Abstract
Retroviruses use unusual recoding strategies to synthesize the Gag-Pol polyprotein precursor of viral enzymes. In human immunodeficiency virus, ribosomes translating full-length viral RNA can shift back by 1 nucleotide at a specific site defined by the presence of both a slippery sequence and a downstream stimulatory element made of an extensive secondary structure. This so-called frameshift mechanism could become a target for the development of novel antiviral strategies. A different recoding strategy is used by other retroviruses, such as murine leukemia viruses, to synthesize the Gag-Pol precursor; in this case, a stop codon is suppressed in a readthrough process, again due to the presence of a specific structure adopted by the mRNA. Development of antiframeshift agents will greatly benefit from the availability of a simple animal and virus model. For this purpose, the murine leukemia virus readthrough region was rendered inactive by mutagenesis and the frameshift region of human immunodeficiency virus was inserted to generate a chimeric provirus. This substitution of readthrough by frameshift allows the synthesis of viral proteins, and the chimeric provirus sequence was found to generate infectious viruses. This system could be a most interesting alternative to study ribosomal frameshift in the context of a virus amenable to the use of a simple animal model.
The genome of retroviruses, such as human immunodeficiency virus type 1 (HIV-1), encodes three polyproteins: the Gag precursor of the viral structural proteins (matrix, capsid, and nucleocapsid), the Pol precursor of the viral enzymes (protease, reverse transcriptase, and integrase), and the Env precursor of the viral envelope glycoproteins (surface and transmembrane) (reviewed in reference 28). The coding sequences of Gag and Pol on the full-length viral RNA are translated from the same initiation codon, the Pol polyprotein being produced as a Gag-Pol fusion protein. In HIV, the Gag- and Pol-encoding sequences overlap and a programmed −1 frameshift allows a minority of ribosomes translating the full-length viral mRNA to slip from the Gag-encoding sequence into the Pol-encoding sequence. This recoding event occurs when translating ribosomes encounter a specific frameshift signal composed of two elements: a slippery heptamer, where the translating ribosomes can slip by 1 nucleotide in the 5′ direction, and a stimulatory structure located a few nucleotides downstream from the slippery sequence (10, 11, 28). While, in most studied cases of the frameshift signal, the RNA secondary structure that acts as a stimulator is well established as being a pseudoknot (20, 30), in HIV-1, the stimulator is generally considered to be a simpler stem-loop located 8 nucleotides downstream of the slippery heptamer (1, 16, 23). Although recent data have raised the possibility that the structure is more complex and could involve downstream nucleotide sequences that influence the efficiency of the slippery sequence (8, 9), it is clearly established that the minimal frameshift signal, defined as the slippery site and classical stem-loop structure, efficiently promotes frameshift (for examples, see references 1, 23, and 29).
Programmed −1 frameshift not only permits synthesis of the Pol precursor but is essential to maintain a specific ratio of Gag-Pol to Gag. An adequate ratio of these two precursors is required for Gag monomers and the Gag portion of Gag-Pol to assemble and incorporate the viral RNA genome. The Gag-Pol precursor is unable to correctly assemble by itself, and mutations that induce changes in the Gag-Pol to Gag ratio preclude the assembly of infectious viruses (17, 22, 27). This importance of the Gag-Pol to Gag ratio in retroviral replication has led some investigators to propose that programmed HIV-1 frameshift could become a target for the development of novel antiviral agents (13, 14). However, there is no adequate animal model to conveniently investigate frameshift and antiframeshift agents in vivo.
At the present time, the best model mimicking HIV infection is clearly the simian immunodeficiency virus model; however, its high cost and limited availability are two of the many reasons that limit its use. This explains the interest in developing small, unexpensive, and easy to handle animal models; the powerful genetic tools available in mice are a further incentive to develop murine models. Different approaches have been investigated in an effort to reproduce HIV replication and HIV-induced pathogenesis in mice (6, 15). Despite their interest and contributions to our understanding of retroviral pathogenesis, none of these models can actually mimic a natural cycle of viral infection, replication, and pathogenesis. A major obstacle remains the unexplained block in viral assembly observed in murine cells (2, 21). An alternative to the development of actual animal models for HIV is the study of animal retroviruses such as murine leukemia viruses (MuLV). This approach will obviously be adequate only when studying the aspects of HIV biology that are shared by these viruses. Another possibility is to produce chimeric viruses in which some elements of the HIV genome are introduced into the MuLV genome (for examples, see references 5, 7, 18, and 24). This last approach was selected in the present study to develop a model that could eventually be used for the in vivo study of ribosomal frameshifting in the context of a replicating virus.
