup) and cells were recovered from LLC-MK2 cell cultures incubated at 37°C for 2 days after infection with SeV18+GFP, SeV18+/ΔF-GFP, or SeV18+/MtsHNtsΔF-GFP at an MOI of 3. Each lane contains the equivalent of 1/10 of the contents of a well of a six-well plate.
Diminished plaque-forming capability of F-deficient SeV vectors carrying temperature-sensitive mutations.
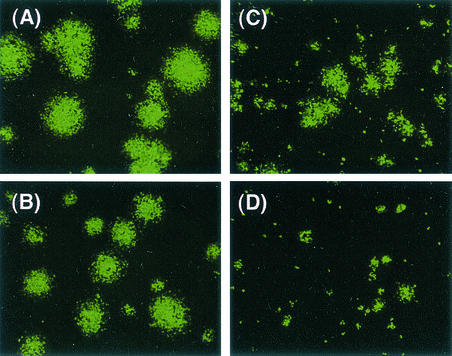
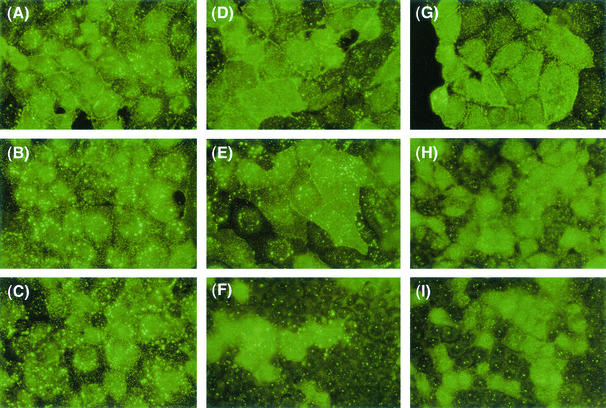
The diameters of plaques formed at 32 and 37°C were also compared for cells expressing F protein (LLC-MK2/F7/A) infected with SeV18+/ΔF-GFP or SeV18+/MtsHNtsΔF-GFP, since they reflect the particle-forming capability of transfected viruses. The diameters observed 6 days after the infection by SeV18+/MtsHNtsΔF-GFP at 37°C was much smaller than that after infection by SeV18+/ΔF-GFP (Fig. 3), suggesting that the virion formation of SeV18+/MtsHNtsΔF-GFP is impaired at 37°C. The diameter after infection by SeV18+GFP was comparable to or a little larger than that after infection by SeV18+/ΔF-GFP at both 32 and 37°C (data not shown).
FIG. 3.
Comparison of plaque-forming capabilities. Microscopic images show the GFP expression in cells (LLC-MK2/F7/A) expressing F protein when cultured at 32°C (A and C) and 37°C (B and D) for 6 days after infection with SeV18+/ΔF-GFP (A and B) or SeV18+/MtsHNtsΔF-GFP (C and D).
Cytotoxicity is not enhanced by suppression of NTVLP formation.
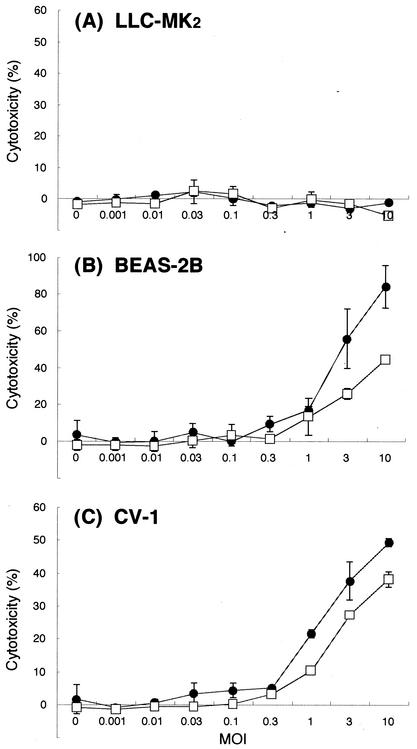
SeV infection causes cytopathic effects in some cell types (2, 7). The effect of the introduction of temperature-sensitive mutations in M and HN in the vectors was thus examined with respect to this cytopathy by using the SeV-sensitive cell lines BEAS-2B and CV-1 as well as LLC-MK2. Cells were grown in 96-well plates, infected with SeV18+/ΔF-GFP or SeV18+/MtsHNtsΔF-GFP, and further incubated in serum-free medium at 37°C. The culture supernatants recovered 3 days after infection were analyzed for LDH to quantify the cytotoxicity. Neither of the SeV vectors was cytotoxic in LLC-MK2 under these conditions (Fig. 4A). Further, the cytotoxicity of SeV18+/MtsHNtsΔF-GFP was comparable to or lower than that of SeV18+/ΔF-GFP in CV-1 and BEAS-2B (Fig. 4B and C). Thus, cytotoxicity was not caused or enhanced by suppressing NTVLP formation via the introduction of temperature-sensitive mutations.
