Abstract
Studies on the replication of hepatitis C virus (HCV) have been facilitated by the development of selectable subgenomic replicons replicating in the human hepatoma cell line Huh-7 at a surprisingly high level. Analysis of the replicon population in selected cells revealed the occurrence of cell culture-adaptive mutations that enhance RNA replication substantially. To gain a better understanding of HCV cell culture adaptation, we characterized conserved mutations identified by sequence analysis of 26 independent replicon cell clones for their effect on RNA replication. Mutations enhancing replication were found in nearly every nonstructural (NS) protein, and they could be subdivided into at least two groups by their effect on replication efficiency and cooperativity: (i) mutations in NS3 with a low impact on replication but that enhanced replication cooperatively when combined with highly adaptive mutations and (ii) mutations in NS4B, -5A, and -5B, causing a strong increase in replication but being incompatible with each other. In addition to adaptive mutations, we found that the host cell plays an equally important role for efficient RNA replication. We tested several passages of the same Huh-7 cell line and found up to 100-fold differences in their ability to support replicon amplification. These differences were not due to variations in internal ribosome entry site-dependent translation or RNA degradation. In a search for cellular factor(s) that might be responsible for the different levels of permissiveness of Huh-7 cells, we found that replication efficiency decreased with increasing amounts of transfected replicon RNA, indicating that viral RNA or proteins are cytopathic or that host cell factors in Huh-7 cells limit RNA amplification. In summary, these data show that the efficiency of HCV replication in cell culture is determined both by adaptation of the viral sequence and by the host cell itself.
The Hepatitis C virus (HCV) is a leading cause of chronic liver disease (37). Most infections are inapparent or initially associated with mild symptoms, but the virus persists in ∼80% of all patients, leading to a high risk of developing severe liver damage such as liver cirrhosis or hepatocellular carcinoma.
HCV is an enveloped positive-strand RNA virus belonging to the genus Hepacivirus in the family Flaviviridae (33). The genome of HCV encompasses a single ∼9,600-nucleotide (nt) RNA molecule carrying one large open reading frame that is flanked by nontranslated regions (NTRs). The 5′ NTR contains an internal ribosome entry site (IRES) that directs translation of the open reading frame (43, 45). In addition, the 5′ NTR is required for RNA replication as is the case with the 3′ NTR (11, 12, 27, 47). All HCV proteins are generated from a polyprotein precursor that is cleaved by cellular and viral proteases into at least 10 different products (for reviews, see references 2 and 38). The structural proteins core, E1, and E2 are located in the amino terminus of the polyprotein (19), followed by p7, a hydrophobic peptide with unknown function, and the nonstructural (NS) proteins NS2, NS3, NS4A, NS4B, NS5A, and NS5B. NS2 and the amino terminus of NS3 comprise the NS2-3 protease responsible for cleavage between NS2 and NS3 (16, 20). NS3 is a multifunctional protein, consisting of an amino-terminal protease domain required for processing of the NS3 to NS5B region (1, 17) and a carboxy-terminal helicase/nucleoside triphosphatase domain (24, 40). NS4A is a cofactor that activates the NS3 protease function by forming a heterodimer (4, 10, 29, 41). The hydrophobic protein NS4B induces the formation of a cytoplasmic vesicular structure, designated membranous web that appears to be the replication complex of HCV (9, 22). NS5A is a phosphoprotein that seems to play an important role in viral replication, since most of the cell culture-adaptive mutations described so far are located within the central region of NS5A (7, 18, 28). It is also supposed to be involved in the resistance of HCV to the antiviral action of alpha interferon (IFN-α) by interaction with the cellular protein kinase R (PKR) via a region including the so-called IFN sensitivity-determining region (14, 15). NS5B is the RNA-dependent RNA polymerase of HCV (6, 30). The NS proteins NS3 to NS5B are necessary and sufficient for HCV replication. They build up a multiprotein complex that, in analogy to other positive-strand RNA viruses, is associated with intracellular membranes (9, 36).
