Abstract
The function of the E5 protein of human papillomaviruses (HPV) is not well characterized, and controversies exist about its role in the viral life cycle. To determine the function of E5 within the life cycle of HPV type 31 (HPV31) we first constructed HPV31 mutant genomes that contained an altered AUG initiation codon or stop codons in E5. Cell lines were established which harbored transfected wild-type or E5 mutant HPV31 genomes. These cell lines all maintained episomal copies of HPV31 and revealed similar phenotypes with respect to growth rate, early gene expression, and viral copy number in undifferentiated monolayer cultures. Following epithelial differentiation, genome amplification and differentiation-dependent late gene expression were observed in mutant cell lines, but at a rate significantly reduced from that observed in cells containing the wild-type genomes. Organotypic raft cultures indicated that E5 does not effect the expression of differentiation markers but does reduce expression of late viral proteins. Western analysis and immunofluorescence staining for cyclins during epithelial differentiation revealed a decreased expression of cyclin A and B in E5 mutant cells compared to HPV wild-type cells. Using a replating assay, a significant reduction in colony-forming ability was detected in the absence of E5 expression when cells containing wild-type or E5 mutant HPV genomes were allowed to proliferate following 24 h in suspension-induced differentiation. This suggests that HPV E5 modifies the differentiation-induced cell cycle exit and supports the ability of HPV31-positive keratinocytes to retain proliferative competence. In these studies, E5 was found to have little effect on the levels of the epidermal growth factor receptor (EGFR) or on its phosphorylation status. This indicates that EGFR is not a target of E5 action. Our results propose a role for high risk HPV E5 in modulation of late viral functions through activation of proliferative capacity in differentiated cells. We suspect that the primary target of E5 is a membrane protein or receptor that then acts to alter the levels or activities of cell cycle regulators.
Human papillomaviruses (HPVs) are small DNA viruses that induce hyperproliferative lesions of cutaneous and mucosal epithelia (35). Half of the more than 100 identified types of HPVs specifically infect the genital epithelium (63). These genital papillomaviruses can be divided into low-risk types, such as HPV type 6 (HPV6) and HPV11, which induce only benign lesions, and high-risk types, such as HPV16, -18, and -31, which are associated with cervical carcinoma (32, 36, 63). The productive life cycle of human papillomaviruses is directly linked to epithelial cell differentiation (25). Following the infection of keratinocytes in the basal layer, HPV genomes are established as episomes at approximately 50 copies per cell and replicate in synchrony with cellular DNA replication (28, 33). The establishment and maintenance of HPV genomes is associated with expression of early HPV transcripts that encode the oncoproteins E6 and E7 as well as the replication proteins E1 and E2. Following cell division, infected daughter cells leave the basal layer, migrate towards the suprabasal regions and begin to differentiate. In contrast to uninfected keratinocytes, which exit the cell cycle as soon as they detach from the basement membrane, HPV-infected cells remain active in the cell cycle and enter into S-phase after reaching the suprabasal layer (12, 48). This entry into S-phase results in amplification of the viral genomes and expression of late transcripts from a differentiation-dependent promoter (11, 16, 28, 48).
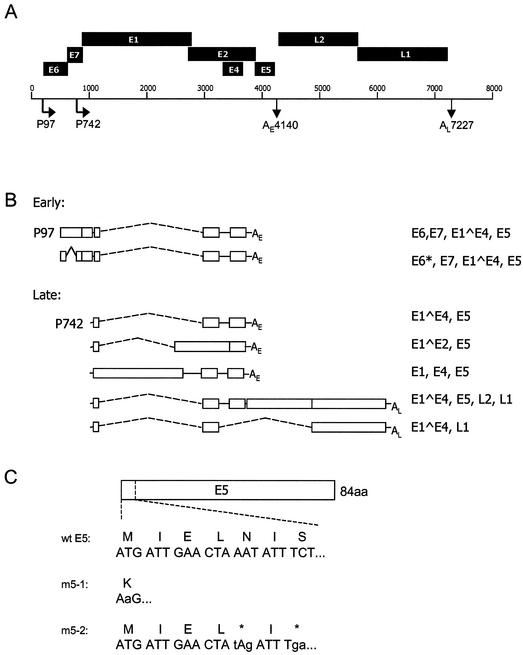
The viral E6 and E7 proteins act as the major oncogenic factors of high-risk HPVs, binding to cell proteins involved in cell cycle regulation. E6 binds the tumor suppressor p53 in a complex with the cellular ubiquitin ligase E6-AP, which leads to its degradation (27, 50, 51, 61). In addition, E6 induces telomerase activity through activation of expression of the catalytic subunit, hTert (18, 44, 60). E7 binds and inactivates the retinoblastoma protein (pRB) (5, 13, 37, 41). Another viral protein, E5, is weakly oncogenic in tissue culture assays and potentiates the transforming activity of E7 (3, 59). HPV E5 proteins are small, extremely hydrophobic, and located mainly at the endosomal membranes, Golgi apparatus, and, to a lesser extent, the plasma membranes (4, 6). In contrast to bovine papillomavirus type 1 E5, which has been shown to encode the primary transforming function, little is known about the biological activity of HPV E5. Abundant mRNA sequences containing the E5 open reading frame (ORF) have been identified in cervical intraepithelial neoplasial lesions and carcinomas (31, 55). In CIN612 cells, which contain episomal copies of HPV31, it has been shown that E5 is encoded in most early and late transcripts (28). However, E5 is transcribed as a part of a polycistronic RNA, and because it is usually the third or fourth ORF, it is not known how efficiently it is translated in undifferentiated cells. In contrast, upon differentiation E5 is the second ORF present on the majority of late transcripts (Fig. 1B). Direct evidence for the presence of E5 is difficult to obtain, as the protein cannot be detected in cells unless it is overexpressed from heterologous promoters (7, 29).
FIG. 1.
Schematic of a linearized HPV31 genome. (A) Diagram representing the HPV31 ORFs, the two major promoters that drive viral expression (P97 and P742), and the two polyadenylation sites (AE 4140 and AL 7227). (B) Diagram depicting the most-abundant spliced viral transcripts from the early and late promoter. (C) Diagram showing the HPV31 E5 mutants used in this study (wt, wild type).
In E5-transfected mouse fibroblasts, E5 has been shown to increase cellular proliferation in the presence of epidermal growth factor (EGF) (34, 57). The molecular basis for this effect is not clear, although it has been suggested that E5 associates with the EGF receptor (EGFR) (29), resulting in increased ligand-dependent activation of the EGFR (9, 57) and enhanced EGFR-mediated mitogen-activated protein (MAP) kinase activity (8, 20). Additionally, E5 binds the 16-kDa subunit of the vacuolar proton-ATPase (6) and inhibits the acidification of endosomes (56). This has been suggested to result in a delay in EGFR degradation and an increased recycling of EGFR to the cell surface (57). However, since all these studies have used heterologous expression systems in absence of other viral proteins, the role of the E5 protein during the productive life cycle of HPV remains unclear.
The study of the HPV life cycle in tissue culture has been facilitated by the development of methods for the genetic analysis of HPV functions in the context of its productive life cycle (15, 16). Differentiation of keratinocytes can be induced in organotypic raft culture (38) or suspension in methylcellulose (14, 19, 48). Upon differentiation these cells induce viral late functions, including activation of late gene expression and genome amplification (16, 48). Using these methods we have examined the effects of E5 in the context of the whole viral life cycle and identified a role in activation of late viral functions.
MATERIALS AND METHODS
Cell culture.
Human foreskin keratinocytes (HFKs) were derived from neonatal human foreskin epithelia as previously described (21) and were maintained in serum-free keratinocyte growth medium (Clonetics, San Diego, Calif.). HPV31 genome transfectants and control HFKs were grown in serum-containing medium (E medium) supplemented with mouse EGF (5 ng/ml; Collaborative Biomedical Products, Bedford, Mass.) in the presence of mitomycin C-treated J2 3T3 fibroblast feeders, kindly provided by the Howard Green laboratory. Prior to harvesting keratinocytes, total cellular DNA, total cellular RNA, or whole-cell lysates, the fibroblast feeders were first removed by EDTA (phosphate-buffered saline [PBS] [Gibco BRL, Grand Island, N.Y.] with 0.5 mM EDTA). To analyze EGFR activation, keratinocytes were cultivated in serum-free E medium for 20 h followed by a 3-min induction with 5 ng of EGF/ml prior to cell lysis.
Plasmids.