FIG. 4.
Quantitative analysis of SeV infection-dependent cytotoxicity. Cytotoxicity was estimated by measuring the LDH released into the cell culture medium. LLC-MK2 (A), BEAS-2B (B), or CV-1 (C) cells were infected with SeV18+/ΔF-GFP (•) or SeV18+/MtsHNtsΔF-GFP (□) at an MOI of 0.01, 0.03, 0.1, 0.3, 1, 3, or 10. The cells were cultured in serum-free medium. The assay was carried out 3 days after the infection. The values were from three experiments. Error bars indicate standard deviations.
The gene expression level is maintained despite the suppression of NTVLP formation.
The low level of NTVLP formation by SeV18+/MtsHNtsΔF-GFP is an advantage with respect to the use of this vector for purposes such as gene therapy. However, if the expression level of the inserted gene(s) was decreased at the same time, the vector would not be attractive and its further modification might be required. Therefore, the expression level of the vector was evaluated by the use of SEAP, an easily detectable marker of protein production. LLC-MK2 cells were infected with SeV18+SEAP/ΔF-GFP or SeV18+SEAP/MtsHNtsΔF-GFP at an MOI of 3, and then the culture supernatant was repeatedly sampled up to 150 h. The SEAP activity in the supernatant was comparable for the two SeV vectors (Fig. 5A). The assay of the same samples for HA activity showed that the HA activity of SeV18+SEAP/MtsHNtsΔF-GFP was about 1/10 of that of SeV18+SEAP/ΔF-GFP GFP at all times postinfection (Fig. 5B). Further, to quantify the NTVLP formation by the vectors carrying the SEAP gene with another indicator, the viral proteins were harvested from viruses in the supernatants by centrifugation and semiquantitatively analyzed by Western blotting with an anti-M antibody (N-39F). The level of M protein, which is representative of SeV proteins, was also reduced in the supernatant (Fig. 5C). These findings indicate that the introduction of temperature-sensitive mutations reduced the NTVLP formation to 1/10 of that of the parental vector without significant reduction of the expression of an inserted gene.
FIG. 5.
Comparison of expression performances of SeV vectors carrying the SEAP gene. (A) Expression level of the inserted gene quantified by SEAP assay. SEAP activity in the culture supernatants of LLC-MK2 cells cultured for 12, 18, 24, 50, or 120 h after infection (p.i.) with SeV18+SEAP/ΔF-GFP or SeV18+SEAP/MtsHNtsΔF-GFP at an MOI of 3 is shown. The relative activities shown are from three experiments. Error bars indicate standard deviations. (B) Kinetics of NTVLP production by HA assay. HA activity in the culture supernatants of LLC-MK2 cells cultured for 24, 50, or 120 h after infection with SeV18+SEAP/ΔF-GFP (•) or SeV18+SEAP/MtsHNtsΔF-GFP (□) at an MOI of 3 is shown. (C) Quantification of NTVLP by Western blotting. The quantity of NTVLP was determined by Western blotting with an anti-M antibody. LLC-MK2 cells were cultured for 24, 50, or 120 h after infection with SeV18+SEAP/ΔF-GFP or SeV18+SEAP/MtsHNtsΔF-GFP at an MOI of 3. The culture supernatant was centrifuged to recover particles. Each lane contains the equivalent of 1/10 of the contents of a well of a six-well plate.