Studies on the replication of HCV have long been hampered by the lack of efficient cell culture systems. Many attempts have been made to identify cell lines that allow efficient infection with HCV and virus production, but all of these systems suffer from low virus yield and poor reproducibility (reviewed in reference 3). We have recently developed a cell culture system that is based on subgenomic selectable replicons consisting of the HCV 5′ NTR directing translation of the neomycin phosphotransferase gene, the IRES of the Encephalomyocarditis virus (EMCV), the HCV NS proteins NS3 to NS5B and the HCV 3′ NTR. Upon transfection of these RNAs into Huh-7 cells and selection with G418, cell clones have been generated that carry persistently replicating HCV replicons (32, 36). Analysis of the viral sequences in these cell clones revealed conserved mutations in different NS proteins. Introduction of these mutations into the original HCV replicon that was derived from the Con-1 sequence (26) significantly increased the replication efficiency of these RNAs (7, 18, 28, 31). Based on these cell culture-adaptive mutations, transient replication assays, as well as efficiently replicating HCV full-length genomes and genomic replicons, could be established (7, 23, 28, 35). However, little is known about the mechanism underlying cell culture adaptation and the role of host cells factors required for HCV replication. The latter is particularly important because Huh-7 hepatoma cells still are the only cell line supporting efficient replication of HCV RNAs.
In the present study we have extended our analysis of cell culture adaptation by using a panel of 26 independent replicon cell clones. We identified a large number of mutations that could be grouped into at least two categories with respect to their cooperativity or incompatibility for RNA replication. We also observed an important role of the host cell for efficient HCV replication and provide evidence that cellular factors or host cell conditions limit replication of HCV RNAs in Huh-7 cells.
MATERIALS AND METHODS
Cell cultures.
Cell monolayers of the human hepatoma cell line Huh-7 (34) were routinely grown in Dulbecco’s modified Eagle medium (DMEM) (Invitrogen, Karlsruhe, Germany) supplemented with 2 mM l-glutamine, nonessential amino acids, 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 10% fetal calf serum. G418 (Geneticin; Invitrogen) was added at a final concentration of 500 to 1,000 μg/ml in the case of cell clones carrying HCV replicons. Passage numbers given for Huh-7 cells are relative and refer to the number of passages in our laboratory. Unless otherwise stated, Huh-7 cells with passage numbers of >90 were used for transient replication assays.
Plasmid constructions.
All numbers given in parentheses refer to the position of the corresponding amino acid or nucleotide of a complete HCV genome cloned by our group (HCV Con-1; EMBL database accession number AJ238799). Construction of the vector pFK and the parental replicon construct pFK-I389neo/NS3-3′/wt (EMBL database accession number AJ242654) (32), as well as pFK-5.1, pFK-E1202G, and pFK-E1202G+T1280I (28), have already been described. The basic construct for transient-replication assays, pFK-rep PI-luc/5.1, originally designated pFK-nt341-sp-PI-lucEI3420-9605/5.1, containing the complete HCV 5′ NTR, a 63-bp spacer element, the poliovirus (PV) IRES element, the firefly luciferase (Photinus vulgaris) gene, the EMCV IRES, the HCV NS proteins NS3 to NS5B, and the HCV 3′ NTR, has been described recently (12).
Standard recombinant DNA technologies were used for all plasmid constructions (39). Plasmids pFK-rep luc/wt, pFK-rep luc/E1202G, and pFK-rep luc/E1202G+T1280I were generated by transferring SfiI fragments (nt 3622 to 8499) from the parental replicons that carried the mutations or the wild-type sequence (see above) into pFK-rep PI-luc/5.1. Mutations T1261S, M1268V, T1287A, G1304S, K1846T, V1897A, V1897L, V1897 M, Ins2041K, D2177G, A2199D, del2202S, S2204R, S2204I, S2197P, Q2933R, I3004T, 2202delS+S2204R, D2177G+S2204R, and S2197P+S2204R were introduced into pFK-rep PI-luc/wt by site-directed mutagenesis (21) by using oligonucleotide primers carrying the necessary nucleotide alterations. Plasmids pFK-rep PI-luc/R2884G and pFK-rep PI-luc/S2197P+R2884G were generated by insertion of an XhoI-SpeI fragment (nt 7186 to 9605) isolated from pFK5B/R2884G (31) into pFK-rep PI-luc/wt or pFK-rep PI-luc/5.1, respectively. Combinations of mutations were achieved by insertion of restriction fragments carrying individual mutations into one plasmid by using the restriction sites NotI (present upstream of the EMCV IRES), BstXI (nt 4319), MluI (nt 6529), XhoI (nt 7186), BssHII (nt 5923), and SfiI (nt 3622 and 8499). pFK-rep