Abstract
For any of the enveloped RNA viruses studied to date, recognition of a specific RNA packaging signal by the virus's nucleocapsid (N) protein is the first step described in the process of viral RNA packaging. In the murine coronavirus a selective interaction between the viral transmembrane envelope protein M and the viral ribonucleoprotein complex, composed of N protein and viral RNA containing a short cis-acting RNA element, the packaging signal, determines the selective RNA packaging into virus particles. In this report we show that expressed coronavirus envelope protein M specifically interacted with coexpressed noncoronavirus RNA transcripts containing the short viral packaging signal in the absence of coronavirus N protein. Furthermore, this M protein-packaging signal interaction led to specific packaging of the packaging signal-containing RNA transcripts into coronavirus-like particles in the absence of N protein. These findings not only highlight a novel RNA packaging mechanism for an enveloped virus, where the specific RNA packaging can occur without the core or N protein, but also point to a new, biologically important general model of precise and selective interaction between transmembrane proteins and specific RNA elements.
The process of packaging the viral genome is a critical step in the assembly of infectious viruses. For any of the enveloped RNA viruses studied to date, the association of an intracellular form of viral genomic RNA with the nucleocapsid protein is the first step in the process of selective genome packaging into virus particles. A specific RNA element(s), usually referred to as a packaging signal (PS) or an encapsidation signal, which is present in intracellular viral genomic RNA, determines the selective and specific binding of viral nucleocapsid protein to the viral genomic RNA. Subsequently, the viral ribonucleoprotein (RNP) complex containing viral RNA and the nucleocapsid protein binds to a viral envelope protein(s) at the virus budding site, which leads to the budding of virus particles containing the viral RNP complex. In some enveloped viruses, an interaction between the viral RNP complex and envelope proteins drives the budding of virus particles (21, 31, 34, 41), while in other enveloped viruses, the viral RNP complex is dispensable for viral envelope formation and production of virus particles. A typical example of the latter phenomenon is observed in coronavirus envelope formation; coronavirus-like particles (VLPs) that are morphologically similar to infectious virus particles are produced in the absence of viral RNP complex (37).
The murine coronavirus mouse hepatitis virus (MHV) contains three envelope proteins, S, M, and E, and a helical nucleocapsid consisting of N protein and a large single-stranded, positive-stranded RNA genome (19, 33). S protein is dispensable for viral nucleocapsid packaging and viral assembly (13, 15, 30). M protein and E protein are essential for viral envelope formation and release of virus particles; VLPs are released from cells that express both M protein and E protein (37). M protein, the most abundant transmembrane envelope glycoprotein in the virus particle and in infected cells, is characterized as having three domains: a short N-terminal ectodomain, a triple-spanning transmembrane domain, and a C-terminal endodomain (1). E protein is present in only minute amounts in infected cells and in the viral envelope, and yet it plays a central role in coronavirus morphogenesis (7). The viral genomic RNA and N protein form the helical nucleocapsid structure, which exists inside the viral envelope (6, 33).
In infected cells, MHV synthesizes the intracellular form of genomic RNA, mRNA 1, and six to seven species of subgenomic mRNAs. These virus-specific mRNAs comprise a nested set with a common 3′ terminus and a common leader sequence of approximately 72 to 77 nucleotides (nt) at the 5′ end (18, 32). All MHV mRNAs associate with N protein to form RNP complexes (2, 27); however, only the mRNA 1-RNP complex is efficiently packaged into the virus particles. Previous studies demonstrated that only mRNA 1 and the viral genomic RNA, but not subgenomic mRNAs, contain a 190-nt-long PS (9, 36). A specific interaction occurs between the viral transmembrane envelope protein M and the mRNA 1-N protein complex at the budding site in infected cells (27), and the 190-nt-long PS mediates the specific interaction between M protein and the mRNA 1-N protein complex (or other RNP complexes containing the PS) to drive the specific packaging of RNA into virus particles (28). How M protein selectively and specifically recognizes the PS-containing RNP complex is unknown. We previously suggested two models to explain the mechanism of specific recognition of PS-containing RNP complexes by M protein (28). One was that M protein recognizes a specific helical nucleocapsid structure formed by the mRNA 1-N protein complex. The binding of N protein to the PS might trigger the formation of helical nucleocapsid structure. In this model, both M protein and N protein contribute to the selective packaging of specific RNA species into virus particles. Another model was that the direct interaction of M protein with the PS in the PS-containing RNP complex is responsible for the selectivity in RNA packaging.