The generation of the E5 mutant genes was performed in a pBR322-derived plasmid, pBRmin. pBRmin was made by cutting out the 1,724-bp fragment between ClaI and Eco47III, filling in the 5′ overhang to form blunt ends, and religating. The plasmid pBRmin-HPV31 contains the HPV31 genome inserted into the HindIII site of the pBR322 min. The mutants m5-1 and m5-2 were constructed via site-directed mutagenesis by overlap extension using PCR. The mutant m5-1 contains a point mutation in the E5 ATG initiation codon, creating a lysine-coding triplet (AAG), and m5-2 possesses point mutations at codons 5 (AAT) and 7 (TCT) in the E5 ORF to generate stop codons (TAG and TGA, respectively; Fig. 1C). The mutated E5 sequences were subsequently exchanged with the wild-type sequence into pBRmin-HPV31 by using the single-cutting restriction endonucleases EcoRI (nucleotide 3361) and PpuMI (nucleotide 4464). pSV2neo carries the neomycin drug resistance gene.
Transfection of HFKs.
Ten micrograms of the pBRmin-HPV31 constructs was digested with HindIII to release the viral genome. The restriction enzyme was heat inactivated, and genomes were unimolecularly ligated in the same buffer with T4 DNA ligase (10 U/900 μl). The DNA was then precipitated with isopropyl alcohol and resuspended in 10 mM Tris-1 mM EDTA (pH 7.5). One microgram of the religated DNA was cotransfected with 1 μg of the selectable marker, pSV2neo, into HFKs with FuGene (Roche Diagnostics, Mannheim, Germany) as described by the manufacturer. At 1 day posttransfection, cells were plated onto mitomycin C-treated fibroblast feeders in E medium. Selection began day 2 posttransfection with G418 (Gibco BRL) as follows: G418 (200 mg/ml) was added every two days for a total of 4 days, and then G418 at a concentration of 100 mg/ml was added every two days for 4 more days. After selection, pooled populations were expanded for analyses.
Differentiation of keratinocytes in semisolid medium.
HFKs and HPV31 transfectants were suspended in 1.5% methylcellulose to induce differentiation. The methylcellulose solution was prepared by adding half of the final volume of E medium containing 5% fetal bovine serum to autoclaved dry methylcellulose (4,000 cps; Sigma, St. Louis, Mo.) and heating in a 60°C water bath for 20 min. The remaining E medium containing 10% fetal bovine serum was added, and the mixture was stirred at 4°C overnight. Approximately 1 × 106 to 2 × 106 keratinocytes were harvested by trypsinization, resuspended in 1 ml of E medium, and added dropwise to a 10-cm-diameter petri dish containing 25 ml of 1.5% methylcellulose. Cells were stirred with a pipette and incubated at 37°C in a CO2-humidified incubator at the indicated times. Cells in methylcellulose were harvested by scraping into four 50-ml conical tubes, washing with PBS (50 ml) three times, combining into a 15-ml conical tube for a final PBS wash, and pelleting by centrifugation. Samples were then subjected to Southern analyses to detect HPV31 genomic DNA and Northern analyses to examine transcripts and Western analysis.
Differentiation of keratinocytes in raft cultures.
HFKs as well as HPV31 wild-type and E5 mutant transfectants were differentiated in raft cultures as previously described (39). Briefly, cells were plated onto a solidified collagen matrix containing J2 3T3 fibroblasts, allowed to grow to confluency, and then transferred to a metal grid which provides an air-liquid interface for differentiation. Cultures were harvested at 14 days, fixed in 4% paraformaldehyde, paraffin embedded, sectioned, and stained with hematoxylin and eosin for visualization of differentiated raft tissue.
Immunohistochemistry.
The expression of E1∧E4 proteins was examined by immunofluorescence of cross sections of raft tissue. Thin sections of paraformaldehyde-fixed, paraffin-embedded tissue on silanized slides were incubated at 50°C for 30 min, and this was followed by three 5-min rinses in xylene to remove residual paraffin. The sections were rehydrated in absolute ethanol and then incubated in 10 mM citrate, pH 6.0, at 95°C for 20 min, followed by an additional 20-min incubation at room temperature. Incubation with the primary antibody rabbit anti-E1∧E4, which has been described previously (47), was performed overnight at 4°C. The sections were subsequently incubated with the secondary anti-rabbit antibody conjugated to fluorescein isothiocyanate (Amersham Pharmacia, Piscataway, N.J.) for 1 h at room temperature. Following 5 min of DNA-counterstaining with 4′,6′-diamidino-2-phenylindole-2HCl (DAPI) (1 μg/ml; Serva, Heidelberg, Germany) the sections were mounted and sealed.
Southern blot analyses.
Total genomic DNA from wild-type and mutant HPV31 transfectants was prepared by resuspension of the cell pellet in lysis buffer (400 mM NaCl, 10 mM Tris-HCl [pH 7.4], 10 mM EDTA); then, RNase A (50 μg/ml), proteinase K (50 μg/ml) and sodium dodecyl sulfate (SDS) (0.2%) were added, and this was followed by incubation at 37°C overnight. DNA was sheared by passing through an 18-gauge needle approximately 10 times, extracted with phenol-chloroform, and then precipitated with ethanol. Total genomic DNA (5 μg) was digested with DpnI to remove any residual input DNA, and another 5 μg of each sample was additionally linearized with XbaI to serve as a copy number control. Digested DNA was separated on a 0.8% agarose gel, treated, and alkaline transferred onto DuPont GeneScreen Plus nylon membrane (NEN Research Products, Boston, Mass.) as described by the manufacturer. The membrane was prehybridized in a solution containing 50% formamide, 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5× Denhardt's solution, 1% SDS, 10% dextran sulfate, and denatured salmon sperm DNA (0.1 mg/ml) for 1 h at 42°C. The HPV31 probe was prepared by gel purification of the entire HPV31 genome from pBRmin-HPV31 digested with HindIII and labeling with the Ready-To-Go DNA labeling kit (Amersham Pharmacia). Labeled probe was purified with ProbeQuant G-50 Micro columns (Amersham Pharmacia), denatured, and added to fresh hybridization solution, which was incubated with membrane at 42°C overnight. Membrane was washed twice with 2× SSC-0.1% SDS for 15 min at room temperature, twice with 0.5× SSC-0.1% SDS for 15 min at room temperature, twice with 0.1× SSC-0.1% SDS for 15 min at room temperature, and once with 0.1× SSC-1% SDS for 30 min at 50°C. Hybridizing species were visualized by autoradiography. Quantitative analysis was done with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
Analyses of HPV31 late transcripts.
Total RNA was isolated from methylcellulose-treated normal HFKs and HPV31 transfectants with TRIzol reagent (Gibco BRL) as described by the manufacturer and examined by Northern analyses as follows. Total RNA (10 μg) was separated on a 1.0% agarose-2.2 M formaldehyde gel in 1× MOPS buffer (10× MOPS buffer is 0.2 M MOPS, 50 mM Na acetate, 10 mM EDTA) and transferred onto a Zeta-Probe membrane (Bio-Rad, Hercules, Calif.). After cross-linking, the membrane was prehybridized in a solution containing 1 mM EDTA, 0.5 M Na2HPO4, and 7% SDS for 10 min at 65°C. The HPV31 probe was prepared by gel purification of the entire HPV31 genome from pBRmin-HPV31 digested with HindIII and labeling with the Ready-To-Go DNA labeling kit (Amersham Pharmacia). Labeled probe was purified with ProbeQuant G-50 Micro columns (Amersham Pharmacia), denatured, added to fresh hybridization solution, and incubated with membrane overnight at 65°C. The membrane was washed twice with 2× SSC-10% SDS for 5 min at room temperature and once with 0.2× SSC-1% SDS for 15 min at 55°C. As a loading control, the levels of 28S and 18S rRNA were examined in the gel after staining with ethidium bromide. Hybridizing species were visualized by autoradiography. Quantitative analysis was done with a PhosphorImager (Molecular Dynamics).
Western analysis.