Abnormal subcellular localization of M protein caused by introduction of temperature-sensitive mutations.
In order to elucidate the mechanism underlying the suppression of NTVLP formation by the introduction of temperature-sensitive mutations, the subcellular localization of M protein was examined. LLC-MK2 cells were infected with each type of SeV vector (SeV18+GFP, SeV18+/ΔF-GFP, and SeV18+/MtsHNtsΔF-GFP) and cultured at 32, 37, or 38°C for 2 days. The cells were then permeabilized and immunostained by using the anti-M antibody (N-39F). In the case of self-replicating SeV18+GFP that expresses normal F and HN proteins, accumulated M protein was detectable on the cell surface at all temperatures tested (Fig. 6A to C). Such accumulation of M protein has been reported, and this had been assumed to be the site of virion formation (37). The normal cell surface accumulation of M protein is consistent with the observation that the virion formation is not significantly affected by such a temperature change. In the case of SeV18+/ΔF-GFP, the degree of accumulation of M protein was reduced at 38°C compared to that in M-positive, that is, SeV-infected, cells (Fig. 6F). A more remarkable reduction of the M protein accumulation was observed for SeV18+/MtsHNtsΔF-GFP. The accumulation was considerably disturbed even at 37°C, at which NTVLP formation was also reduced (Fig. 6H).
FIG. 6.
Subcellular localization of M protein analyzed by fluorescence microscopy. The subcellular localization of M protein (represented by green) in LLC-MK2 cells cultured at 32°C (A, D, and G), 37°C (B, E, and H), or 38°C (C, F, and I) for 2 days after infection with SeV18+GFP (A, B, and C), SeV18+/ΔF-GFP (D, E, and F), or SeV18+/MtsHNtsΔF-GFP (G, H, and I) at an MOI of 1, observed by immunostaining with an anti-M antibody, is shown. Magnification, ×400.
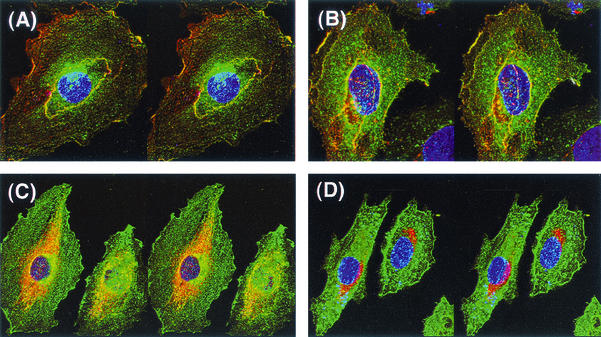
In order to study the subcellular localization of SeV proteins in more detail, analysis by confocal laser microscopy was performed. In this case, A-10 cells were used, as they are large and thick when cultured in limited numbers and thus are suitable for confocal microscopic analysis. A-10 cells were infected with SeV18+SEAP/ΔF-GFP or SeV18+SEAP/MtsHNtsΔF-GFP and then cultured at 32 or 37°C. After 1 day, the cells were immunostained with anti-M and anti-HN antibodies and observed under the microscope. In the case of SeV18+SEAP/ΔF-GFP, cell surface colocalization of M and HN proteins was seen under these culture conditions at both 32 and 37°C (Fig. 7A and B). On the other hand, for SeV18+SEAP/MtsHNtsΔF-GFP, the localization of each protein was significantly different from that for SeV18+SEAP/ΔF-GFP. M protein was hardly localized on the cell surface but was localized at a site that appeared to be the vicinity of the Golgi complex, leading to the separate subcellular location of M and HN proteins when the SeV18+SEAP/MtsHNtsΔF-GFP-infected cells were cultured at 37°C (Fig. 7D). When these cells were cultured at 32°C, M protein seemed to be localized with the subcellular component, which would be the microtubule-like fibriform structure (Fig. 7C). Similar localizations were observed for the cells 2 days after the infection (data not shown). These results indicate that the transport of the M protein in the SeV18+SEAP/MtsHNtsΔF-GFP-infected cells might be greatly suppressed. This may lead to the reduction of NTVLP formation by the temperature-sensitive mutations. In contrast, the subcellular localization of HN protein was similar in the cells infected with SeV18+SEAP/ΔF-GFP and SeV18+SEAP/MtsHNtsΔF-GFP (Fig. 7).