neo-Δ GDD was obtained by insertion of a SfiI fragment from pFK-rep PI-luc/ET, carrying the adaptive mutations into pFKI389neo/NS3-3′/Δ (32). pFK-rep neo-stop was derived from pFK-rep neo-Δ GDD by introducing a stop codon 70 amino acids downstream of the NS3 initiation codon. The vector pTM was used to generate in vitro transcripts of different HCV genes under translational control of the EMCV IRES (13). The plasmid pTM luc was generated by transferring a PCR fragment derived from the vector pGL-2 (Promega, Mannheim, Germany) and spanning the complete luciferase gene of the firefly into a modified pTM1 vector (4) via a newly generated NcoI site at the 5′ and an SpeI site at the 3′ end of the gene. Plasmid pTM lacZ was obtained by using a newly generated NcoI restriction site encompassing the start codon of the lacZ gene and a BamHI site downstream of the gene to transfer the lacZ gene from pCH 110 (Amersham-Pharmacia, Freiburg, Germany) into pTM1. Plasmid pTM 3-5Bcon is described elsewhere (pTM3-5BC [26]); pTM 3-5B ET was generated by transferring a SfiI fragment from rep PI-luc/ET containing all adaptive mutations into pTM 3-5Bcon. pBSK lacZ was used to generate capped in vitro transcripts covering the lacZ gene. A HindIII/BamHI fragment harboring the gene encoding β-galactosidase was obtained from the vector pCH110 (Amersham-Pharmacia) and transferred into pBluescript (Stratagene, Amsterdam, The Netherlands).
Preparation of total RNA and quantification of HCV RNA by Northern hybridization.
The methods for RNA preparation and quantification have been described recently (31). In brief, total RNA was prepared by a single-step isolation method (8), denatured by treatment with 5.9% glyoxal in 50% dimethyl sulfoxide and 10 mM sodium phosphate buffer (pH 7.0), and analyzed after denaturing agarose gel electrophoresis by Northern hybridization. Prior to hybridization the membrane was stained with methylene blue, cut ∼1 cm below the 28S rRNA band, and the upper strip containing the HCV replicon RNA was hybridized with a 32P-labeled negative-sense riboprobe complementary to the 3′ end of the NS5B region and part of the 3′ NTR (nt 8362 to 9408). The lower strip that was hybridized with a β-actin-specific antisense riboprobe was used to correct for total RNA amounts loaded in each lane of the gel. Specific bands were quantitated by phosphorimaging with a BAS 2500 scanner (Fuji), and the number of replicon molecules was determined by comparison with a serial dilution of in vitro transcripts loaded in parallel onto the gel.
Amplification of replicon RNA by RT-PCR and cloning of amplified DNA fragments.
Amplification of replicon RNAs was done by long-distance reverse transcription-PCR (RT-PCR) as described recently (28, 31) by using Expand RT and the Expand Long Template PCR system (Roche Biochemicals). PCR products were purified by preparative agarose gel electrophoresis and cloned into a pCR-Topo vector according to the instructions of the manufacturer (Invitrogen).
Sequence analysis.
Sequences were verified by using a Thermo Sequenase fluorescent-labeled primer cycle sequencing kit with 7-deaza-dGTP (Amersham-Pharmacia Biotech) and IRD-41-labeled primers (MWG-Biotech, Ebersberg, Germany) according to the instructions of the manufacturer. Reactions were analyzed on a Licor DNA sequencer 4000 (MWG-Biotech).
In vitro transcription.
In vitro transcripts were generated by using the protocol described recently (31). In brief, plasmid DNA was restricted with AseI and ScaI (New England Biolabs, Bad Schwalbach/Taunus, Germany), extracted with phenol and chloroform, precipitated with ethanol, and dissolved in RNase-free water. In vitro transcription reaction mixtures contained 80 mM HEPES (pH 7.5), 12 mM MgCl2, 2 mM spermidine, 40 mM dithiothreitol (DTT), 3.125 mM concentrations of each nucleoside triphosphate, 1 U of RNasin (Promega)/μl, 0.1 μg of restricted plasmid DNA/μl, and 0.6 U of T7 RNA polymerase (Promega)/μl. After 2 h at 37°C, an additional 0.3 U of T7 RNA polymerase/μl was added, and the reaction was incubated for another 2 h. Transcription was terminated by the addition of 1.2 U of RNase-free DNase (Promega) per μg of plasmid DNA and a 30-min incubation at 37°C. After extraction with acidic phenol and chloroform, DNA was precipitated with isopropanol and dissolved in RNase-free water. The concentration was determined by measurement of the optical density at 260 nm, and the RNA integrity was checked by denaturing agarose gel electrophoresis. Capped RNAs were transcribed by using the protocol described above except that nucleotide concentrations were 1 mM for ATP, CTP, and UTP and 0.5 mM for GTP and that the cap analog m7G(5′)ppp(5′)G (New England Biolabs, Frankfurt, Germany) was included at a final concentration of 1 mM.