The programmed frameshift of HIV-1 was reproduced in different expression systems, by means of either reporter genes, expression of Gag and Gag-Pol-encoding constructs, or transfection of cloned HIV proviral DNA (1, 3, 4, 22, 23, 25, 27). The frameshift phenomenon relies on fundamental mechanisms that are conserved in cells of diverse origins. However, despite the fact that frameshift signals, as found in HIV-1, can act in cells from diverse mammalian species, retroviruses such as MuLV have evolved a programmed readthrough, rather than frameshifting, to generate a Gag-Pol fusion protein (28). For such retroviruses, the coding sequences of Gag and Pol do not overlap but rather are juxtaposed in the same reading frame. A minority of ribosomes translating the full-length viral RNA bypass the termination codon at the 3′ end of the gag gene and continue the translation of the mRNA, producing the Pol portion of the Gag-Pol fusion protein. A pseudoknot, located 8 nucleotides downstream from the termination codon, is required for the readthrough process to occur (11, 28, 30-33).
Since MuLVs do not rely on frameshifting to produce their pol-encoded proteins, these viruses could not be used directly for the purpose of studying frameshift and its putative inhibitors. However, it was reasoned that replacing MuLV readthrough by the HIV-1 frameshift could make the production of the Pol precursor of the murine retrovirus dependent on the HIV-1 frameshift signal to create a simple chimeric retrovirus able to replicate and propagate in mice. Moloney MuLV strain (Mo-MuLV) was chosen for this study since this viral strain replicates at a high titer in murine cell lines in vitro and has been extensively studied. It induces the rapid appearance of a T-cell leukemia in experimentally inoculated mice or rats; a thermosensitive mutant was also shown to induce neuropathogenesis (reviewed in reference 26). Altogether, it thus appears as an ideal polyvalent virus for the development of a murine model to study frameshift.
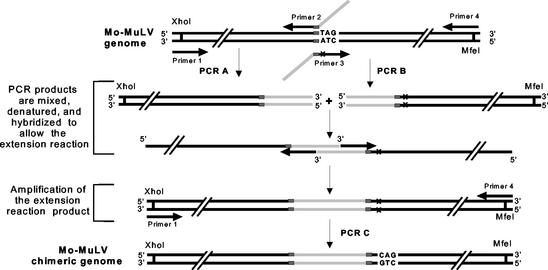
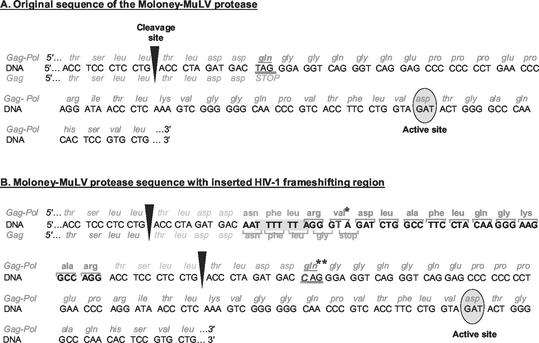
By using the two-step PCR extension method (12) schematized in Fig. 1, the HIV-1 frameshift signal, encompassing both the slippery heptamer and stem-loop, was thus inserted at the junction of the gag and pol gene in Mo-MuLV, upstream from the termination codon of gag that is normally suppressed in the readthrough process (Fig. 2). A complete infectious cloned proviral DNA of Mo-MuLV, flanked by EcoRI sites (19), was first subcloned at the EcoRI site of the pGEM-7z(−) plasmid vector (Promega) and used as a template to construct the chimeric MuLV-HIV DNA fragment as schematized in Fig. 1 and 2. The sequence of oligonucleotide primers 1 (5′-gtc gat gcc gct ttt ccc ctc gag cgc cca gac t-3′) and 4 (5′-tgt ggg agt ctg gtc cag gtc aat tgt cct gag att-3′) is completely homologous to the Mo-MuLV sequence and encompasses, respectively, the underlined XhoI and MfeI sites. The sequence of primers 2 (5′-CCT TCC CTT GTA GGA AGG CCA GAT CTA CCC TAA AAA ATT gtc atc tag ggt cag gag gga ggt ctg ggg tct tgg tcc ccg-3′) and 3 (5′-GGG TAG ATC TGG CCT TCC TAC AAG GGA AGG CCA Gga cct ccc tcc tga ccc tag atg acc agg gag gtc agg gtc ag-3′) is also homologous to the genome of Mo-MuLV at the 3′ ends (lowercase letters) of the two primers, except for one base in primer 3 (underlined), corresponding to the first nucleotide in the stop codon, that is normally subject to readthrough in MuLV. These two oligonucleotides possess long 5′-end extensions (uppercase letters), corresponding to the two halves of the HIV sequences to be inserted in the final construct with an overlap of 30 nucleotides. To ensure that the production of Pol proteins was dependent