FIG. 7.
Subcellular localization of M and HN proteins analyzed by confocal laser scanning microscopy. Stereo three-dimensional images of the subcellular localization of M and HN proteins (represented by red and green, respectively) observed under a confocal laser microscope are shown. A-10 cells were cultured at 32°C (A and C) or 37°C (B and D) for 1 day after infection with SeV18+SEAP/ΔF-GFP (A and B) or SeV18+SEAP/MtsHNtsΔF-GFP (C and D) at an MOI of 1. The image was obtained by immunostaining with anti-M (red) and anti-HN (green) antibodies, and the colocalization of M and HN proteins is shown in yellow. The nucleus is represented by blue. Magnification, ×600.
Additive effect of temperature-sensitive mutations in M and HN for suppression of NTVLP formation.
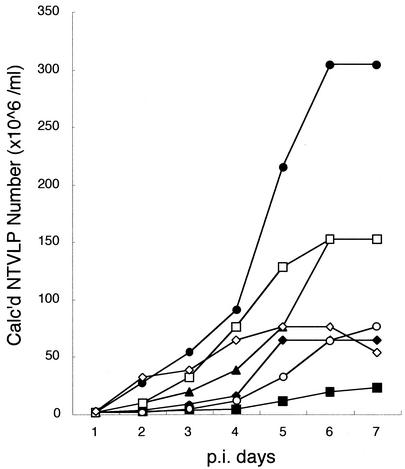
To clarify the roles of individual substitutions, we constructed SeV vectors having single mutations or combinations, and the kinetics of NTVLP formation by these vectors in LLC-MK2 cells at 37°C were quantified together with those of SeV18+SEAP/ΔF-GFP and SeV18+SEAP/MtsHNtsΔF-GFP. All of the single mutations in the M protein (G69E, T116A, or A183S) resulted in the reduction of NTVLP formation (Fig. 8), in other words, there was no substitution which did not affect the NTVLP reduction. However, the level of NTVLP formation by SeV18+SEAP/M(G69E)ΔF-GFP was similar to that by SeV18+SEAP/MtsΔF-GFP; thus, the G69E substitution might be central among them (Mts). The temperature-sensitive mutations in HN protein (HNts) reduced the level in both cases, i.e., from SeV18+SEAP/ΔF-GFP to SeV18+SEAP/HNtsΔF-GFP and from SeV18+SEAP/MtsΔF-GFP to SeV18+SEAP/MtsHNtsΔF-GFP, even though the subcellular localization of HN protein showed little alteration in cells infected with SeV18+SEAP/MtsHNtsΔF-GFP; in contrast, that of M protein did show a clear alteration (Fig. 7). The NTVLP formation of SeV18+SEAP/MtsHNtsΔF-GFP was the smallest among those of SeV vectors constructed in this study (Fig. 8). That is, both of the mutations in the M and HN proteins contribute to the reduction of NTVLP formation, and the combination of them is effective for extensive suppression of NTVLP formation.
FIG. 8.
Effect of individual temperature-sensitive mutations on suppression of NTVLP formation. The kinetics of NTVLP formation from LLC-MK2 cells cultured at 37°C after infection with SeV vectors at an MOI of 3 were quantified by measuring HA activity in the culture supernatant over time (at the time of sampling, the medium was replaced with fresh medium). NTVLP formation of F-deficient SeV vectors having the single mutation G69E (○), T166A (□), or A183S (◊) or all of these mutations (Mts) (⧫) in M protein or having temperature-sensitive mutations only in HN protein (HNts) (▴) with a SEAP gene insertion upstream of the NP ORF was compared with that of SeV18+SEAP/ΔF-GFP (•) and SeV18+SEAP/MtsHNtsΔF-GFP (▪).