Electroporation and transient HCV replication assay.
For electroporation, single-cell suspensions of Huh-7 cells were prepared by trypsinization of monolayers, detaching the cells from the culture dish by rinsing with complete DMEM, one wash with phosphate-buffered saline, counting, and resuspension at 107 cells per ml in Cytomix (44) containing 2 mM ATP and 5 mM glutathione. Unless otherwise stated, 5 μg of in vitro-transcribed RNA was mixed with 400 μl of the cell suspension by pipetting, electroporated, and immediately transferred to 8 ml of complete DMEM. Electroporation conditions were 960 μF and 270 V by using a Gene Pulser system (Bio-Rad, Munich, Germany) and a cuvette with a gap width of 0.4 cm (Bio-Rad). Cells were seeded in six-well plates and harvested 4, 24, 48, and 72 h after electroporation, unless otherwise stated. For assaying the luciferase activity, cells were washed twice with phosphate-buffered saline and scraped off the plate into 350 μl of ice-cold lysis buffer (1% Triton X-100, 25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA, 1 mM DTT). Then, 100 μl of cleared lysate was mixed with 360 μl of assay buffer (25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA, 1 mM DTT, 2 mM ATP, 15 mM K2PO4; pH 7.8) and, after the addition of 200 μl of a 200 μM luciferin solution, was measured in a luminometer (Lumat LB9507; Berthold, Freiburg, Germany) for 20 s. Values obtained with cells harvested 4 h after electroporation were used to normalize the data for transfection efficiency. All luciferase assays were done in duplicate measurements. Since the absolute number of relative light units (RLU) differed between individual experiments due to fluctuations of transfection efficiency, luciferase activity was expressed as the percent RLU. These values represent the percentage of luciferase activity determined at a given time point relative to the one measured 4 h after transfection. Unless otherwise stated, the RLU values determined 48 h after transfection are given.
Generation of G418-resistant replicon cell clones and removal of the replicon by IFN-α treatment.
Selection of G418-resistant replicon cell clones was performed with 1 mg of G418/ml as described elsewhere (31, 32). To cure cell clones from HCV replicons, cells were treated with 1,000 U of IFN-α2 (Roche, Basel, Switzerland)/ml in the absence of G418 and kept at high confluency to decrease HCV replication further (36). After 2 weeks, the absence of HCV RNA was determined by Northern hybridization, and cells were passaged at least two times in the absence of IFN prior to transfection with luciferase reporter replicons.
Determination of β-galactosidase activity.
Cells transfected with in vitro transcripts encoding the lacZ gene were lysed by using luciferase lysis buffer to measure β-galactosidase activity. Briefly, 30 μl of cell extract was incubatedfor 16 h in the presence of o-nitrophenyl-β-d-galactopyranoside (Sigma, Taufkirchen, Germany) under conditions described elsewhere (39). β-Galactosidase activity was determined by measurement of the optical density at 420 nm.
RESULTS
Identification of conserved amino acid substitutions in 26 independent replicon cell clones.