Whole-cell extracts were prepared with NP-40 lysis buffer (150 mM NaCl, 50 mM Tris-HCl [pH 8], 5 mM EDTA [pH 8], 0.5 mM dithiothreitol, 100 mM sodium fluoride, 200 μM sodium-orthovanadate, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride) containing a cocktail of protease inhibitors (Complete, Mini; Roche Diagnostic) and quantitated with the Bradford assay (Bio-Rad). Equal amounts of protein were electrophoresed on a SDS-polyacrylamide gel and subsequently transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore, Bedford, Mass.). The membrane was blocked in wash solution (0.1% Tween 20 in PBS) containing 5% nonfat dry milk. The following antibodies were used: anti-EGFR (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.), anti-p-Tyr (-horseradish peroxidase (Santa Cruz), anti-pERK (Santa Cruz), anti-cyclin E (Santa Cruz), anti-cyclin B (Pharmingen, San Diego, Calif.), anti-p21 (Pharmingen), anti-p27 (Transduction Laboratories, Lexington, Ky.), anti-p57 (Oncogene Research, Uniondale, N.Y.), rabbit anti-cyclin A (a gift of R. Assoian). Western analysis employing anti-p-Tyr-horseradish peroxidase was performed by using block solution containing 0.25% gelatin instead of 5% nonfat dry milk. Proteins were visualized via enhanced chemiluminescence (Amersham Pharmacia).
Replating assay.
Normal HFKs as well as HPV31 wild-type and E5 mutant-positive keratinocytes grown in the presence of fibroblast feeders were harvested at 80% confluence and suspended in 1.5% methylcellulose. Following 24 h of differentiation the cells were collected, washed, and replated onto fibroblast feeders. After 5 days in culture colonies were visible and counted from at least five different randomly chosen fields.
RESULTS
In order to study the role of the E5 protein in the productive life cycle of HPVs, two mutant HPV31 genomes were constructed in the context of the plasmid pBRmin-HPV31 (Fig. 1A). One of the mutant genomes contains a mutated E5 ATG initiation codon (m5-1), which is the only ATG present in the E5 ORF, and the other one possesses translation termination codons inserted at amino acids 5 and 7 in the E5 ORF (m5-2). Both of these mutations inhibit E5 translation (Fig. 1C). Wild-type as well as E5 mutant HPV genomes were then used to transfect HFKs. After selection for neomycin resistance, pooled colonies were analyzed approximately 1 month after transfection.
E5 knockout HPV31 genomes are stably maintained in HFKs and replicate in a manner similar to that of HPV wild-type genomes.
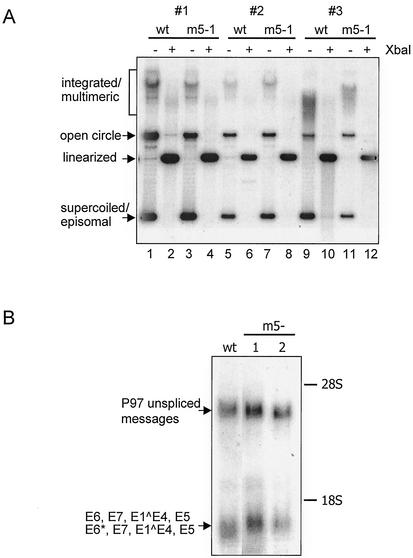
We first investigated whether E5 was necessary for stable maintenance and replication of HPV31 genomes. Southern blot analysis of total genomic DNA derived from three matched sets of independently transfected primary HFK isolates from different donors is shown in Fig. 2A. Harvested DNA from both wild-type and E5 mutant HPV31 cell lines were DpnI digested to remove any residual input DNA. An aliquot of each sample was additionally digested with XbaI, which cuts the HPV genome once to linearize the viral DNA and to facilitate copy number analysis (Fig. 2A, lanes 2, 4, 6, 8, 10, and 12). The cells transfected with wild-type HPV31 genomes were found to contain significant amounts of episomal copies of HPV31 DNA (Fig. 2A, lanes 1, 5, and 9). Similar amounts of episomal forms of viral DNA were detected in cells transfected with E5 mutant genomes (Fig. 2A, lanes 3, 7, and 11). The amount of linearized viral DNA copies was also comparable in matched wild-type and E5 mutant-containing cells (Fig. 2A, lanes 2 and 4, 6 and 8, and 10 and 12), although there were slight variations in viral copy numbers dependent on individual primary HFK isolates. No significant differences concerning episomal maintenance or viral copy number could be observed between cells transfected with one or the other E5 knockout mutant HPV genome. These experiments were repeated a total of six times using different primary HFK isolates with comparable results. Southern analysis of transfectants at later passages as well as cells that had been frozen and thawed showed no differences in the state of viral DNA.
FIG. 2.
Replication of HPV31 and E5 mutant HPV genomes in monolayer cultures. (A) Southern blot analysis of three independent transfections (#1, #2, and #3) of HFKs derived from different donors stably transfected with HPV31 (wt) and E5 mutant (m5-1) DNA. Total genomic DNA was harvested from monolayer cultures and digested with DpnI to remove residual input DNA. XbaI was additionally used to linearize the HPV31 genomes in lanes 2, 4, 6, 8, 10, and 12. The Southern blot was hybridized with a probe, which includes the complete HPV31 genome. (B) Northern blot analysis of mRNA from monolayer cultures of HPV31 and two E5 mutant-positive cell lines (m5-1 and m5-2). The Northern blot was hybridized with a probe, which includes the complete HPV31 genome. 26S and 18S markers correspond to molecular sizes of approximately 4.7 and 1.8 kb, respectively. Equal loading was monitored by comparing the levels of 28S and 18S rRNA in ethidium bromide-stained gels. The slight change in mobility of the smaller HPV messages observed in lane wt was not seen in repeated experiments.
Cell lines immortalized by E5 mutant or wild-type HPV genomes show similar expression patterns of early viral transcripts.
We next compared the amount of viral expression in wild-type and E5 mutant HPV-containing undifferentiated keratinocytes. The result of a representative Northern analysis is shown in Fig. 2B. Two major sets of transcripts of approximately 1.5 and 4.3 kb were observed in cells containing either wild-type or E5 mutant genomes. The larger transcript represents the unspliced mRNAs initiated at the early promoter P97, whereas the 1.5-kb messages probably encode two of the most abundant spliced mRNAs: E6, E7, E1∧E4, E5 and E6*, E7, E1∧E4, E5 (Fig. 1B). HPV wild-type as well as both of the E5 mutant cell lines revealed a similar pattern and level of early expression.
E5-deficient HPV containing keratinocytes did not reveal any phenotype relative to the wild-type cells in monolayer culture.
Since it was possible that E5 affected the growth properties of cells, we next compared the morphology of E5 knockout keratinocytes to wild-type HPV transfected keratinocytes as well as the relative growth rates and life spans of these cells. No differences concerning these criteria could be detected in undifferentiated monolayer cultures. In heterologous expression systems E5 has been shown to enhance mitogenic signaling through the EGFR in the presence of EGF (8, 34, 46, 57) by various mechanisms. Based on these observations we wanted to evaluate the influence of EGF on the growth rates of the transfectants. HPV31 wild-type and E5 mutant transfected cells were split in E medium in absence or presence of 5 ng of EGF per ml, and cell growth was monitored by counting cells. The results revealed that the growth rates of both HPV31 wild-type and E5 mutant-containing cells were reduced by approximately 50% when cultivated in EGF-free medium compared to medium supplemented with EGF (data not shown). Surprisingly, the growth rates of HPV31 wild-type and E5 mutant-transfected cells in the presence or absence of EGF were similar (data not shown), suggesting that E5 does not influence the growth of HPV-immortalized cells in monolayer culture.
The hyperphosphorylation of EGFR is not due to E5 in HPV31-containing cells.
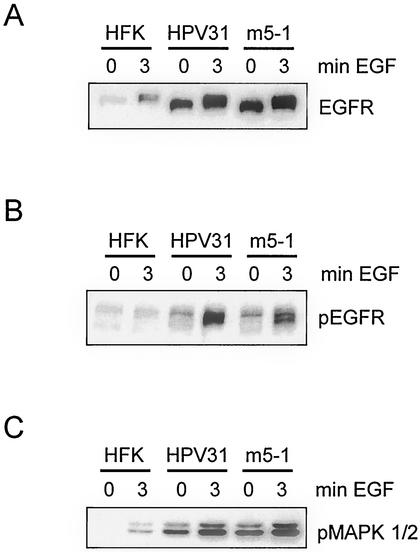
While we did not observe a synergistic effect of EGF on cell growth mediated by E5, it was still possible E5 had an influence on EGFR expression and activation. To investigate this possibility, untransfected HFKs as well as wild-type and E5 mutant HPV31-containing cells were starved in EGF- and serum-free medium for 20 h. Total cell lysates were then harvested following a 3-min induction with 5 ng of EGF per ml, and the levels of EGFR were examined by Western analysis. Keratinocytes stably transfected with HPV31 wild-type or E5 mutant genomes were found to contain levels of EGFR increased over those in HFKs (Fig. 3A). The levels of EGFR expression varied moderately between normal HFKs isolated from different donors (data not shown). However, the expression level of EGFR was found to be consistently higher in HPV-containing cells. No differences concerning EGFR levels or their degree of phosphorylation were detected between wild-type and E5 mutant-containing cells (Fig. 3B). The downstream targets of EGFR signaling, the MAP kinases 1 and 2, were also found phosphorylated to the same extent in both HPV wild-type and E5 mutant-containing cell lines (Fig. 3C). These results suggest that inhibition of HPV31 E5 in the context of all other viral proteins does not effect EGFR hyperphosphorylation in undifferentiated cells growing in monolayer culture.