DISCUSSION
The M proteins of negative-strand RNA viruses are known to play a central role in virion formation (4, 8, 13). SeV M is also thought to play key roles in both virus assembly and budding (24, 25, 31, 32), since it binds not only to the SeV RNP (5) but also to the spike proteins (1, 27) and self-aggregates to form molecular clusters (10). In concert with M, the spike proteins are involved in virion formation. The cytoplasmic tails of F and HN of SeV bind to M through the secretory pathway (1, 27), indicating that they might transport M to the cell surface for effective virus assembly. A few reports on negative-strand RNA viruses have described the effect of the modulation of these proteins on virus particle assembly. Particle formation of spike protein (G)-deficient rabies virus is reduced to 1/30 of that of G-possessing wild-type virus (22). Alterations of the cytoplasmic tails of spike proteins (F and H) of measles virus decrease virus assembly, resulting in enhanced cell-cell fusion (3). In the case of SeV, however, the lack of HN (30) or F (20) seems to result in normal NTVLP formation. Although further analysis is necessary to understand the roles of the SeV M and spike proteins in viral assembly, we tried in this study to determine the effects of manipulating those proteins.
F-deficient SeV was found to have temperature sensitivity in NTVLP formation. In the cells infected with SeV18+/ΔF-GFP, the accumulation of M protein at the cell surface was reduced at a temperature at which NTVLP formation was suppressed. M protein has been thought to be localized on the cell surface, binding to the cytoplasmic tails of both F and HN proteins (1, 28). Because the F gene had been deleted in SeV18+/ΔF-GFP, the lack of F protein in the infected cells is assumed to cause some problem with the localization and/or intracellular trafficking of M protein. It is also possible that the binding of M protein to the cytoplasmic tail of HN protein might be affected by temperature.
The formation of NTVLP by SeV/ΔF was decreased by introducing temperature-sensitive mutations in M and HN proteins, as expected from the results of F-possessing SeV having the same mutations in M (17) or HN (33). Also, our data suggest that the abnormal subcellular localization of M protein brings about the suppressed NTVLP formation by SeV/MtsHNtsΔF. In this SeV vector, a total of six amino acid substitutions have been introduced into the M (G69E, T116A, and A183S) and HN (A262T, G264R, and K461G) proteins. Individual substitutions affect NTVLP formation, and they contribute in combination for its extensive suppression. However, the mechanism for suppression of NTVLP formation is still unknown. Further analysis of this issue would also clarify the molecular details of the formation of SeV particles. The subcellular localization of M protein in cells infected with SeV18+SEAP/MtsHNtsΔF-GFP was quite different from that in cells infected with SeV18+SEAP/ΔF-GFP. Temperature-sensitive M protein was found to be localized at a subcellular site presumed to be the vicinity of the Golgi complex at the nonpermissive temperature of 37°C. Even at the permissive temperature of 32°C, M protein localization of SeV18+SEAP/MtsHNtsΔF-GFP was different from that of SeV18+SEAP/ΔF-GFP and showed a microtubule-like structure. The M protein carrying temperature-sensitive mutations might anchor on the pathway of intracellular transport in the SeV18+SEAP/MtsHNtsΔF-GFP-infected cells, and therefore its NTVLP formation was still suppressed even at 32°C, as shown in Fig. 1. The spike protein (G) of vesicular stomatitis virus has been shown to move along microtubule tracks from the Golgi apparatus to the plasma membrane (12). Thus, characterization of the role of microtubules not only in the transportation of spike proteins of SeV to the cell surface but also in that of M protein is of special interest and is now in progress.
In this study, we have succeeded in the reduction of NTVLP formation by F-deficient SeV vector, which had already been modified to be non-self-transmissible, by introducing temperature-sensitive mutations in the M and HN proteins. The production of the generated SeV18+/MtsHNtsΔF was as high as 108 CIU/ml at 32°C. Further, the reduction of NTVLP formation by SeV/MtsHNtsΔF was not accompanied by either enhanced cytotoxicity or reduced gene expression in infected cells. This vector is thus a good candidate for use as an attenuated SeV vector for gene transfer. The combination of modifications, i.e., deletion of F and temperature-sensitive mutations in M and HN proteins, used here is very effective in improvement of the SeV vector. However, this SeV18+/MtsHNtsΔF is still capable of releasing NTVLP from infected cells, although the quantity of the release is remarkably small. In order to abolish NTVLP formation completely, it would be effective to delete the M gene from the SeV genomic RNA. We already have succeeded in recovering an M-deficient SeV vector (SeV18+/ΔM) at high titer by using a new helper cell expressing SeV-M protein (unpublished data). The characterization of this vector is now in progress.