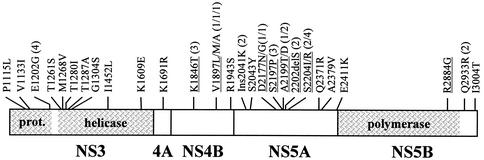
Upon transfection of our original selectable subgenomic replicons into Huh-7 cells and selection with G418, we found that only a small number of colonies formed that supported persistent replication of the replicons (32). Subsequent studies showed that each of them had acquired cell culture-adaptive mutations that enhanced replication to various extents (28, 31). Every cell clone contained at least one mutation that was conserved in the whole replicon population, and this mutation seemed to be the prerequisite for the survival of the corresponding cell clone since it enhanced replication to a level sufficient to confer G418 resistance. In order to get a deeper understanding of the mechanisms underlying adaptation to Huh-7 cells, we amplified replicons from 26 independent cell clones that were selected after transfection of our parental replicon (32). From each cell clone, two amplicons covering the coding sequence of the HCV NS proteins were cloned and sequenced. Amino acid substitutions present in both amplicons were assumed to be conserved in the replicon population of this particular cell clone. Each amplicon contained 3 to 16 amino acid substitutions (11 on the average), of which one to four were conserved, depending on the particular cell clone (Table 1 and Fig. 1). Conserved mutations were found in every NS protein and arranged in several clusters. One cluster was located at the amino terminus of the NS3 helicase domain, and one was in the center of NS5A in a region presumably involved in hyperphosphorylation (42). Mutations at two positions in NS4B (K1846 and V1897), five in NS5A (D2177, S2197, A2199, 2202delS, and S2204) and one in NS5B (R2884) were each found as single conserved mutations in a particular cell clone. Interestingly, most of these alterations either affect serine residues in NS5A that are potential phosphorylation sites (S2197, S2204, and 2202delS) or a charged residue (K1846, D2177, and R2884). In the case of A2199, a hydrophobic residue was changed either to a potential phosphorylation site (A2199T) or to a negatively charged residue (A2199D). Only the exchange of valine at position 1897 for leucine, alanine, or methionine was conservative. The fact that these alterations were present as the only conserved substitution in the replicon population of a given cell clone suggested that they substantially contribute to RNA replication. In contrast, mutations in NS3 and in NS4A, as well as some amino acid substitutions in NS4B, NS5A, and NS5B, were only found in conjunction with other conserved mutations, indicating that they had a minor effect on HCV replication.
TABLE 1.
Conserved mutations in replicons isolated from 26 cell clones
| Cell clone | Conserved mutation(s) | NS protein(s) affected |
|---|---|---|
| 5-1a | K1846T | 4B |
| 5-5a | E1202G, K1846T, Q2933R | 3, 4B, 5B |
| 5-9a | M1268V, ins2041K, I3004T | 3, 5A, 5B |
| 5-15a,b | S2197P | 5A |
| 5-15-9-2-3b | E1202G, T1280I, S2197P | 3, 3, 5A |
| 9-12a | 2202delS | 5A |
| 9-13a,c | R2884G | 5B |
| 11-7a | T1261S, K1846T | 3, 4B |
| 261198/1-1 | G1304S, S2204I | 3, 5A |
| 261198/1-2 | S2204I | 5A |
| 261198/1-3 | V1897L | 4B |
| 261198/3-1 | S2204R | 5A |
| 261198/9-2 | S2204R | 5A |
| 261198/9-4 | K1609E, V1897M | 3, 4B |
| 040199/5 | V1133I, 2202delS, Q2371R | 3, 5A, 5A |
| 040199/6 | ins2041K, A2379V | 5A, 5A |
| 040199/14-2 | S2204R | 5A |
| 040199/14-3 | S2204R | 5A |
| 040199/14-5 | D2177N | 5A |
| 111199/51-1 | R1943S, A2199D | 4B, 5A |
| 111199/52-1 | E1202G, V1897A, Q2933R | 3, 4B, 5B |
| 111199/52-2 | K1691R, S2043Y | 4A, 5A |
| 020999/1-1 | E1202G, I1452L, D2177G, E2411K | 3, 3, 5A, 5A |
| 020999/1-2 | A2199T | 5A |
| 020999/1-3 | T1287A, A2199D | 3, 5A |
| 020999/21-1 | P1115L, S2197P | 3, 5A |
FIG. 1.
Conserved mutations in replicons isolated from 26 independently selected replicon cell clones. A schematic drawing of the HCV NS proteins as present in the subgenomic replicons is shown. Regions encoding known enzymatic functions are shaded. Mutations conserved in the replicon population of single cell clones are given above. Numbers refer to the amino acid position in the complete polyprotein of the HCV Con-1 isolate. The frequency with which a given mutation was found in these cell clones is indicated in parentheses. Mutations identified as the only conserved alteration in at least one cell clone are underlined. A summary of these data is given in Table 1. Ins, insertion; del, deletion; prot., proteinase.
Improvement of the luciferase-based HCV transient replication assay by using the PV IRES.