The present study investigated the mechanism by which MHV M protein selectively recognizes PS-containing RNP complexes. We found that expressed M protein specifically interacted with coexpressed noncoronavirus RNA transcripts containing the PS in the absence of N protein. Furthermore, this M protein-PS interaction led to specific packaging of the PS-containing RNA transcripts into VLPs in the absence of N protein. Our study revealed that MHV employs a novel mechanism of specific and selective RNA packaging, in which the specific interaction between M protein and the PS determines the selectivity and specificity of RNA packaging in the absence of the core or N protein. Furthermore, MHV M protein is the first viral transmembrane protein that binds to a specific viral RNA element in the absence of any other viral proteins.
MATERIALS AND METHODS
Cells and viruses.
DBT cells were used for DNA transfection and production of VLPs (12), while BHK cells were used for the preparation of Sindbis virus pseudovirions. RK13 cells were used for the production and titer determination of recombinant vaccinia virus vTF7-3 (10).
Preparation of Sindbis virus pseudovirions.
The Sindbis virus vector expressing MHV S protein (pSinS) was constructed by inserting the entire open reading frame of MHV type 2 (MHV-2) S protein (39) into the StuI site of a Sindbis virus pseudovirion expression vector, pSinRep5 (5) (Invitrogen, San Diego, Calif.). Sindbis virus pseudovirions, SinM (22), SinE (22), SinN (27), SinLacZ, and SinS were produced as described previously (22).
DNA transfection.
Subconfluent monolayers of DBT cells were infected with vTF7-3 at a multiplicity of infection of 5 for 1 h at 37°C. At 1 h postinfection, the cells were transfected with 20 μg of plasmid DNA by a lipofection procedure (14), and at 4 h postinfection the cells were superinfected with Sindbis virus pseudovirions.
Labeling of proteins, immunoprecipitation, and SDS-PAGE.
Infected cells were labeled with 100 μCi of Tran35S-label/ml of medium for 5 h from 7 to 12 h post-Sindbis virus pseudovirion infection. Cell lysates were prepared at 12 h post-Sindbis virus pseudovirion infection with lysis buffer (1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS] in phosphate-buffered saline) (23), and the intracellular MHV-specific proteins were immunoprecipitated with anti-MHV N monoclonal antibody (MAb) J3.3, anti-MHV M MAb J1.3 (8), anti-E protein peptide-2 antibody (40), and the non-MHV MAb H2KkDk (H2K), as described previously (15). For protein analysis, the immunoprecipitated proteins were incubated at 37°C for 30 min in sample buffer to prevent M protein aggregation (33) and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE). RNAs were extracted from immunoprecipitated samples as described previously (27).
Purification of VLPs.
The cell culture medium from infected cells was collected at 12 h post-Sindbis virus pseudovirion infection and briefly centrifuged to remove cell debris. Released radiolabeled VLPs were partially purified by ultracentrifugation on a discontinuous sucrose gradient consisting of 50, 30, and 20% sucrose prepared in NTE buffer (0.1 M NaCl, 0.01 M Tris-HCl [pH 7.5], 0.001 M EDTA) (22). After centrifugation at 26,000 rpm for 16 h at 4°C in a Beckman SW28 rotor, VLPs at the interface of 30 and 50% sucrose were collected. In the case of RNase A treatment, the samples (in sucrose prepared in NTE buffer) were treated with 0.5 ng of RNase A per ml of interface for 30 min at room temperature. The samples were further purified on a continuous sucrose gradient of 20 to 60% sucrose at 26,000 rpm for 18 h at 4°C. The VLPs, in the fractions corresponding to the reported density of coronavirus VLPs (1.14 to 1.16 g/cm3) (22, 37), were collected and pelleted by ultracentrifugation in a Beckman SW28 rotor at 26,000 rpm for 3 h at 4°C. The pellets were suspended in the lysis buffer.
Analysis of VLP RNA and intracellular RNA.