The SeV vector is expected to have advantages for gene therapy, as it infects and replicates in most mammalian cells, including human cells, and directs high-level expression of the genes it carries. Indeed, it has been shown to mediate effective gene transfer to the respiratory tracts of mice, ferrets, and sheep (36); the vascular system of rabbits (21); and the skeletal muscle (29) as well as the central nervous system (19) of rats. The attenuation of this vector would contribute not only to understanding the basic biology of SeV but also to improving the vector for use in gene therapy.
Acknowledgments
We acknowledge B. Boss for supplying vTF7-3; D. Kolakofsky for supplying pGEM-NP, pGEM-P, and pGEM-L; H. Iba for supplying pCALNdLw; N. Miura for supplying anti-HN antibody; T. Yoshida for supplying the Cl.151 strain of SeV; H. Hirata for psoralen and long-wave UV treatment of vTF7-3; and A. Kato, M. Nakanishi, M. Okayama, Y. F. Zhu, K. Kitazato, and Y. Ueda for helpful discussions.
REFERENCES
- 1.Ali, A., and D. P. Nayak. 2000. Assembly of Sendai virus: M protein interacts with F and HN proteins and with the cytoplasmic tail and transmembrane domain of F protein. Virology 276:289-303. [DOI] [PubMed] [Google Scholar]
- 2.Bitzer, M., F. Prinz, M. Bauer, M. Spiegel, W. J. Neubert, M. Gregor, K. Schulze-Osthoff, and U. Lauer. 1999. Sendai virus infection induces apoptosis through activation of caspase-8 (FLICE) and caspase-3 (CPP32). J. Virol. 73:702-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cathomen, T., H. Y. Naim, and R. Cattaneo. 1998. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J. Virol. 72:1224-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coronel, E. C., K. G. Murti, T. Takimoto, and A. Portner. 1999. Human parainfluenza virus type 1 matrix and nucleoprotein genes transiently expressed in mammalian cells induce the release of virus-like particles containing nucleocapsid-like structures. J. Virol. 73:7035-7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coronel, E. C., T. Takimoto, K. G. Murti, N. Varich, and A. Portner. 2001. Nucleocapsid incorporation into parainfluenza virus is regulated by specific interaction with matrix protein. J. Virol. 75:1117-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcin, D., G. Taylor, K. Tanebayashi, R. Compans, and D. Kolakofsky. 1998. The short Sendai virus leader region controls induction of programmed cell death. Virology 243:340-353. [DOI] [PubMed] [Google Scholar]
- 8.Garoff, H., R. Hewson, and D. J. Opstelten. 1998. Virus maturation by budding. Microbiol. Mol. Biol. Rev. 62:1171-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasan, M. K., A. Kato, T. Shioda, Y. Sakai, D. Yu, and Y. Nagai. 1997. Creation of an infectious recombinant Sendai virus expressing the firefly luciferase gene from the 3′ proximal first locus. J. Gen. Virol. 78:2813-2820. [DOI] [PubMed] [Google Scholar]
- 10.Heggeness, M. H., P. R. Smith, and P. W. Choppin. 1982. In vitro assembly of the nonglycosylated membrane protein (M) of Sendai virus. Proc. Natl. Acad. Sci. USA 79:6232-6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirata, T., A. Iida, T. Shiraki-Iida, K. Kitazato, A. Kato, Y. Nagai, and M. Hasegawa. 2002. An improved method for recovery of F-defective Sendai virus expressing foreign genes from cloned cDNA. J. Virol. Methods 104:125-133. [DOI] [PubMed] [Google Scholar]