FIG. 3.
Expression of EGFR in HFKs, HPV31, and E5 mutant-positive cells. Equivalent amounts of whole-cell extracts of HFK, HPV31, and E5 mutant-containing cells, not treated (lanes 0) or treated (lanes 3) with EGF for the indicated times were separated by SDS-polyacrylamide gel electrophoresis and examined by Western blot analysis with anti-EGFR (A), anti-pTyr (phosphorylated EGFR) (B), and anti-pERK (phosphorylated MAP kinases 1 and 2) (C) antibodies. Proteins were visualized by chemiluminescence.
Amplification of the E5 mutant HPV31 genomes is reduced upon epithelial differentiation.
Since we were not able to detect any phenotype due to the lack of E5 in monolayer culture, we next examined whether E5 plays a role in the later phases of the HPV life cycle. It is not possible to determine in which phase of the viral life cycle E5 is expressed by measuring protein levels, because efficient E5 antibodies are not available. While most HPV31 early mRNAs (Fig. 1B) contain the E5 ORF, it is most often the fourth ORF in these transcripts and unlikely to be efficiently translated. In contrast, E5 is the second ORF present in the major late E1∧E4, E5 transcripts, which may allow it to be more efficiently translated. Thus, it seemed plausible that E5 would exert some effects on late viral functions.
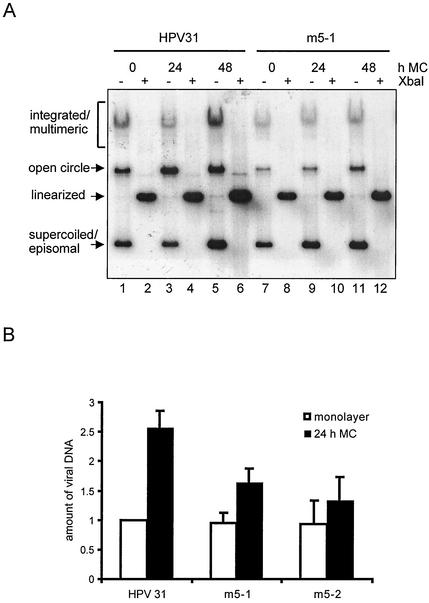
We first studied the ability of the E5 mutant HPV31 genomes to undergo productive replication upon epithelial differentiation following suspension in semisolid medium. It has been previously shown that growth in semisolid medium induces the differentiation of HPV-containing keratinocytes, resulting in activation of viral late functions (49). Total genomic DNA from wild-type and E5 mutant HPV31 transfected cells cultivated in monolayer culture or in methylcellulose were harvested and Southern analysis was performed. Following incubation in methylcellulose for a total of 48 h wild-type HPV31 DNA was found to increase to over 3-fold in contrast to 1.6-fold increase seen in mutant E5 HPV m5-1 (Fig. 4A). Figure 4B shows a graph of the average results of five independent Southern blot analyses. After 24 h of suspension in methylcellulose, the wild-type HPV31 DNA was amplified on average 2.6-fold in contrast to the two mutant E5 HPV DNAs m5-1 and m5-2, which were increased 1.6- and 1.3-fold, respectively. The reduced amplification of E5 mutant HPV upon differentiation compared to wild-type HPV suggests that at least some of the effects of E5 are directed against the late phases in the viral life cycle.
FIG. 4.
Differentiation-dependent amplification of HPV31 and E5 mutant HPV DNA following suspension in methylcellulose. (A) Autoradiogram of Southern analysis of HFKs stably transfected with HPV31 (wt) and E5 mutant (m5-1) DNA. Total genomic DNA was harvested from cells cultured in methylcellulose (MC) for the indicated times and prepared as described before. The Southern blot was hybridized with a probe corresponding to the complete HPV31 genome. (B) Southern blot analysis was performed on total genomic DNA of HPV31 and E5 mutant cells (m5-1 and m5-2), cultured in monolayer culture or in methylcellulose (MC) for 24 h. Equal amounts of total genomic DNA were digested with DpnI to remove residual input DNA and XbaI to linearize the HPV31 genomes. Differentiation-dependent viral amplification was quantified using a PhosphorImager. Results are the means + standard deviations (error bars) of data from five experiments.
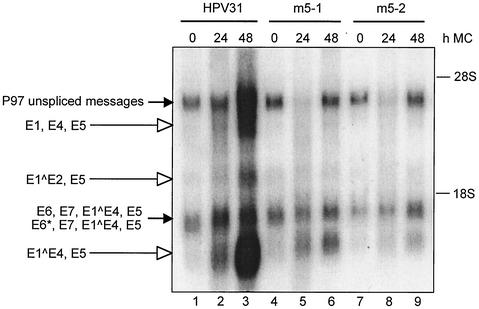
Late gene transcription is decreased upon differentiation in E5 mutant HPV transfected keratinocytes.
It was next important to analyze the patterns of viral expression upon differentiation in cells maintaining wild-type and E5 mutant genomes. RNA was harvested from cell lines cultured for 24 and 48 h in methylcellulose, and Northern analysis was performed using a probe encompassing the whole HPV genome. The probe allows for the identification of all early and late transcripts (Fig. 5). Following differentiation, three transcripts appeared in HPV31-positive cells which were not seen in undifferentiated cells (Fig. 5, lanes 1 to 3). Previous analyses indicate that these encode the late transcripts E1, E4, E5 (3.8 kb), E1∧E2, E5 (2.2 kb) and E1∧E4, E5 (1.3 kb) (Fig. 1B). While the expression of transcripts from the early promoter P97 in monolayer culture is similar in both E5 mutant and wild-type HPV-transfected cell lines (Fig. 5, dark arrows), expression of differentiation-dependent transcripts from the late promoter P742 is significantly impaired in E5 mutant cell lines (Fig. 5, open arrowheads). Unspliced P97 messages were also upregulated upon differentiation in wild-type HPV containing cells in contrast to E5 mutant-containing cell lines. Similar results were seen in three separate experiments. This result further supports the observation that keratinocytes that stably maintain HPV31 genomes lacking a functional E5 are significantly impaired in their ability to induce late viral functions.
FIG. 5.
Northern blot analysis of differentiation-dependent transcripts in HPV31 and E5 mutant positive cell lines following suspension in methylcellulose (MC). Total RNA was isolated at various times from HPV31 and two E5 mutant-positive cell lines (m5-1 and m5-2). Bands of early transcripts are labeled with a black arrow and bands of late transcripts are labeled with arrows with open heads. The Northern blot was hybridized with a probe corresponding to the complete HPV31 genome. 26S and 18S markers correspond to molecular sizes of approximately 4.7 and 1.8 kb, respectively. Uniform loading was monitored by comparing levels of 28S and 18S rRNA in ethidium bromide-stained gels. The reduced levels of P97 unspliced messages in lanes 5 and 8 were not seen in other experiments.
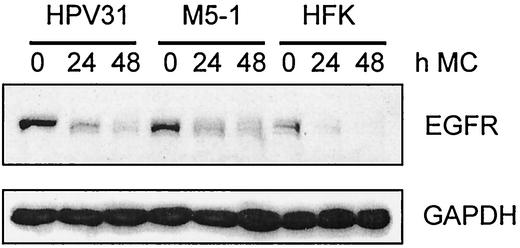
Low levels of EGFR expression are maintained upon differentiation in HPV immortalized cells, but not in HFKs.
Upon epithelial differentiation induced by suspension in methylcellulose, expression of EGFR decreased dramatically (Fig. 6). However, in HPV wild-type and E5 mutant-positive cells significant levels of EGFR were observed for periods up to 48 h. In contrast, no EGFR expression was detected in HFKs upon differentiation. This result indicates that the less pronounced decrease of EGFR expression upon differentiation in HPV-positive cells is not due to E5 but to the presence of other viral proteins.