Purified VLPs were suspended in the lysis buffer, and RNA was extracted from VLP lysates by established methods (24). The intracellular RNA was extracted from cytoplasmic lysates as described previously (25). After DNase treatment (38), RNAs were denatured and separated on a 1% agarose gel containing formaldehyde (23). After electrophoresis, Northern blot analysis was performed with a digoxigenin-labeled random-primed probe (Boehringer) specific to the chloramphenicol acetyltransferase (CAT) gene (27, 38); the RNAs were visualized with the DIG luminescent detection kit (Boehringer).
RESULTS
Envelope M protein selectively interacts with PS-containing non-MHV RNA transcript in the absence of N protein.
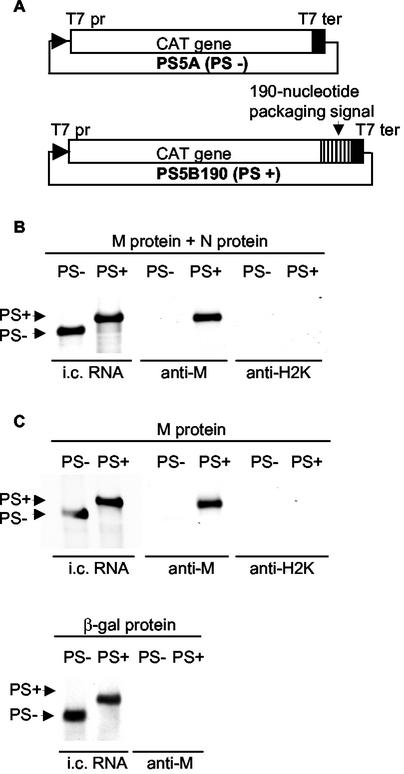
The specific and selective interaction between M protein and PS-containing RNP complexes drives the specific packaging of PS-containing RNAs into MHV particles (28). In all known enveloped RNA viruses studied thus far, N protein or capsid protein plays an essential role in viral RNA packaging. However, the role(s) of N protein in MHV RNA packaging is unknown. In coexpression experiments testing packaging of non-MHV RNA with inserted PS in the presence of combinations of various expressed MHV proteins, we attempted to understand how M protein selectively recognizes PS-containing RNP complexes. We were particularly interested in determining whether N protein is essential for this selective interaction. DBT cells were infected with a recombinant vaccinia virus, vTF7-3, which encodes the T7 RNA polymerase (10). One hour later, the cells were independently transfected with either plasmid PS5A, which contains the entire CAT gene under the dual controls of the T7 promoter and the T7 terminator, or plasmid PS5B190, which carries the MHV 190-nt PS positioned downstream of the CAT gene (Fig. 1A). RNA transcripts are expressed from transfected PS5A and PS5B190 in vTF7-3-infected cells (28, 38). At 4 h post-vTF7-3 infection, which was 3 h post-plasmid transfection, cultures from both plasmid transfections were superinfected with one or combinations of three Sindbis virus expression vectors: SinM pseudovirion (expressing MHV M protein), SinN pseudovirion (expressing MHV N protein), and SinLacZ pseudovirion (encoding the β-galactosidase protein) (22, 27). Cell extracts were prepared at 12 h post-Sindbis virus pseudovirion infection and used for coimmunoprecipitation analysis with anti-M MAb or control anti-H2K MAb. RNA was extracted from the immunoprecipitated samples and treated with DNase (28, 38). Northern blot analysis with a CAT sequence-specific probe showed that PS5A and PS5B190 RNA transcripts were expressed at similar levels (Fig. 1). Strikingly, anti-M MAb coimmunoprecipitated PS5B190 RNA transcript from cells coexpressing PS5B190 RNA transcript and only the M protein as well as from cells coexpressing M protein and N protein with the same transcript (Fig. 1B and C). Anti-M MAb did not coprecipitate PS5A transcripts from coexpressing cells, nor did it coprecipitate PS5B190 transcripts from cells coexpressing β-galactosidase. Anti-H2K MAb did not coimmunoprecipitate either PS5B190 or PS5A transcript, establishing that the coimmunoprecipitation with anti-M MAb was specific. Consistent with a previous study (27), SDS-PAGE analysis showed that only M MAb and not H2K MAb immunoprecipitated M protein (data not shown). These data demonstrated that coexpressed M protein bound to expressed PS-containing RNA transcripts in the absence of other MHV functions, including N protein. Furthermore, these data strongly suggested that the PS was a signal for binding the envelope transmembrane protein, M.