- 12.Hirschberg, K., C. M. Miller, J. Ellenberg, J. F. Presley, E. D. Siggia, R. D. Phair, and J. Lippincott-Schwartz. 1998. Kinetic analysis of secretory protein traffic and characterization of Golgi to plasma membrane transport intermediates in living cells. J. Cell Biol. 143:1485-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Justice, P. A., W. Sun, Y. Li, Z. Ye, P. R. Grigera, and R. R. Wagner. 1995. Membrane vesiculation function and exocytosis of wild-type and mutant matrix proteins of vesicular stomatitis virus. J. Virol. 69:3156-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanegae, Y., K. Takamori, Y. Sato, G. Lee, M. Nakai, and I. Saito. 1996. Efficient gene activation system on mammalian cell chromosomes using recombinant adenovirus producing Cre recombinase. Gene 181:207-212. [DOI] [PubMed] [Google Scholar]
- 15.Kato, A., Y. Sakai, T. Shioda, T. Kondo, M. Nakanishi, and Y. Nagai. 1996. Initiation of Sendai virus multiplication from transfected cDNA or RNA with negative or positive sense. Genes Cells 1:569-579. [DOI] [PubMed] [Google Scholar]
- 16.Kawana-Tachikawa, A., M. Tomizawa, J. I. Nunoya, T. Shioda, A. Kato, E. E. Nakayama, T. Nakamura, Y. Nagai, and A. Iwamoto. 2002. An efficient and versatile mammalian viral vector system for major histocompatibility complex class I/peptide complexes. J. Virol. 76:11982-11988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kondo, T., T. Yoshida, N. Miura, and M. Nakanishi. 1993. Temperature-sensitive phenotype of a mutant Sendai virus strain is caused by its insufficient accumulation of the M protein. J. Biol. Chem. 268:21924-21930. [PubMed] [Google Scholar]
- 18.Lamb, R. A., and D. Kolakofsky. 1996. Paramyxoviridae: the viruses and their replication, p. 1177-1204. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology. Lippincott-Raven, Philadelphia, Pa.
- 19.Li, H. O., Y. F. Zhu, M. Asakawa, H. Kuma, T. Hirata, Y. Ueda, Y. S. Lee, M. Fukumura, A. Iida, A. Kato, Y. Nagai, and M. Hasegawa. 2000. A cytoplasmic RNA vector derived from nontransmissible Sendai virus with efficient gene transfer and expression. J. Virol. 74:6564-6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, H. O., Y. F. Zhu, Y. Ueda, M. Hasegawa, A. Iida, F. Tokito, T. Hirata, T. Tokusumi, H. Kuma, and M. Asakawa. May 2000. Paramyxoviridae virus vector defective in envelope gene. PCT/WO 00/70070.
- 21.Masaki, I., Y. Yonemitsu, K. Komori, H. Ueno, Y. Nakashima, K. Nakagawa, M. Fukumura, A. Kato, M. K. Hasan, Y. Nagai, K. Sugimachi, M. Hasegawa, and K. Sueishi. 2001. Recombinant Sendai virus-mediated gene transfer to vasculature: a new class of efficient gene transfer vector to the vascular system. FASEB J. 28:28.. [DOI] [PubMed] [Google Scholar]
- 22.Mebatsion, T., M. Konig, and K. K. Conzelmann. 1996. Budding of rabies virus particles in the absence of the spike glycoprotein. Cell 84:941-951. [DOI] [PubMed] [Google Scholar]
- 23.Miura, N., T. Uchida, and Y. Okada. 1982. HVJ (Sendai virus)-induced envelope fusion and cell fusion are blocked by monoclonal anti-HN protein antibody that does not inhibit hemagglutination activity of HVJ. Exp. Cell Res. 141:409-420. [DOI] [PubMed] [Google Scholar]
- 24.Mottet, G., A. Muhlemann, C. Tapparel, F. Hoffmann, and L. Roux. 1996. A Sendai virus vector leading to the efficient expression of mutant M proteins interfering with virus particle budding. Virology 221:159-171. [DOI] [PubMed] [Google Scholar]