on the frameshift signal while allowing the synthesis of Gag proteins, a few changes in the frameshift and readthrough sequences had to be made. To shut down the readthrough mechanism of MuLV, a mutation (T→C) was introduced into the stop codon of gag, changing it to a glutamine codon (Fig. 2B). Since the production of the Gag precursor is essential to viral replication, a new stop codon was created in the sequence, located between the slippery heptamer and the stem-loop, and in the same reading frame as the gag initiation codon (Fig. 2B). In the chimeric proviral construct, ribosomes should translate the full-length viral mRNA until they encounter the stop codon in order to generate the Gag precursor. A minority of ribosomes are, however, expected to slip in the −1 reading frame at the HIV slippery sequence and then continue translation of the mRNA until they reach the pol stop codon, producing the Gag-Pol precursor of pol-encoded enzymes. In Mo-MuLV, the junction between the Gag and Pol proteins includes a specific sequence recognized as a cleavage site by the viral protease to process the Gag precursor into mature proteins (Fig. 2A); in the chimeric proviral construct, this sequence was duplicated to flank the extra amino acid sequence generated by the introduction of additional nucleotides that form the frameshift signal (Fig. 2B). During maturation of the Gag and Gag-Pol precursors, it is expected that the viral protease cuts the polyprotein at these sites, removing the extra amino acids encoded by the HIV-1 sequence. The resulting proteins, nucleocapsid and protease, should thus be identical to those of an authentic wild-type Mo-MuLV. Two independently obtained chimeric proviral plasmid constructs were sequenced to verify the inserted HIV fragment (data not shown).
FIG. 1.
Construction of chimeric virus sequence. The PCR strategy used to introduce the HIV-1 frameshift signal at the junction of gag and pol in Mo-MuLV is schematized. The Mo-MuLV sequence is represented by black lines, and the HIV-1 sequence is represented by light gray lines. Gray boxes represent the sequence of the cleavage site recognized by the viral protease. The target sequence of Mo-MuLV, where the HIV-1 frameshift was inserted, was flanked by restriction sites XhoI, in 5′, and MfeI, in 3′. The X symbol, in primer 3, indicates the single mutation (T→C) that replaced the gag stop codon (TAG) with a glutamine codon (CAG). The sequence to insert was encompassed in primers 2 and 3 and included in two separate PCR products, the 3′-end sequence of the PCR A product overlapping with the 5′-end sequence of PCR B product. The two PCR products were mixed and denatured, allowing one strand from each fragment to act as a primer on the other fragment. The new DNA product resulting from this extension reaction was then amplified in a third PCR (PCR C) using primers 1 and 4. The final PCR product was then digested with XhoI and MfeI and inserted into pGMo.
FIG. 2.
Sequence of the chimeric virus. (A) Original sequence at the gag-pol junction in Mo-MuLV. (B) Insertion of the HIV-1 frameshift region at the Mo-MuLV gag-pol junction. The Mo-MuLV DNA sequence is given in standard uppercase letters, and the HIV-1 DNA sequence is shown in boldface. The slippery sequence of the frameshift site is enclosed in light gray. A nucleotide was changed (G→T) in the sequence between the slippery sequence and the sequence coding for the stem-loop, producing an UAG stop codon in the 0 reading frame and a codon coding for a val* in the −1 reading frame. The first base of the gag stop codon was substituted, replacing the stop codon with a glutamine codon (gln**). The cleavage sites recognized by the viral protease are indicated by arrowheads.