FIG. 1.
Specific interaction of M protein with the PS-containing RNA transcripts in the absence of N protein. (A) Schematic diagrams of the structures of plasmids PS5A (PS−) and PS5B190 (PS+). T7 pr, T7 promoter; T7 ter, T7 terminator. (B and C) Northern blot analysis of expressed RNA transcripts from RNA-expressing cells coinfected with SinM and SinN pseudovirions (M protein + N protein) (B) or infected with either SinM pseudovirion (M protein) or SinLacZ pseudovirion (β-galactosidase protein) (C). Equal volumes of cell lysates were immunoprecipitated with anti-M protein MAb (anti-M) and a control MAb (anti-H2K). Intracellular (i.c.) RNAs and coimmunoprecipitated RNAs were analyzed by Northern blot analysis with a probe that binds to the CAT gene sequence. The arrows indicate expressed RNA transcripts. Each panel shows representative data from triplicate experiments. RNAs extracted from 105 cells and 106 cells were analyzed for the intracellular RNA lanes and the anti-M and anti-H2K lanes, respectively. The anti-M and anti-H2K lanes were exposed eight times longer than the intracellular RNA lanes.
PS-containing RNAs are selectively packaged into VLPs in the absence of N protein.
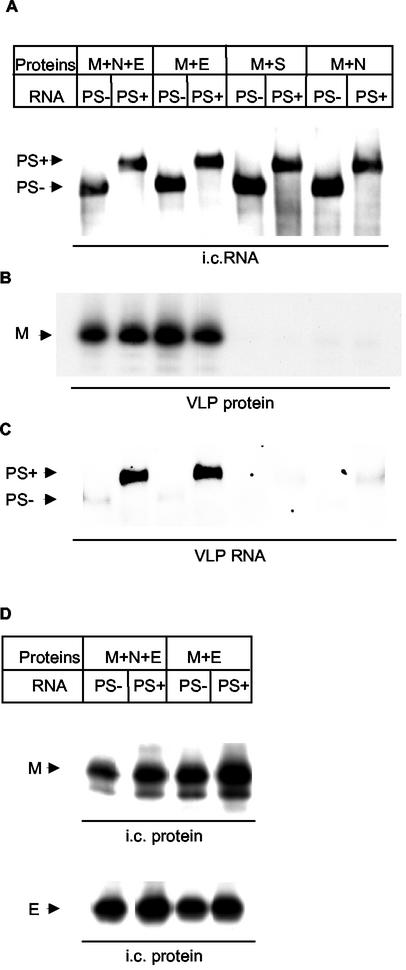
The finding that M protein bound to PS-containing RNA transcripts in the absence of N protein led us to investigate whether the PS-containing RNA transcripts could be packaged into VLPs in the absence of N protein. Expression of two coronavirus envelope proteins, M and E, results in the production of VLPs (3, 37), which are indistinguishable from authentic coronavirus virions in size and shape (37); S protein is nonessential for coronavirus assembly (13, 15, 30, 37). vTF7-3-infected DBT cells were independently transfected with the PS5B190 plasmid or the PS5A plasmid. The cells were superinfected with a mixture of Sindbis virus pseudovirions. PS5A transcript served as a negative control for testing the specificity of RNA packaging. Because coexpression of M and E protein is required for VLP production (37), omission of E protein expression served as a negative control for the VLP production. Cells were radiolabeled, and culture fluids and cell extracts from coexpressing cells were collected at 12 h post-Sindbis virus pseudovirion infection. The PS5B190 and PS5A RNA transcripts were expressed similarly in all the samples (Fig. 2A). The released VLPs were purified by sucrose gradient centrifugation, and the fractions corresponding to VLP density (22, 37) were collected. The corresponding fractions in the negative control samples were also collected. VLP production was measured through detection of M protein in the sucrose fractions (22, 37); similar amounts of VLPs were produced from the cells coexpressing M and E proteins or M, N, and E proteins. As expected, VLPs were not produced from cells coexpressing M and S proteins or coexpressing M and N proteins (Fig. 2B). A similar amount of PS5B190 transcript was easily detected in the released VLPs from the cells coexpressing M and E proteins and the PS5B190 RNA transcripts, as well as from cells additionally coexpressing N protein (Fig. 2C). In contrast, only a very low level of PS5A transcript was detected in the released VLPs (Fig. 2C). Also, only trace amounts of expressed RNA transcripts were detected in the supernatant from cells coexpressing M and N proteins or M and S proteins (Fig. 2C). Analysis of the intracellular proteins showed that both M and E proteins accumulated to similar levels in the expressing cells (Fig. 2D). N and S proteins also accumulated to similar levels in the expressing cells (data not shown). These data demonstrated that coexpression of M, N, and E proteins and the RNA containing the PS resulted in the production of VLPs containing the RNA transcripts. Most importantly, our data convincingly demonstrated that coexpression of M and E proteins and RNA containing the PS resulted in the production of VLPs containing the RNA transcripts; surprisingly, N protein was dispensable for RNA packaging.