- 25.Mottet, G., V. Muller, and L. Roux. 1999. Characterization of Sendai virus M protein mutants that can partially interfere with virus particle production. J. Gen. Virol. 80:2977-2986. [DOI] [PubMed] [Google Scholar]
- 26.Sakai, Y., K. Kiyotani, M. Fukumura, M. Asakawa, A. Kato, T. Shioda, T. Yoshida, A. Tanaka, M. Hasegawa, and Y. Nagai. 1999. Accommodation of foreign genes into the Sendai virus genome: sizes of inserted genes and viral replication. FEBS Lett. 456:221-226. [DOI] [PubMed] [Google Scholar]
- 27.Sanderson, C. M., N. L. McQueen, and D. P. Nayak. 1993. Sendai virus assembly: M protein binds to viral glycoproteins in transit through the secretory pathway. J. Virol. 67:651-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanderson, C. M., H. H. Wu, and D. P. Nayak. 1994. Sendai virus M protein binds independently to either the F or the HN glycoprotein in vivo. J. Virol. 68:69-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shiotani, A., M. Fukumura, M. Maeda, X. Hou, M. Inoue, T. Kanamori, S. Komaba, K. Washizawa, S. Fujikawa, T. Yamamoto, C. Kadono, K. Watabe, H. Fukuda, K. Saito, Y. Sakai, Y. Nagai, J. Kanzaki, and M. Hasegawa. 2001. Skeletal muscle regeneration after insulin-like growth factor I gene transfer by recombinant Sendai virus vector. Gene Ther. 8:1043-1050. [DOI] [PubMed] [Google Scholar]
- 30.Stricker, R., and L. Roux. 1991. The major glycoprotein of Sendai virus is dispensable for efficient virus particle budding. J. Gen. Virol. 72:1703-1707. [DOI] [PubMed] [Google Scholar]
- 31.Takimoto, T., K. G. Murti, T. Bousse, R. A. Scroggs, and A. Portner. 2001. Role of matrix and fusion proteins in budding of Sendai virus. J. Virol. 75:11384-11391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tashiro, M., N. L. McQueen, J. T. Seto, H. D. Klenk, and R. Rott. 1996. Involvement of the mutated M protein in altered budding polarity of a pantropic mutant, F1-R, of Sendai virus. J. Virol. 70:5990-5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson, S. D., and A. Portner. 1987. Localization of functional sites on the hemagglutinin-neuraminidase glycoprotein of Sendai virus by sequence analysis of antigenic and temperature-sensitive mutants. Virology 160:1-8. [DOI] [PubMed] [Google Scholar]
- 34.Tokusumi, T., A. Iida, T. Hirata, A. Kato, Y. Nagai, and M. Hasegawa. 2002. Recombinant Sendai viruses expressing different levels of a foreign reporter gene. Virus Res. 86:33-38. [DOI] [PubMed] [Google Scholar]
- 35.Tsung, K., J. H. Yim, W. Marti, R. M. Buller, and J. A. Norton. 1996. Gene expression and cytopathic effect of vaccinia virus inactivated by psoralen and long-wave UV light. J. Virol. 70:165-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yonemitsu, Y., C. Kitson, S. Ferrari, R. Farley, U. Griesenbach, D. Judd, R. Steel, P. Scheid, J. Zhu, P. K. Jeffery, A. Kato, M. K. Hasan, Y. Nagai, I. Masaki, M. Fukumura, M. Hasegawa, D. M. Geddes, and E. W. Alton. 2000. Efficient gene transfer to airway epithelium using recombinant Sendai virus. Nat. Biotechnol. 18:970-973. [DOI] [PubMed] [Google Scholar]
- 37.Yoshida, T., S. Nagai, Y. Yoshii, K. Maeno, and T. Matsumoto. 1976. Membrane (M) protein of HVJ (Sendai virus): its role in virus assembly. Virology 71:143-161. [DOI] [PubMed] [Google Scholar]
- 38.Yu, D., T. Shioda, A. Kato, M. K. Hasan, Y. Sakai, and Y. Nagai. 1997. Sendai virus-based expression of HIV-1 gp120: reinforcement by the V(−) version. Genes Cells 2:457-466. [DOI] [PubMed] [Google Scholar]