In order to determine if the frameshift mechanism could functionally replace readthrough in an MuLV background, resulting in the production of replication-competent viruses, proviral DNA was introduced by transfection in NIH 3T3 mouse fibroblasts permissive for Mo-MuLV replication. The chimeric virus was able to replicate, since levels of reverse transcriptase activity associated with particles that were released in the cell culture supernatant increased with time upon cell passages (Fig. 3). Maximal virus production was reached about 2 weeks posttransfection in both the wild-type and chimeric viruses and was about 10 to 20% lower at all time points for the chimeric virus than for the wild type (Fig. 3). These results suggest that both viruses replicate until all cells become infected, generating a chronically infected cell line; the chimeric virus appears to replicate normally, although a slight reduction in virus production was observed.
FIG. 3.
Replication of chimeric virus. Control wild-type Mo-MuLV proviral DNA (▴) and chimeric (•) constructs were introduced by transfection in murine NIH 3T3 fibroblasts by the calcium-phosphate coprecipitation procedure. Transfected cells were each passaged 3 days, and the supernatant from 24 h of incubation was harvested at day 2 of each passage. Virus-containing supernatants were subjected to a low-speed centrifugation (15 min at 1,000 × g in a Sorval SH 3000 rotor at 4°C) to remove cell debris and ultracentrifuged at 100,000 × g for 1 h in a Beckman 70.1 Ti rotor at 4°C. Viral pellets were then resuspended overnight in 200 μl of TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0]), and 50 μl of each sample was mixed with 50 μl of a cocktail reaction {final concentrations of 50 mM Tris-HCl [pH 8.2], 20 mM dithiothreitol, 50 mM NaCl, 0.05% Nonidet P-40, 0.6 mM MnCl2, 10 μM dTTP, 20 μg of poly(A) · oligo(dT)15 (Roche Molecular Biochemicals)/ml, 0.4 μCi [3H]dTTP (1,000 Ci/mmol; ICN)}. The reaction was incubated for 2 h at 37°C, and reaction products were precipitated by the addition of 1 ml of 10% trichloroacetic acid and left on ice for 2 h. The trichloroacetic acid-precipitable material was filtered onto 0.45-μm-pore-size Millipore filters, filters were dried, and radioactivity was measured in a Beckman LS 600 SC scintillation counter. The results are summarized from three separate experiments using independent plasmid preparations.
To confirm that the viruses produced in chronically infected cells actually harbored the HIV sequence and to verify the stability of this sequence in the background Mo-MuLV genome, sequencing of the viral genome by duplex reverse transcription-PCR was performed. Viruses were recovered from chronically infected cells approximately 30 days posttransfection, at which time viral production had reached its peak and was stably maintained. Briefly, the viral genomic RNA of chimeric or wild-type Mo-MuLV was isolated from resuspended viral particles and treated with fast-performance liquid chromatography pure DNase I (Amersham Pharmacia Biotech) before being used for the synthesis of a first DNA strand by using MuLV reverse transcriptase (Gibco-BRL) primed with an oligonucleotide located downstream of the MfeI site (5′-gcc gta gga cag agg atg ag-3′). Amplification of double-stranded DNA was then achieved by using the same primer in combination with a second one located 5′ of the XhoI site (5′-gcc cca ttg gtc cca taa cc-3′). One-tenth of the obtained PCR product was finally amplified in a second PCR by using two internally located primers, 5′-gga ggt ccc aac tcg atc gcg-3′ and 5′-cag cga tac cgc ttt cct cca g-3′; these same two primers were also used to sequence directly the resulting PCR product by automated DNA sequencing. The replicating virus retained the expected HIV-1 insertion without any additional change in either the introduced HIV sequence or the adjacent sequences (data not shown). The experiment was repeated on two separately established cell lines with the same result. Sequencing of the PCR product obviously revealed only the predominant sequence, and it could not be completely excluded that there were minor variants in the viral population. Closer examination of the sequence did not reveal consistent minor peaks that would be indicative of the presence of such a viral subpopulation, suggesting that, if they do exist, such minor variants do not possess a strong selective advantage, at least not under tissue culture conditions. Production of pol-encoded protein is thus clearly dependent upon the use of the introduced frameshift signal in the chimeric virus.