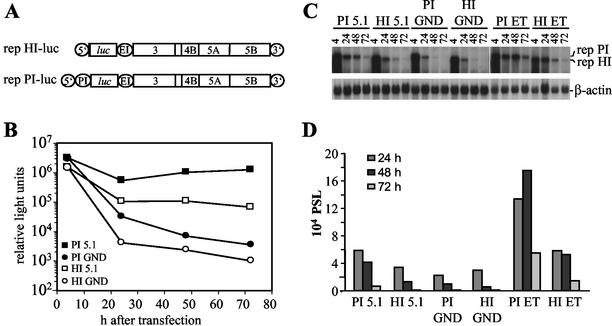
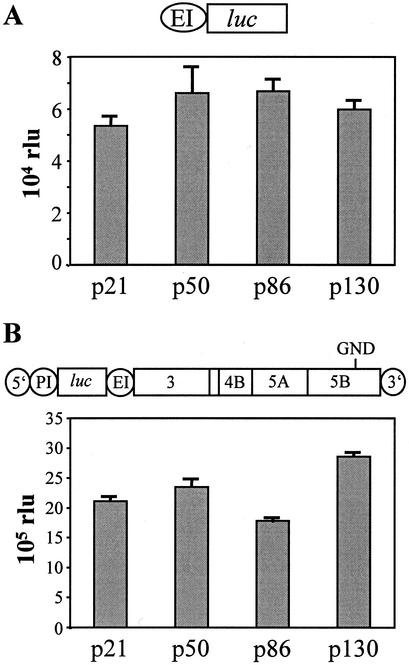
In order to analyze the effect of defined mutations on HCV RNA replication, we used a transient replication assay based on the expression of firefly luciferase (28). Since luciferase activities were consistently higher when translation of this reporter was directed by the PV IRES instead of the HCV IRES (12), we first compared two different replicons carrying the authentic HCV 5′ NTR or an HCV PV chimeric one (Fig. 2A) with respect to their impact on assay sensitivity and replication efficiency. Figure 2B shows the result of a representative experiment in which a comparison of these two replicons, each carrying three cell culture-adaptive mutations, and the corresponding negative controls carrying an inactivating mutation in the GDD motif of the NS5B RdRp was performed. At 4 h after electroporation luciferase activity was ∼3-fold higher in the case of the replicons carrying the PV IRES compared to the ones with the HCV IRES, which most likely was due to a higher translational activity of the PV IRES. More interestingly, at 72 h after transfection, luciferase activity obtained with the replicon carrying the PV IRES was ca. 500-fold above the background level as determined with the negative control, but only 100-fold greater in case of the replicon with the HCV IRES. The analogous result was obtained when we directly analyzed the RNA levels after transfection by a Northern hybridization, demonstrating that luciferase activities correlate well with RNA replication (Fig. 2C and D). Moreover, we compared the impact of several adaptive mutations on replication of replicons carrying only the HCV IRES at the 5′ end or the HCV-PV chimeric one (see below). By using luciferase assays and Northern hybridizations in parallel, we found in all cases that the luciferase activity accurately reflected RNA replication and that the PV IRES constructs yielded higher RNA replication and more reproducible results (data not shown). Therefore, we used replicons with the PV IRES throughout the present study. Since HCV replication and host cell growth are linked (36) and cells often reached confluency 72 h after transfection, HCV replication was determined 48 h after transfection in all subsequent analyses unless otherwise indicated.
FIG. 2.
Transient replication of replicons with different IRES elements at the 5′ end. (A) Structure of the replicons rep HI-luc and rep PI-luc used for transient replication assays. 5′, HCV 5′ NTR; 3′, HCV 3′ NTR; PI, PV IRES; luc, firefly luciferase; EI, EMCV IRES. (B) Transient replication of the replicons carrying the luciferase reporter gene under translational control of the HCV IRES or the PV IRES. Huh-7 cells were transfected with 5 μg of rep PI-luc/5.1, rep PI-luc/GND, rep HI-luc/5.1, or rep HI-luc/GND, and luciferase activity was determined in cell lysates that were prepared at given time points posttransfection. Data were normalized for transfection efficiency among constructs with identical IRES elements as determined by measurement of the luciferase activity 4 h after transfection. Electroporations and luciferase assays were performed in duplicates. Note the logarithmic scale of the ordinate. (C) Analysis of transient replication of different luciferase replicons by Northern hybridization. Huh-7 cells were transfected with 5 μg of luciferase replicons 5.1 that carry two cell culture-adaptive mutations in NS3 and one in NS5A (28) or with the ET replicons carrying the same NS3 mutations and one in NS4B instead of NS5A (Table 2). Cells were harvested at different time points postelectroporation (pE) as indicated above each lane. Then, 10 μg of total RNA was analyzed by Northern hybridization with a 32P-labeled negative-sense riboprobe spanning the NS3 to NS5B region. The positions of replicon RNAs with PV IRES (rep PI) or HCV IRES (rep HI) and β-actin mRNA are given on the right. (D) Quantification of the Northern hybridization shown in panel C by phosphorimaging. Values were corrected for different RNA amounts by using the β-actin signal and normalized for the PSL obtained at 4 h after transfection.