FIG. 2.
Nucleocapsid-independent RNA packaging into VLPs. [35S]methionine-cysteine-labeled VLPs were purified from culture fluid of the cells expressing PS5B190 (PS+) or PS5A (PS−) RNA transcripts and the MHV-specific proteins. (A) Intracellular (i.c.) RNAs were extracted from the cytoplasmic lysates and analyzed by Northern blot analysis as described in the legend to Fig. 1. The intracellular RNAs extracted from 3 × 105 cells were applied to each lane. (B) Release of M protein in VLPs. A part of the purified VLP lysate was immunoprecipitated with anti-M protein MAb and analyzed by SDS-PAGE. Only the section of the autoradiogram with M protein is shown. (C) Northern blot analysis of VLP RNAs. VLP RNA was extracted from purified VLPs and analyzed by Northern blot analysis as described above. VLPs released from 107 cells were used for analysis of VLP RNAs. Panel C was exposed eight times longer than panel A. (D) Intracellular expression levels of M protein and E protein. Cytoplasmic lysates were immunoprecipitated with anti-M protein MAb and anti-E protein peptide-2 antibody and analyzed by SDS-PAGE. Only the sections of the autoradiogram with M protein and E protein are shown. Each panel shows representative data from triplicate experiments.
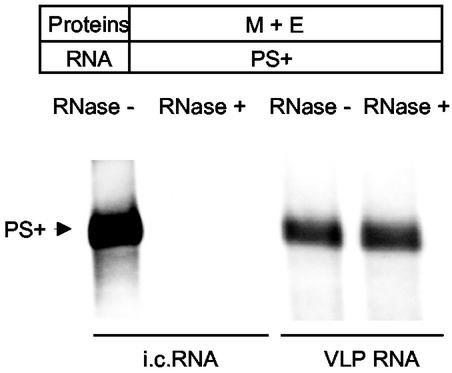
To further confirm that the PS5B190 RNA transcripts were indeed packaged within the VLPs, the samples containing the released VLPs were treated with RNase A. If the RNA transcripts are present within the VLPs, then the RNAs should be inaccessible to RNase A and hence resistant to RNase A treatment. Partially purified VLPs, released from cells expressing PS5B190 RNA transcripts and M protein and E protein, were incubated in the presence of RNase A. Subsequently the VLPs were purified by ultracentrifugation, and the RNAs were extracted from the purified VLPs. As a control, the intracellular RNAs extracted from the same cells were also subjected to the same RNase A treatment under the same buffer conditions. Northern blot analysis revealed that the intracellular RNAs extracted from the same cells were completely degraded by the same RNase A treatment (Fig. 3), demonstrating that the experimental condition for RNase treatment was appropriate. In contrast, no degradation of PS5B190 RNA transcripts occurred after RNase A treatment of the VLPs (Fig. 3), demonstrating that PS5B190 RNA transcripts were indeed selectively packaged into VLPs.
FIG. 3.
Northern blot analysis of VLP-associated RNAs after RNase A treatment. Partially purified VLPs, released from cells coexpressing PS5B190 (PS+) RNA transcripts, M protein, and E protein, were incubated in the presence (RNase +) or absence (RNase −) of RNase A and subsequently purified by ultracentrifugation. VLP-associated RNAs were extracted from purified VLPs. Intracellular (i.c.) RNAs were extracted from the cytoplasmic lysates of the same cells and incubated in the presence (RNase +) or absence (RNase −) of RNase A. Partially purified VLPs released from 2 × 107 cells and 3 μg of intracellular RNA were used for RNase A digestion. Northern blot analysis was performed as described in the legend to Fig. 1 to examine the susceptibility of VLP-associated RNAs and intracellular RNAs to RNase A treatment. Each panel shows representative data from triplicate experiments.