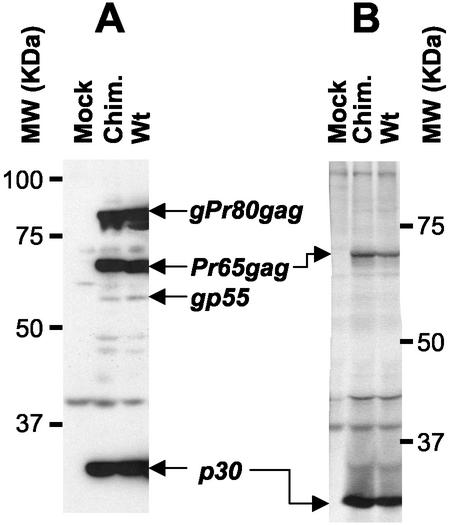
Having proven that the frameshift signal is conserved upon viral propagation, viral proteins were finally examined. There were no obvious differences between the nature and amount of viral proteins detected in cells chronically infected by either the wild-type or chimeric virus (Fig. 4A). The three main proteins recognized by the anti-p30(CA) antiserum were the Gag precursor Pr65Gag, its glycosylated form, gPr80Gag, and the viral capsid protein p30(CA) that is the main product of Gag precursor proteolytic maturation. Partially processed intermediates and fully processed proteins could also be seen, especially the amino-terminal cleavage fragment of gPr80Gag, gp55. Similar observations were made in five independent experiments using two independently established cell lines. This shows that synthesis and processing of viral proteins does occur normally in the chimeric virus despite the introduction of the HIV frameshift signal and that of an additional cleavage site for the viral protease. The proteins incorporated into the viral particles were also examined by the metabolic radiolabeling and immunoprecipitation of semipurified viruses (Fig. 4B). Again, the protein profile was virtually indistinguishable between wild-type and chimeric viruses by this procedure; the two viral proteins were recognized by the anti-p30 antiserum being the remaining unprocessed Gag precursor (Pr65Gag) and p30, the major protein of the viral capsid.
FIG. 4.
Analysis of viral proteins in chimeric viruses. (A) Chronically infected NIH 3T3 cells were lysed in RIPA buffer (10 mM sodium phosphate [pH 7.2], 140 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate) containing a cocktail of protease inhibitors. Cell debris was removed by centrifugation (15,000 × g for 45 min in a Sorvall F-20 rotor at 4°C); samples of cell lysates were then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting using a goat anti-p30 antiserum (National Cancer Institute) and horseradish peroxidase-conjugated swine anti-goat secondary antibody (Cedarlane). Antigen-antibody complexes were detected by enhanced chemiluminescence and autoradiography. (B) Subconfluent cells were subjected to metabolic radiolabeling (200 μCi of ICN Tran35S-Label per 10-cm-diameter petri dish); cell supernatants were collected 8 h later and clarified by centrifugation (1,000 × g for 15 min in a Sorvall SH 3000 rotor at 4°C) followed by filtration onto 0.45-μm-pore-size nitrocellulose filters. Viruses were then semipurified by ultracentrifugation of clarified supernatants through a 3-ml 20% sucrose cushion in phosphate-buffered saline for 2 h at 100,000 × g in a 70.1 rotor (Beckman) at 4°C. Pelleted viral particles were resuspended in 100 μl of RIPA buffer and centrifuged for 30 min at 15,000 ×g in a Sorvall F-20 microrotor. These samples containing viral proteins were incubated overnight at 4°C with the anti-p30 antiserum, and the immune complexes were collected by incubation with protein G-Sepharose (Amersham) for 3 h at 4°C. Sepharose beads were briefly pelleted in a microcentrifuge and washed extensively in RIPA buffer. Bound antigen-antibody complexes were analyzed on sodium dodecyl sulfate-10% polyacrylamide gels, fixed, dried, and exposed on Biomax MR films (Kodak). The positions of the molecular weight markers and selected viral proteins in the wild type (Wt) and the chimeric virus (Chim.) are indicated in both panels A and B.
In conclusion, replacement of readthrough in MuLV by frameshift, consecutive to the introduction of the RNA sequence of HIV-1, was achieved and resulted in the production of infectious virions at an efficiency that closely mimics that of wild-type MuLV. The small reduction in viral production with the chimeric virus could be explained if the frameshift efficiency was slightly different from the frequency of ribosomal readthrough. This could be intrinsic to the two mechanisms, the exact frequency in vivo of frameshifting and readthrough remaining difficult to establish, or, simply, this could result from the introduction of the frameshift element in a different sequence context.