FIG. 6.
Expression of EGFR in HFKs, HPV31, and E5 mutant-positive cells upon differentiation. Equivalent amounts of whole-cell extracts of HFKs, HPV31, and E5 mutant-positive cells cultured in methylcellulose (MC) for the indicated times were separated by SDS-polyacrylamide gel electrophoresis and examined by Western blot analysis with an anti-EGFR antibody. An anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody was used as a loading control. Proteins were visualized by chemiluminescence.
HPV31 and E5 mutant HPV transfected keratinocytes exhibit the same morphological differentiation pattern in organotypic raft cultures.
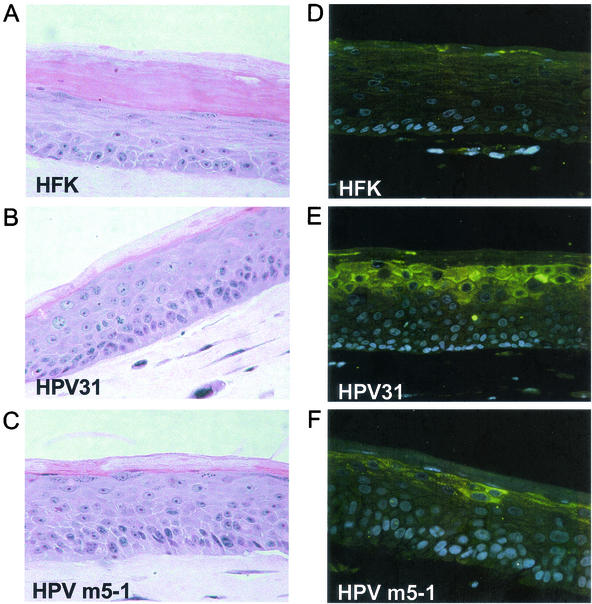
The next set of experiments examined whether the cells containing E5 mutant genomes induced any histological changes upon differentiation that could correlate with a reduced ability to induce late viral functions. Normal keratinocytes rapidly lose nuclei upon differentiation, while cells that express high-risk HPVs maintain nuclei throughout the suprabasal layers. To examine putative effects of E5 on epithelial morphology we performed organotypic raft culture analysis with HPV31 and HPV E5 mutant transfectants. As shown in Fig. 7A untransfected cells demonstrated a normal differentiation pattern in the raft cultures, with nuclear staining predominantly localized to cells in the basal layer. In contrast, HPV31-positive cells showed an altered differentiation pattern, with a thickening of the basal layer and nuclear staining throughout all layers (Fig. 7B). Organotypic raft cultures from HPV E5 mutant-containing cells revealed a morphology similar to that of HPV wild-type transfectants (Fig. 7C). We further examined if cells with HPV E5 mutant genes exhibited altered expression of differentiation markers such as keratin 10 (a differentiation-specific keratin), involucrin, and filaggrin, which are expressed beginning in the spinous layer (17). No significant difference in staining of any of these markers could be detected in HPV wild-type and E5 mutant transfectants in organotypic raft culture (data not shown).
FIG. 7.
Stained sections of organotypic raft cultures. Normal and transfected HFKs were induced to differentiate in raft cultures as described in Materials and Methods. Sections were stained with hematoxylin and eosin for visualization of differentiation (A to C) or stained with an antibody to HPV31 E1∧E4 protein and examined by immunofluorescence (D to F). As a secondary antibody a fluorescein-conjugated anti-rabbit antibody was used and the nuclei were counterstained with DAPI. (A and D) Normal HFKs; (B and E) HPV31-transfected HFKs; (C and D) E5 mutant-transfected HFKs.
E1∧E4 protein expression is remarkably reduced in E5 mutant HPV-transfected organotypic raft cultures compared to wild-type transfectants.
Since Northern blot analysis indicated that late viral gene expression is reduced in E5 mutant-containing cell lines (Fig. 5), we next wanted to analyze the distribution pattern of the most-abundant viral late E1∧E4 protein in organotypic raft cultures. Immunohistochemical analysis using antibodies against E1∧E4 protein showed it was expressed predominantly in the suprabasal cells of HPV31 containing raft cultures, confirming previous reports (Fig. 7E) (47). In contrast, rafts of cells containing HPV E5 mutants exhibited a consistent reduction in the number of E1∧E4 positive cells (Fig. 7F). This result was confirmed by immunofluorescence staining for E1∧E4 of cells suspended in semisolid medium for 24 h. On average 24% of HPV31 wild-type-containing cells were positive for E1∧E4 expression, in contrast to 14.5% of positive cells in E5 mutant cell lines (data not shown). It was not possible, however, to determine in this assay, if the level of staining per cell was also reduced.
Cyclin A and B expression is slightly reduced in HPV31 E5 mutant-containing cells compared to that in wild type.
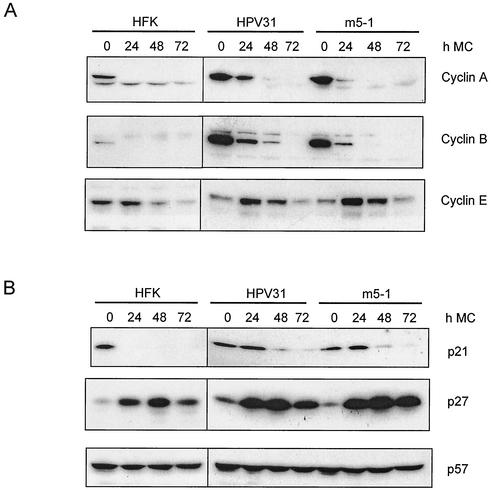
Next we wanted to investigate the mechanism by which the E5 protein stimulates viral amplification and late viral gene expression. One possibility is that E5 assists E6 and E7 in progression into S-phase to enhance viral late functions. Previous studies indicated that activation of late gene expression as well as amplification occurs in S-phase (49). To address the E5 effects on cell cycle progression we first compared the expression of cyclins in untransfected HFKs, HPV31 wild type, and E5 mutant cell lines upon differentiation. Cells were harvested as a function of time following suspension in methylcellulose, and Western blot analyses for the cyclins A, B, and E were performed. The HPV31 wild-type and E5 mutant-containing cell lines both expressed levels of cyclin A and B about two to three times higher than HFKs when growing as monolayer culture (Fig. 8A). Following suspension in methylcellulose for 24 h, cyclin A levels were dramatically reduced in HFKs, whereas HPV31-positive cells retained cyclin A expression. E5 mutant cells also retained cyclin A expression, but to a lesser extent than HPV31 wild-type cells. The pattern of cyclin B expression during suspension culture was found to be similar to that of cyclin A. HPV31 containing cells retained significant levels of cyclin B at 48 h, whereas in HFKs cyclin B is undetectable after 24 h. E5 mutant cells exhibited an intermediate phenotype. These effects were consistently seen in four independent experiments. When cyclin E expression during differentiation was examined, it was found to increase in the first 24 h and then rapidly decrease in all HPV cells tested, but at rates less rapid than for cyclin A and B. For cyclin E no differences between wild-type and E5 mutant cell lines were observed. From these studies, we conclude that E5 moderately influences the levels of cyclins A and B but not E in differentiated HPV positive cells.
FIG. 8.
Expression of cell cycle proteins in HFK, HPV31, and E5 mutant cells. Western blot analyses were performed on whole-cell extracts of HFK, HPV 31, and E5 mutant cells cultured in methylcellulose (MC) for the indicated times. Equivalent amounts of whole-cell extracts were separated by SDS-polyacrylamide gel electrophoresis and examined by Western blot analysis with anti-cyclin A, B, and E antibodies (A) or anti-CKI p21, p27, and p57 antibodies (B) as indicated. Proteins were visualized by chemiluminescence.
We next examined the expression of the cyclin-cyclin-dependent kinase complex inhibitors (CKI) p21, p27, and p57 in these lines after suspension in methylcellulose. CKI have been shown to be upregulated during the differentiation of keratinocytes (40, 43). By Western blot analysis we found that p27 was dramatically induced in methylcellulose in both HFKs and HPV-positive cells. (Fig. 8B). In contrast, the p53-regulated CKI p21 was not induced upon differentiation of HFKs but was retained for up to 24 h in HPV-positive cells. There was no change in the level of p57 expression following suspension culture in either HFKs or HPV-containing cells. These experiments confirm the induction of CKI during suspension-induced cell cycle exit in keratinocytes (2, 24, 42, 48) but demonstrate that E5 does not influence this process. These experiments were repeated five times with similar results. In these studies we observed a modest but consistent reduction of cyclin A and B expression in E5 mutant-containing keratinocytes (Fig. 8A) compared to HPV31 wild-type cells which is not due to increased CKI expression.