DISCUSSION
The present study revealed a novel paradigm for viral genome packaging, in which the selective and specific interaction between a viral envelope protein and a viral cis-acting RNA element (PS) determines the exclusivity of viral RNA packaging. MHV N protein binds to all MHV mRNAs as well as expressed non-MHV RNA transcripts in infected cells (2, 27), which makes it difficult to explain how the formation of N protein-mRNA 1 RNP complex might lead to the specific packaging of RNA into virus particles. The present study convincingly demonstrated the selective interaction of M protein with PS-containing RNA in the absence of N protein, thereby indicating that the mechanism of recognition of PS-containing RNA by M protein does not require the formation of RNP complex with N protein. Furthermore, remarkably, N protein was not necessary for RNA packaging. A specific interaction of a viral envelope protein with a viral RNA element, like the PS, that occurs independently of a nucleocapsid protein with subsequent specific RNA packaging into VLP or virus particles, also in the absence of a nucleocapsid protein, has not been described for any RNA virus. Furthermore, MHV M protein is the first example of a viral transmembrane protein that binds to a specific viral RNA element in the absence of any other viral structural proteins. Our data were consistent with the observation that MHV M protein cosediments with MHV genomic RNA, but not MHV N protein, in Renografin density gradient centrifugation of NP-40-solubilized MHV particles (33). Currently, it is not clear whether M protein directly interacted with the PS. If M protein directly binds to PS, then MHV M protein may be the first example of a transmembrane protein that binds to a specific RNA element; M-PS binding represents a novel type of macromolecular interaction with a clear biological significance. The nature of binding of M protein to PS deserves further study.
Bos et al. (3) reported the production of infectious MHV defective-interfering (DI) particles in vTF7-3-infected cells that were transfected with five different plasmids expressing the synthetic MHV DI RNA containing the PS and all four (M, N, S, and E) MHV structural proteins (3). In that study, culture fluid was collected from expressing cells, and a mixture of the culture fluid and MHV was used to infect MHV-susceptible cells. Following overnight incubation, supernatant was collected for subsequent passages. After several undiluted passages of the supernatant, accumulation of DI RNA was demonstrated, implying the packaging of expressed DI RNA into VLPs in the coexpressing cells. The present data were consistent with the observation by Bos et al. that MHV nonstructural proteins are not necessary for RNA packaging into VLPs. However, neither the specificity and selectivity of DI RNA packaging nor the role of N protein in DI RNA packaging was examined in the study reported by Bos et al. (3).
Using the Semliki Forest virus (SFV) expression system, others have shown that infectious enveloped particles or vesicles containing the vesicular stomatitis virus envelope G glycoprotein and vector RNA can be produced after expression of this protein (29), but in that system, the vector RNA is randomly incorporated into the vesicles; there is no selectivity in RNA packaging. With use of the SFV expression system, the random packaging of SFV-derived mRNAs into SFV-encoded murine leukemia virus Gag VLPs was also reported. The SFV-derived mRNAs compensated for the absence of retroviral mRNAs in the VLPs (26). These mRNAs were packaged despite the lack of any retroviral PS sequences. In a sharp contrast, our study demonstrated an absolutely specific and selective nucleocapsid-independent packaging of the PS-containing RNA into VLPs.
Based on this study and other studies, we propose a model to elucidate the mechanism of RNA packaging in MHV. Since MHV N protein binds to all MHV mRNAs in infected cells (2, 27), probably N protein binds to the intracellular form of genomic RNA, mRNA 1, during nascent mRNA 1 synthesis or as soon as mRNA 1 is synthesized on intracellular membranes (4, 11). M protein, which accumulates and probably oligomerizes in the intermediate compartment between the endoplasmic reticulum and the Golgi complex (16, 20, 35), binds to the PS present in mRNA 1; this binding determines the selective genomic RNA packaging and excludes the packaging of MHV subgenomic mRNAs lacking the PS. After the binding of M protein to the PS, N protein that is associated with mRNA 1 interacts with the oligomerized M protein (6, 27). Subsequently, the M protein-mRNA 1 RNP complex undergoes virion morphogenesis in concert with E protein.