The construction of an infectious chimeric MuLV encompassing the HIV ribosomal frameshift signal opens novel avenues of research on frameshifting. A recent report indicated that the sequence of the frameshift signal is quite variable among HIV isolates (29); however, the biological significance of this variability is unclear. It will clearly be of interest to look at the replication of the chimeric virus developed in the present study to determine if, upon prolonged viral replication in an experimental animal, the sequence of the frameshift signal could evolve, even though this was not the case in vitro. Finally, perhaps more important in the context of our constant need for novel therapies to control HIV as the AIDS etiological agent, this experimental system could be used for the study of frameshift inhibitors in the context of in vivo viral replication and disease development.
Acknowledgments
We thank Carole Danis for expert technical support conducing to the smooth operation of the Lemay laboratory at the Département de Microbiologie et Immunologie. We thank all the members of the Brakier-Gingras laboratory at the Département de Biochimie for their constant support and numerous discussions in the course of this project. We also thank Éric A. Cohen (from the Département de Microbiologie et Immunologie) for generously letting us use some of his laboratory facilities and Guy Boileau and Luc Des Groseillers (both from the Département de Biochimie) for numerous helpful discussions.
This work was supported by a Canadian Institute for Health Research (CIHR) operating grant (to L.B.-G. and G.L.), a pilot study grant from the Fonds de la Recherche en Santé du Québec (FRSQ) via the AIDS and Infectious Diseases Network (Réseau SIDA-MI to L.B.-G. and G.L.), and a Fonds pour les Chercheurs et l'Aide à la Recherche (FCAR) group grant (to L.B.-G. and G.L). G.L. was also the recipient of an FRSQ senior scholarship award, and M.-N.B. was the recipient of a studentship from the International Council for Canadian Studies.
REFERENCES
- 1.Bidou, L., G. Stahl, B. Grima, H. Liu, M. Cassan, and J. P. Rousset. 1997. In vivo HIV-1 frameshifting efficiency is directly related to the stability of the stem-loop stimulatory signal. RNA 3:1153-1158. [PMC free article] [PubMed] [Google Scholar]
- 2.Bieniasz, P. D., and B. R. Cullen. 2000. Multiple blocks to human immunodeficiency virus type 1 replication in rodent cells. J. Virol. 74:9868-9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunelle, M. N., C. Payant, G. Lemay, and L. Brakier-Gingras. 1999. Expression of the human immunodeficiency virus frameshift signal in a bacterial cell-free system: influence of an interaction between the ribosome and a stem-loop structure downstream from the slippery site. Nucleic Acids Res. 27:4783-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cassan, M., N. Delaunay, C. Vaquero, and J. P. Rousset. 1994. Translational frameshifting at the gag-pol junction of human immunodeficiency virus type 1 is not increased in infected T-lymphoid cells. J. Virol. 68:1501-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, B. K., I. Rousso, S. Shim, and P. S. Kim. 2001. Efficient assembly of an HIV-1/MLV Gag-chimeric virus in murine cells. Proc. Natl. Acad. Sci. USA 98:15239-15244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, J. 2001. Building a small animal model for AIDS, block by block. Science 293:1034-1036. [DOI] [PubMed] [Google Scholar]
- 7.Deminie, C. A., and M. Emerman. 1994. Functional exchange of an oncoretrovirus and a lentivirus matrix protein. J. Virol. 68:4442-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dinman, J. D., S. Richter, E. P. Plant, R. C. Taylor, A. B. Hammell, and T. M. Rana. 2002. The frameshift signal of HIV-1 involves a potential intramolecular triplex RNA structure. Proc. Natl. Acad. Sci. USA 99:5331-5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dulude, D., M. Baril, and L. Brakier-Gingras. 2002. Characterization of the frameshift stimulatory signal controlling a programmed −1 ribosomal frameshift in the human immunodeficiency virus type 1. Nucleic Acids Res. 30:5094-5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farabaugh, P. J. 2000. Translational frameshifting: implication for the mechanism of translational frame maintenance. Prog. Nucleic Acid Res. Mol. Biol. 64:131-170. [DOI] [PubMed] [Google Scholar]
- 11.Gesteland, R. F., and J. F. Atkins. 1996. Recoding: dynamic reprogramming of translation. Annu. Rev. Biochem. 65:741-768. [DOI] [PubMed] [Google Scholar]