The HPV E5 protein supports ongoing mitosis and cell division following differentiation-induced cell cycle arrest.
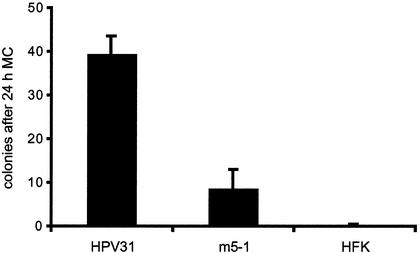
From the above studies, E5 was suggested to have a modulatory effect on cell cycle regulators, and we wanted to test if E5 had functional consequences in cell cycle progression in differentiating cells. For these studies we performed a replating assay that measured the ability of cells to remain replication competent following differentiation. In this assay HPV-positive cell lines were first induced to undergo epithelial differentiation by suspension culture in methylcellulose. After 24 h the cells were washed and an aliquot of cells were replated into tissue culture dishes containing fibroblast feeders. After 5 days colonies appeared and random fields of cells were counted (Fig. 9). Cells containing HPV31 wild-type genomes were able to form colonies in an efficient manner, with about 40 (mean ± standard deviation, 39.25 ± 4.15) colonies visible per field at day 5. After an additional week in culture, the plates became confluent. In contrast, cells containing the E5 mutant genomes formed significantly reduced numbers of colonies, with about 8 (mean ± standard deviation, 8.55 ± 4.48) colonies detectable after 5 days. These cells grew poorly and did not become confluent after 2 weeks. HFKs did not form colonies after methylcellulose-induced differentiation (mean ± standard deviation, 0.2 ± 0.42). This suggests that HPV E5 modulates the differentiation-induced cell cycle exit and supports the ability of HPV31-positive keratinocytes to remain active in the cell cycle.
FIG. 9.
Replating of HPV31- and E5 mutant HPV-positive cells following suspension in methylcellulose (MC). Monolayer keratinocytes were induced to undergo epithelial differentiation by suspension culture in methylcellulose. After 24 h the cells were harvested and replated onto a feeder layer of J2s. Colonies were visible after a few days and were counted on day 5 from five random fields. Results are the means + standard deviations (error bars) of data from four experiments. Approximately 10% of the replated wild-type HPV31 cells grew into colonies in these assays.
DISCUSSION
Our studies have examined the function of the E5 protein of the high-risk HPV types during the productive viral life cycle. In bovine papillomavirus type 1, E5 encodes a primary transforming activity, but in high risk types the E6 and E7 proteins provide this function (10, 52). In heterologous expression systems E5 can act as a weak-transforming protein, and it has been suggested that E5 acts to augment E6/E7 action (3). However, it is not clear whether this is a physiologically relevant activity during viral pathogenesis. In our studies we analyzed the role of E5 in the productive life cycle by monitoring the effects of loss of E5 function in the context of the complete viral genome. We examined two different mutations in E5 and observed similar effects. The lack of E5 expression was found to have little effect on the growth properties, the ability to maintain episomes or the expression of viral genes in HFKs transfected with HPV 31 genomes in undifferentiated cells. Wild-type or E5 mutant genomes were equally effective at immortalization, and transfected cells grew at comparable rates either in the absence or presence of added EGF. This was consistent with Western blot analysis that showed that the loss of E5 had no effect on the number or phosphorylation status of the EGFRs. Our observations were surprising and are in contrast to previous reports that expression of E5 from heterologous promoters can lead to increased levels of EGFR (57). In our studies, increased levels of EGFR were detected in cells that maintain complete and E5 mutant viral genomes, which we believe is due to the action of E6 and E7 rather than E5. These observations are consistent with previous reports on the effects of E6 and E7 on EGFR levels (1, 26, 30, 53). We conclude that E5 in undifferentiated cells has no effect on any of the aspects of viral pathogenesis assays we tested. However, it is possible E5 has effects in other aspects of the life cycle in undifferentiated cells that we have not examined. Previous studies have analyzed the effects of HPV16 E5 in heterologous overexpression systems, whereas our studies have focused on presumably natural expression levels of the HPV31 protein. While these proteins are 77% homologous, it remains possible that the differential effects on EGFR levels observed with HPV16 E5 could be due to either expression levels or different functional properties attributed to this sequence variation.
In contrast to the lack of effects due to E5 observed in undifferentiated monolayer cultures, we detected a significant increase in differentiation-dependent viral amplification and late gene transcription when E5 was present. The decrease of late viral transcripts observed in the absence of E5 was not due to reduced stability of the late RNAs resulting from the mutations in the E5 ORF since we observed no difference in the levels of early transcripts which also contain the E5 ORF. Viral amplification and late gene transcription are strictly dependent on two apparently opposing processes: epithelial differentiation and DNA synthesis in suprabasal cells. First, it is possible that E5 has an effect on epithelial differentiation, a process that is necessary to induce efficient transcription from the differentiation-dependent late viral promoter and to activate viral DNA amplification. Alternatively, E5 could act to augment the activity of E6 and E7 in modulating progression through cell cycle in differentiated cells. Similar analyses have been performed using HPV16 genomes containing mutant E5 genes (17a) and yielded a quantitative reduction in ability of the E5 mutant HPV16 genome to reprogram differentiated cells to support DNA synthesis. In our study, we observed effects of E5 on late viral functions that were more severe than those seen in the study on HPV16. This difference may be due to the different HPV types examined or to the assays used.
We examined whether E5 has any effect on differentiation of cells containing HPV31 wild-type and HPV E5 mutant genomes in organotypic raft cultures. No differences in morphology of cells or expression of differentiation markers such as keratin 10, filaggrin, and involucrin were observed. However, we did observe a reduction in E1∧E4 expression in keratinocytes that contain E5 mutant genomes, consistent with our findings from Northern blot analysis. Our studies support the idea that E5 does not act to modulate late functions through alteration of epithelial differentiation.
Differentiation of HPV-positive cells results in retention of high levels of S-phase cyclins, such as cyclins A, E, and B. The levels of these cyclins decrease with differentiation, but the presence of HPV gene products reduces the rate of loss. Analysis of HPV-positive cells lacking E5 consistently revealed a moderately increased rate of loss of cyclin A and B levels compared to that observed in HPV31 wild-type cells. While we do not believe E5 directly targets cyclin A and B expression, it is likely that E5's interaction with a membrane-bound cellular receptor indirectly leads to altered levels of the cell cycle regulators. The reduced levels of cyclins A and B may be sufficient to contribute to the reduced capability of HPV E5 mutant transfectants to amplify viral genomes and induce late gene expression. We examined the number of cyclin A-positive cells by immunohistochemistry following suspension on methylcellulose and found minimal differences between keratinocytes containing wild-type and E5 mutant genomes (F. Fehrmann and L. A. Laimins, unpublished data). Since HPV late functions are activated in S-phase, our observations suggest that E5 does not significantly alter the number of cells activating late functions but increases the levels of activation in each cell. Other potential targets of E5 action include the CKI, which have been shown to be upregulated during differentiation in vivo (22, 23, 40, 45, 54, 62). In our study, we observed no difference in the expression pattern of p21, p27, and p57 between cells transfected with HPV31 wild-type or HPV E5 mutant genomes following differentiation. It was previously reported using an inducible heterologous system that HPV16 and -11 E5 suppressed p21 expression in mouse fibroblasts and immortalized human keratinocytes (58). Our work indicates that when E5 is expressed from its natural promoter in the context of the complete viral genome, it does not significantly repress p21 transcription. We conclude that effects of E5 on activation of late viral functions are most likely not mediated through altered expression of CKI.
More direct support for the hypothesis that E5 has a role in the modulation of cell cycle progression during differentiation of HPV positive cells comes from a replating assay. This assay measures the ability of cells to remain competent for proliferation after being induced to differentiate. We observed that cells that expressed the complete viral genome were able to reinitiate proliferation at four times the rate of cells that lack E5. This further suggests that E5 is involved in overcoming the differentiation-induced cell cycle arrest and on maintaining proliferative potential. It is possible that the moderate reduction in cyclin A and B expression in the absence of E5 can explain this effect or alternatively other cell cycle regulators maybe more significantly impaired. Furthermore, the moderate reduction in cyclin A and B levels may be a consequence of targeting another regulator of the cell cycle. The search for such a regulatory target for E5 is ongoing.