These data provoke a question about the biological role of N protein in MHV. As shown here, N protein appears to be dispensable for MHV RNA packaging. M-N interaction, however, might compensate for defects in viral envelope formation due to mutation in M protein (17). N protein may play a crucial role early in infection; for example, after virus uncoating, one of the functions of N protein may be to deliver the viral RNP complex to the appropriate compartment for initiation of viral replication.
Acknowledgments
We thank Fumihiro Taguchi and Yasuko Yamada for a plasmid encoding the MHV-2 S gene. We also thank Julian Leibowitz for anti-E protein antibody.
This work was supported by Public Health Service grant AI29984 from the National Institutes of Health to S.M. K.N. was supported by a McLaughlin fellowship.
REFERENCES
- 1.Armstrong, J., H. Niemann, S. Smeekens, P. Rottier, and G. Warren. 1984. Sequence and topology of a model intracellular membrane protein, E1 glycoprotein, from a coronavirus. Nature 308:751-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baric, R. S., G. W. Nelson, J. O. Fleming, R. J. Deans, J. G. Keck, N. Casteel, and S. A. Stohlman. 1988. Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J. Virol. 62:4280-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bos, E. C., W. Luytjes, H. V. van der Meulen, H. K. Koerten, and W. J. Spaan. 1996. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology 218:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bost, A. G., E. Prentice, and M. R. Denison. 2001. Mouse hepatitis virus replicase protein complexes are translocated to sites of M protein accumulation in the ERGIC at late times of infection. Virology 285:21-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bredenbeek, P. J., I. Frolov, C. M. Rice, and S. Schlesinger. 1993. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J. Virol. 67:6439-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Escors, D., J. Ortego, H. Laude, and L. Enjuanes. 2001. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J. Virol. 75:1312-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischer, F., C. F. Stegen, P. S. Masters, and W. A. Samsonoff. 1998. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 72:7885-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fleming, J. O., R. A. Shubin, M. A. Sussman, N. Casteel, and S. A. Stohlman. 1989. Monoclonal antibodies to the matrix (E1) glycoprotein of mouse hepatitis virus protect mice from encephalitis. Virology 168:162-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fosmire, J. A., K. Hwang, and S. Makino. 1992. Identification and characterization of a coronavirus packaging signal. J. Virol. 66:3522-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirano, N., K. Fujiwara, S. Hino, and M. Matumoto. 1974. Replication and plaque formation of mouse hepatitis virus (MHV-2) in mouse cell line DBT culture. Arch. Gesamte Virusforsch. 44:298-302. [DOI] [PubMed] [Google Scholar]
- 13.Holmes, K. V., E. W. Doller, and L. S. Sturman. 1981. Tunicamycin resistant glycosylation of coronavirus glycoprotein: demonstration of a novel type of viral glycoprotein. Virology 115:334-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joo, M., S. Banerjee, and S. Makino. 1996. Replication of murine coronavirus defective interfering RNA from negative-strand transcripts. J. Virol. 70:5769-5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, K. H., K. Narayanan, and S. Makino. 1997. Assembled coronavirus from complementation of two defective interfering RNAs. J. Virol. 71:3922-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klumperman, J., J. K. Locker, A. Meijer, M. C. Horzinek, H. J. Geuze, and P. J. Rottier. 1994. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 68:6523-6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuo, L., and P. S. Masters. 2002. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J. Virol. 76:4987-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai, M. M., R. S. Baric, P. R. Brayton, and S. A. Stohlman. 1984. Characterization of leader RNA sequences on the virion and mRNAs of mouse hepatitis virus, a cytoplasmic RNA virus. Proc. Natl. Acad. Sci. USA 81:3626-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai, M. M., and S. A. Stohlman. 1978. RNA of mouse hepatitis virus. J. Virol. 26:236-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Locker, J. K., D. J. Opstelten, M. Ericsson, M. C. Horzinek, and P. J. Rottier. 1995. Oligomerization of a trans-Golgi/trans-Golgi network retained protein occurs in the Golgi complex and may be part of its retention. J. Biol. Chem. 270:8815-8821. [DOI] [PubMed] [Google Scholar]