- 12.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 13.Hung, M., P. Patel, S. Davis, and S. R. Green. 1998. Importance of ribosomal frameshifting for human immunodeficiency virus type 1 particle assembly and replication. J. Virol. 72:4819-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Irvine, J. H., J. A. Horsfield, C. Z. McKinney, and W. P. Tate. 1998. A novel strategy to interfere with human immunodeficiency virus type 1 propagation. New Zealand Med. J. 111:222-224. [PubMed] [Google Scholar]
- 15.Jamieson, B. D., and J. A. Zack. 1999. Murine models for HIV disease. AIDS 13(Suppl. A):S5-S11. [PubMed] [Google Scholar]
- 16.Kang, H. 1998. Direct structural evidence for formation of a stem-loop structure involved in ribosomal frameshifting in human immunodeficiency virus type 1. Biochim. Biophys. Acta 1397:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karacostas, V., E. J. Wolffe, K. Nagashima, M. A. Gonda, and B. Moss. 1993. Overexpression of the HIV-1 gag-pol polyprotein results in intracellular activation of HIV-1 protease and inhibition of assembly and budding of virus-like particles. Virology 193:661-671. [DOI] [PubMed] [Google Scholar]
- 18.Kondo, E., F. Mammano, E. A. Cohen, and H. G. Göttlinger. 1995. The p6gag domain of human immunodeficiency virus type 1 is sufficient for the incorporation of Vpr into heterologous viral particles. J. Virol. 69:2759-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemay, G., and P. Jolicoeur. 1984. Rearrangement of a DNA sequence homologous to a cell-virus junction fragment in several Moloney murine leukemia virus-induced rat thymomas. Proc. Natl. Acad. Sci. USA 81:38-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marczinke, B., R. Fisher, M. Vidakovic, A. J. Bloys, and I. Brierley. 1998. Secondary structure and mutational analysis of the ribosomal frameshift signal of Rous sarcoma virus. J. Mol. Biol. 284:205-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mariani, R., G. Rutter, M. E. Harris, T. J. Hope, H.-G. Krausslich, and N. R. Landau. 2000. A block to human immunodeficiency virus type 1 assembly in murine cells. J. Virol. 74:3859-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park, J., and C. D. Morrow. 1991. Overexpression of the gag-pol precursor from human immunodeficiency virus type 1 proviral genomes results in efficient proteolytic processing in the absence of virion production. J. Virol. 65:5111-5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parkin, N. T., M. Chamorro, and H. E. Varmus. 1992. Human immunodeficiency virus type 1 gag-pol frameshifting is dependent on downstream mRNA secondary structure: demonstration by expression in vivo. J. Virol. 66:5147-5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reed, M., R. Mariani, L. Sheppard, K. Pekrun, N. R. Landau, and N. W. Soong. 2002. Chimeric human immunodeficiency virus type 1 containing murine leukemia virus matrix assembles in murine cells. J. Virol. 76:436-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reil, H., H. Kollmus, U. H. Weidle, and H. Hauser. 1993. A heptanucleotide sequence mediates ribosomal frameshifting in mammalian cells. J. Virol. 67:5579-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenberg, N., and P. Jolicoeur. 1997. Retroviral pathogenesis, p. 475-586. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, New York, N.Y. [PubMed]
- 27.Shehu-Xhilaga, M. S., M. Crowe, and J. Mak. 2001. Maintenance of the Gag/Gag-Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J. Virol. 75:1834-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, New York, N.Y. [PubMed]
- 29.Telenti, A., R. Martinez, M. Munoz, G. Bleiber, G. Greub, D. Sanglard, and S. Peters. 2002. Analysis of natural variants of the human immunodeficiency virus type 1 gag-pol frameshift stem-loop structure. J. Virol. 76:7868-7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.ten Dam, E. B., C. W. Pleij, and L. Bosch. 1990. RNA pseudoknots: translational frameshifting and readthrough on viral RNAs. Virus Genes 4:121-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wills, N. M., R. F. Gesteland, and J. F. Atkins. 1991. Evidence that a downstream pseudoknot is required for translational read-through of the Moloney murine leukemia virus gag stop codon. Proc. Natl. Acad. Sci. USA 88:6991-6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wills, N. M., R. F. Gesteland, and J. F. Atkins. 1994. Pseudoknot-dependent readthrough of retroviral gag termination codons: importance of sequence in the spacer and loop 2. EMBO J. 13:4137-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshinaka, Y., I. Katoh, T. D. Copeland, and S. Oroszlan. 1985. Murine leukemia virus protease is encoded by the gag-pol gene and is synthesized through suppression of an amber termination codon. Proc. Natl. Acad. Sci. USA 82:1618-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]