The E5 protein has been shown to be a membrane protein that is localized either to the Golgi, endoplasmic reticulum, or cytoplasmic membranes (4, 6). The question arises as to how a membrane protein can alter cell cycle activities that are primarily localized to the nucleus. We suspect that the primary target of E5 is a membrane protein or receptor that then acts to alter the levels or activities of cell cycle regulators. Our studies suggest that this target is not the EGFR; however, it is possible that another member of the EGFR family or some other growth factor receptor is the primary target. Elucidation of the target of E5 action will require screening for altered levels or activities of membrane associated proteins in differentiated cells.
We conclude from our studies that the E5 protein from high risk HPVs acts in the late phases of the viral life cycle to modulate differentiation-induced functions like viral amplification and late gene expression. Additionally, E5 may play a major role in retaining proliferative activity following differentiation, a process that is essential for high levels of viral production.
Acknowledgments
We thank Kathy Rundell, Richard Longnecker, and Lawrence Banks for critical reviews of the manuscript. We gratefully acknowledge Walter Hubert and the members of the Laimins laboratory for helpful discussions.
This work was supported by a grant from the Penny Severns Breast and Cervical Cancer Research Fund from the Illinois Department of Public Health and a Gramm Fellowship Award from Northwestern University to F.F. as well as by a grant from the National Cancer Institute (CA74202) to L.A.L.
REFERENCES
- 1.Akerman, G. S., W. H. Tolleson, K. L. Brown, L. L. Zyzak, E. Mourateva, T. S. Engin, A. Basaraba, A. L. Coker, K. E. Creek, and L. Pirisi. 2001. Human papillomavirus type 16 E6 and E7 cooperate to increase epidermal growth factor receptor (EGFR) mRNA levels, overcoming mechanisms by which excessive EGFR signaling shortens the life span of normal human keratinocytes. Cancer Res. 61:3837-3843. [PubMed] [Google Scholar]
- 2.Alani, R. M., J. Hasskarl, and K. Munger. 1998. Alterations in cyclin-dependent kinase 2 function during differentiation of primary human keratinocytes. Mol. Carcinog. 23:226-233. [DOI] [PubMed] [Google Scholar]
- 3.Bouvard, V., G. Matlashewski, Z. M. Gu, A. Storey, and L. Banks. 1994. The human papillomavirus type 16 E5 gene cooperates with the E7 gene to stimulate proliferation of primary cells and increases viral gene expression. Virology 203:73-80. [DOI] [PubMed] [Google Scholar]
- 4.Burkhardt, A., M. Willingham, C. Gay, K. T. Jeang, and R. Schlegel. 1989. The E5 oncoprotein of bovine papillomavirus is oriented asymmetrically in Golgi and plasma membranes. Virology 170:334-339. [DOI] [PubMed] [Google Scholar]
- 5.Cheng, S., D. C. Schmidt-Grimminger, T. Murant, T. R. Broker, and L. T. Chow. 1995. Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes Dev. 9:2335-2349. [DOI] [PubMed] [Google Scholar]
- 6.Conrad, M., V. J. Bubb, and R. Schlegel. 1993. The human papillomavirus type 6 and 16 E5 proteins are membrane-associated proteins which associate with the 16-kilodalton pore-forming protein. J. Virol. 67:6170-6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conrad, M., D. Goldstein, T. Andresson, and R. Schlegel. 1994. The E5 protein of HPV-6, but not HPV-16, associates efficiently with cellular growth factor receptors. Virology 200:796-800. [DOI] [PubMed] [Google Scholar]
- 8.Crusius, K., E. Auvinen, and A. Alonso. 1997. Enhancement of EGF- and PMA-mediated MAP kinase activation in cells expressing the human papillomavirus type 16 E5 protein. Oncogene 15:1437-1444. [DOI] [PubMed] [Google Scholar]
- 9.Crusius, K., E. Auvinen, B. Steuer, H. Gaissert, and A. Alonso. 1998. The human papillomavirus type 16 E5-protein modulates ligand-dependent activation of the EGF receptor family in the human epithelial cell line HaCaT. Exp. Cell Res. 241:76-83. [DOI] [PubMed] [Google Scholar]
- 10.DiMaio, D., D. Guralski, and J. T. Schiller. 1986. Translation of open reading frame E5 of bovine papillomavirus is required for its transforming activity. Proc. Natl. Acad. Sci. USA 83:1797-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dollard, S. C., J. L. Wilson, L. M. Demeter, W. Bonnez, R. C. Reichman, T. R. Broker, and L. T. Chow. 1992. Production of human papillomavirus and modulation of the infectious program in epithelial raft cultures. OFF. Genes Dev 6:1131-1142. [DOI] [PubMed] [Google Scholar]
- 12.Doorbar, J., C. Foo, N. Coleman, L. Medcalf, O. Hartley, T. Prospero, S. Napthine, J. Sterling, G. Winter, and H. Griffin. 1997. Characterization of events during the late stages of HPV16 infection in vivo using high-affinity synthetic Fabs to E4. Virology 238:40-52. [DOI] [PubMed] [Google Scholar]
- 13.Dyson, N., P. M. Howley, K. Munger, and E. Harlow. 1989. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243:934-937. [DOI] [PubMed] [Google Scholar]
- 14.Flores, E. R., and P. F. Lambert. 1997. Evidence for a switch in the mode of human papillomavirus type 16 DNA replication during the viral life cycle. J. Virol. 71:7167-7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frattini, M. G., H. B. Lim, J. Doorbar, and L. A. Laimins. 1997. Induction of human papillomavirus type 18 late gene expression and genomic amplification in organotypic cultures from transfected DNA templates. J. Virol. 71:7068-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frattini, M. G., H. B. Lim, and L. A. Laimins. 1996. In vitro synthesis of oncogenic human papillomaviruses requires episomal genomes for differentiation-dependent late expression. Proc. Natl. Acad. Sci. USA 93:3062-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuchs, E. 1995. Keratins and the skin. Annu. Rev. Cell Dev. Biol. 11:123-153. [DOI] [PubMed] [Google Scholar]
- 17a.Genther, S. M., S. Sterling, S. Duensing, K. Münger, C. Sattler, and P. F. Lambert. 2003. Quantitative role of the human papillomavirus type 16 E5 gene during the productive stage of the viral life cycle. J. Virol. 77:2832-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gewin, L., and D. A. Galloway. 2001. E box-dependent activation of telomerase by human papillomavirus type 16 E6 does not require induction of c-myc. J. Virol. 75:7198-7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green, H. 1977. Terminal differentiation of cultured human epidermal cells. Cell 11:405-416. [DOI] [PubMed] [Google Scholar]
- 20.Gu, Z., and G. Matlashewski. 1995. Effect of human papillomavirus type 16 oncogenes on MAP kinase activity. J. Virol. 69:8051-8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halbert, C. L., G. W. Demers, and D. A. Galloway. 1992. The E6 and E7 genes of human papillomavirus type 6 have weak immortalizing activity in human epithelial cells. J. Virol. 66:2125-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halevy, O., B. G. Novitch, D. B. Spicer, S. X. Skapek, J. Rhee, G. J. Hannon, D. Beach, and A. B. Lassar. 1995. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science 267:1018-1021. [DOI] [PubMed] [Google Scholar]
- 23.Hauser, P. J., D. Agrawal, M. Flanagan, and W. J. Pledger. 1997. The role of p27kip1 in the in vitro differentiation of murine keratinocytes. Cell Growth Differ. 8:203-211. [PubMed] [Google Scholar]
- 24.Hauser, P. J., D. Agrawal, and W. J. Pledger. 1998. Primary keratinocytes have an adhesion dependent S phase checkpoint that is absent in immortalized cell lines. Oncogene 17:3083-3092. [DOI] [PubMed] [Google Scholar]
- 25.Howley, P. M., and D. R. Lowy. 2001. Papillomaviruses and their replication, p. 2197-2229. In P. M. Howley (ed.), Virology, vol. 2. Lippincott/The Williams & Wilkins Co, Philadelphia, Pa.