- 21.Lopez, S., J. S. Yao, R. J. Kuhn, E. G. Strauss, and J. H. Strauss. 1994. Nucleocapsid-glycoprotein interactions required for assembly of alphaviruses. J. Virol. 68:1316-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maeda, J., A. Maeda, and S. Makino. 1999. Release of coronavirus E protein in membrane vesicles from virus-infected cells and E protein-expressing cells. Virology 263:265-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makino, S., M. Joo, and J. K. Makino. 1991. A system for study of coronavirus mRNA synthesis: a regulated, expressed subgenomic defective interfering RNA results from intergenic site insertion. J. Virol. 65:6031-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makino, S., C. K. Shieh, J. G. Keck, and M. M. Lai. 1988. Defective-interfering particles of murine coronavirus: mechanism of synthesis of defective viral RNAs. Virology 163:104-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makino, S., F. Taguchi, N. Hirano, and K. Fujiwara. 1984. Analysis of genomic and intracellular viral RNAs of small plaque mutants of mouse hepatitis virus, JHM strain. Virology 139:138-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 98:5246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narayanan, K., A. Maeda, J. Maeda, and S. Makino. 2000. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 74:8127-8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narayanan, K., and S. Makino. 2001. Cooperation of an RNA packaging signal and a viral envelope protein in coronavirus RNA packaging. J. Virol. 75:9059-9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rolls, M. M., P. Webster, N. H. Balba, and J. K. Rose. 1994. Novel infectious particles generated by expression of the vesicular stomatitis virus glycoprotein from a self-replicating RNA. Cell 79:497-506. [DOI] [PubMed] [Google Scholar]
- 30.Rottier, P. J., M. C. Horzinek, and B. A. van der Zeijst. 1981. Viral protein synthesis in mouse hepatitis virus strain A59-infected cells: effect of tunicamycin. J. Virol. 40:350-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simons, K., and H. Garoff. 1980. The budding mechanisms of enveloped animal viruses. J. Gen. Virol. 50:1-21. [DOI] [PubMed] [Google Scholar]
- 32.Spaan, W., H. Delius, M. Skinner, J. Armstrong, P. Rottier, S. Smeekens, B. A. van der Zeijst, and S. G. Siddell. 1983. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 2:1839-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sturman, L. S., K. V. Holmes, and J. Behnke. 1980. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J. Virol. 33:449-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suomalainen, M., P. Liljestrom, and H. Garoff. 1992. Spike protein-nucleocapsid interactions drive the budding of alphaviruses. J. Virol. 66:4737-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tooze, J., S. Tooze, and G. Warren. 1984. Replication of coronavirus MHV-A59 in sac− cells: determination of the first site of budding of progeny virions. Eur. J. Cell Biol. 33:281-293. [PubMed] [Google Scholar]
- 36.van der Most, R. G., P. J. Bredenbeek, and W. J. Spaan. 1991. A domain at the 3′ end of the polymerase gene is essential for encapsidation of coronavirus defective interfering RNAs. J. Virol. 65:3219-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vennema, H., G. J. Godeke, J. W. Rossen, W. F. Voorhout, M. C. Horzinek, D. J. Opstelten, and P. J. Rottier. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 15:2020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woo, K., M. Joo, K. Narayanan, K. H. Kim, and S. Makino. 1997. Murine coronavirus packaging signal confers packaging to nonviral RNA. J. Virol. 71:824-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamada, Y. K., K. Takimoto, M. Yabe, and F. Taguchi. 1997. Acquired fusion activity of a murine coronavirus MHV-2 variant with mutations in the proteolytic cleavage site and the signal sequence of the S protein. Virology 227:215-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu, X., W. Bi, S. R. Weiss, and J. L. Leibowitz. 1994. Mouse hepatitis virus gene 5b protein is a new virion envelope protein. Virology 202:1018-1023. [DOI] [PubMed] [Google Scholar]
- 41.Zhao, H., and H. Garoff. 1992. Role of cell surface spikes in alphavirus budding. J. Virol. 66:7089-7095. [DOI] [PMC free article] [PubMed] [Google Scholar]