- 26.Hu, G., W. Liu, J. Mendelsohn, L. M. Ellis, R. Radinsky, M. Andreeff, and A. B. Deisseroth. 1997. Expression of epidermal growth factor receptor and human papillomavirus E6/E7 proteins in cervical carcinoma cells. J. Natl. Cancer Inst. 89:1271-1276. [DOI] [PubMed] [Google Scholar]
- 27.Huibregtse, J. M., M. Scheffner, and P. M. Howley. 1991. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 10:4129-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hummel, M., J. B. Hudson, and L. A. Laimins. 1992. Differentiation-induced and constitutive transcription of human papillomavirus type 31b in cell lines containing viral episomes. J. Virol. 66:6070-6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang, E. S., T. Nottoli, and D. Dimaio. 1995. The HPV16 E5 protein: expression, detection, and stable complex formation with transmembrane proteins in COS cells. Virology 211:227-233. [DOI] [PubMed] [Google Scholar]
- 30.Johnston, D., H. Hall, T. P. DiLorenzo, and B. M. Steinberg. 1999. Elevation of the epidermal growth factor receptor and dependent signaling in human papillomavirus-infected laryngeal papillomas. Cancer Res. 59:968-974. [PubMed] [Google Scholar]
- 31.Kell, B., R. J. Jewers, J. Cason, F. Pakarian, J. N. Kaye, and J. M. Best. 1994. Detection of E5 oncoprotein in human papillomavirus type 16-positive cervical scrapes using antibodies raised to synthetic peptides. J. Gen. Virol. 75:2451-2456. [DOI] [PubMed] [Google Scholar]
- 32.Laimins, L. A. 1993. The biology of human papillomaviruses: from warts to cancer. Infect. Agents Dis. 2:74-86. [PubMed] [Google Scholar]
- 33.Lambert, P. F. 1991. Papillomavirus DNA replication. J. Virol. 65:3417-3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leechanachai, P., L. Banks, F. Moreau, and G. Matlashewski. 1992. The E5 gene from human papillomavirus type 16 is an oncogene which enhances growth factor-mediated signal transduction to the nucleus. Oncogene 7:19-25. [PubMed] [Google Scholar]
- 35.Lowy, D. R., and P. M. Howley. 2001. Papillomaviruses, p. 2231-2264. In P. M. Howley (ed.), Virology, vol. 2. Lippincott/The Williams & Wilkins Co, Philadelphia, Pa.
- 36.Lowy, D. R., R. Kirnbauer, and J. T. Schiller. 1994. Genital human papillomavirus infection. Proc. Natl. Acad. Sci. USA 91:2436-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin, L. G., G. W. Demers, and D. A. Galloway. 1998. Disruption of the G1/S transition in human papillomavirus type 16 E7-expressing human cells is associated with altered regulation of cyclin E. J. Virol. 72:975-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCance, D. J., R. Kopan, E. Fuchs, and L. A. Laimins. 1988. Human papillomavirus type 16 alters human epithelial cell differentiation in vitro. Proc. Natl. Acad. Sci. USA 85:7169-7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meyers, C., and L. A. Laimins. 1994. In vitro systems for the study and propagation of human papillomaviruses. Curr. Top. Microbiol. Immunol. 186:199-215. [DOI] [PubMed] [Google Scholar]
- 40.Missero, C., E. Calautti, R. Eckner, J. Chin, L. H. Tsai, D. M. Livingston, and G. P. Dotto. 1995. Involvement of the cell-cycle inhibitor Cip1/WAF1 and the E1A-associated p300 protein in terminal differentiation. Proc. Natl. Acad. Sci. USA 92:5451-5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munger, K., B. A. Werness, N. Dyson, W. C. Phelps, E. Harlow, and P. M. Howley. 1989. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 8:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noya, F., W. M. Chien, T. R. Broker, and L. T. Chow. 2001. p21cip1 Degradation in differentiated keratinocytes is abrogated by costabilization with cyclin E induced by human papillomavirus E7. J. Virol. 75:6121-6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oh, H., C. Mammucari, A. Nenci, S. Cabodi, S. N. Cohen, and G. P. Dotto. 2002. Negative regulation of cell growth and differentiation by TSG101 through association with p21(Cip1/WAF1). Proc. Natl. Acad. Sci. USA 99:5430-5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oh, S. T., S. Kyo, and L. A. Laimins. 2001. Telomerase activation by human papillomavirus type 16 E6 protein: induction of human telomerase reverse transcriptase expression through Myc and GC-rich Sp1 binding sites. J. Virol. 75:5559-5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parker, S. B., G. Eichele, P. Zhang, A. Rawls, A. T. Sands, A. Bradley, E. N. Olson, J. W. Harper, and S. J. Elledge. 1995. p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells. Science 267:1024-1027. [DOI] [PubMed] [Google Scholar]
- 46.Pim, D., M. Collins, and L. Banks. 1992. Human papillomavirus type 16 E5 gene stimulates the transforming activity of the epidermal growth factor receptor. Oncogene 7:27-32. [PubMed] [Google Scholar]
- 47.Pray, T. R., and L. A. Laimins. 1995. Differentiation-dependent expression of E1-E4 proteins in cell lines maintaining episomes of human papillomavirus type 31b. Virology 206:679-685. [DOI] [PubMed] [Google Scholar]
- 48.Ruesch, M. N., and L. A. Laimins. 1998. Human papillomavirus oncoproteins alter differentiation-dependent cell cycle exit on suspension in semisolid medium. Virology 250:19-29. [DOI] [PubMed] [Google Scholar]
- 49.Ruesch, M. N., F. Stubenrauch, and L. A. Laimins. 1998. Activation of papillomavirus late gene transcription and genome amplification upon differentiation in semisolid medium is coincident with expression of involucrin and transglutaminase but not keratin-10. J. Virol. 72:5016-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75:495-505. [DOI] [PubMed] [Google Scholar]
- 51.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129-1136. [DOI] [PubMed] [Google Scholar]
- 52.Schiller, J. T., W. C. Vass, K. H. Vousden, and D. R. Lowy. 1986. E5 open reading frame of bovine papillomavirus type 1 encodes a transforming gene. J Virol. 57:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sizemore, N., C. K. Choo, R. L. Eckert, and E. A. Rorke. 1998. Transcriptional regulation of the EGF receptor promoter by HPV16 and retinoic acid in human ectocervical epithelial cells. Exp. Cell Res. 244:349-356. [DOI] [PubMed] [Google Scholar]
- 54.Steinman, R. A., B. Hoffman, A. Iro, C. Guillouf, D. A. Liebermann, and M. E. el-Houseini. 1994. Induction of p21 (WAF-1/CIP1) during differentiation. Oncogene 9:3389-3396. [PubMed] [Google Scholar]
- 55.Stoler, M. H., C. R. Rhodes, A. Whitbeck, S. M. Wolinsky, L. T. Chow, and T. R. Broker. 1992. Human papillomavirus type 16 and 18 gene expression in cervical neoplasias. Hum. Pathol. 23:117-128. [DOI] [PubMed] [Google Scholar]
- 56.Straight, S. W., B. Herman, and D. J. McCance. 1995. The E5 oncoprotein of human papillomavirus type 16 inhibits the acidification of endosomes in human keratinocytes. J. Virol. 69:3185-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Straight, S. W., P. M. Hinkle, R. J. Jewers, and D. J. McCance. 1993. The E5 oncoprotein of human papillomavirus type 16 transforms fibroblasts and effects the downregulation of the epidermal growth factor receptor in keratinocytes. J. Virol. 67:4521-4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsao, Y. P., L. Y. Li, T. C. Tsai, and S. L. Chen. 1996. Human papillomavirus type 11 and 16 E5 represses p21(WafI/SdiI/CipI) gene expression in fibroblasts and keratinocytes. J. Virol. 70:7535-7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Valle, G. F., and L. Banks. 1995. The human papillomavirus (HPV)-6 and HPV-16 E5 proteins co-operate with HPV-16 E7 in the transformation of primary rodent cells. J. Gen. Virol. 76:1239-1245. [DOI] [PubMed] [Google Scholar]
- 60.Veldman, T., I. Horikawa, J. C. Barrett, and R. Schlegel. 2001. Transcriptional activation of the telomerase hTERT gene by human papillomavirus type 16 E6 oncoprotein. J. Virol. 75:4467-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Werness, B. A., A. J. Levine, and P. M. Howley. 1990. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science 248:76-79. [DOI] [PubMed] [Google Scholar]
- 62.Yan, Y., J. Frisen, M. H. Lee, J. Massague, and M. Barbacid. 1997. Ablation of the CDK inhibitor p57Kip2 results in increased apoptosis and delayed differentiation during mouse development. Genes Dev. 11:973-983. [DOI] [PubMed] [Google Scholar]
- 63.zur Hausen, H., and E. M. de Villiers. 1994. Human papillomaviruses. Annu. Rev. Microbiol. 48:427-447. [DOI] [PubMed] [Google Scholar]