Enhancement of transient HCV replication by single adaptive mutations.
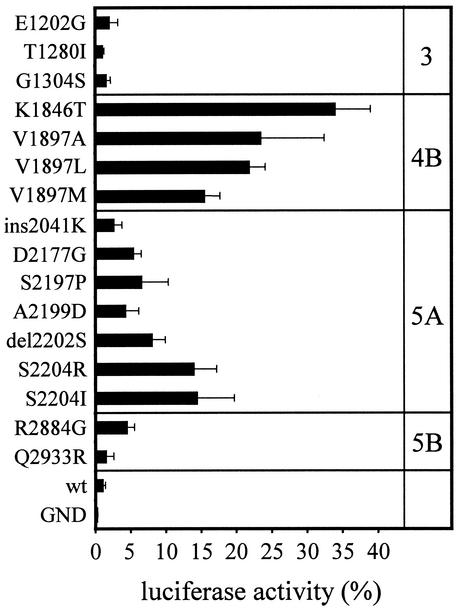
Based on the assumption that conserved mutations found with high frequency would have the greatest impact on replication, these mutations, as well as some NS3 substitutions, were introduced independently into the luciferase reporter replicon with the PV IRES (Fig. 2A) and analyzed for RNA replication (Fig. 3 and Table 2). We found that mutations in NS4B were most efficient, with K1846T enhancing replication ∼30-fold and with V1897A or V1897L enhancing replication ∼20-fold compared to the wild type. Among the conserved mutations in NS5A, S2204R and S2204I caused the strongest increase (∼13-fold), whereas the insertion of lysine at position 2041 had the least effect. The conserved mutations in NS3 and Q2933R in NS5B had only a a minor effect on replication, although the latter was found in two and E1202G was in four independent cell clones. However, these mutations were always associated with other adaptive mutations (Table 1). To differentiate these mutations from the mutations in NS4B, NS5A, and NS5B that strongly increased replication efficiency, we termed all mutations resulting in a >4-fold increase in RNA replication compared to the wild-type replicon as “highly” adaptive (Table 2).
FIG. 3.
Effect of single amino acid substitutions in the HCV coding sequence on transient replication. Values represent the ratio of RLU measured at 48 h and at 4 h after electroporation. The 4-h value was set at 100%. Data are means and the standard deviations of at least three independent experiments. For all experiments, Huh-7 cells with passage numbers of >90 were used.
TABLE 2.
Transient replication efficiency of the parental and mutated replicons
| Mutation(s) | Luciferase activity (%)a | Designation |
|---|---|---|
| None | 1.1 ± 0.3 | rep PI-luc/wt |
| D2737N | 0.2 ± 0.1 | rep PI-luc/GND |
| E1202G | 2.1 ± 1.1 | |
| T1280Ib | 1.1 ± 0.2 | |
| G1304S | 1.6 ± 0.6 | |
| K1846T | 34.0 ± 4.9 | |
| V1897A | 23.5 ± 9.0 | |
| V1897L | 21.8 ± 2.2 | |
| V1897M | 15.5 ± 2.2 | |
| ins2041K | 2.7 ± 1.1 | |
| D2177G | 5.4 ± 1.0 | |
| S2197P | 6.6 ± 3.8 | |
| A2199D | 4.3 ± 1.8 | |
| del2202S | 8.0 ± 1.8 | |
| S2204R | 14.0 ± 3.1 | |
| S2204I | 14.5 ± 5.2 | |
| R2884G | 4.5 ± 1.0 | |
| Q2933R | 1.6 ± 1.0 | |
| I3004T | 1.4 ± 0.6 | |
| E1202G+T1280I | 3.5 ± 0.7 | |
| E1202G+T1280I+K1846T | 210.9 ± 74.6 | rep PI-luc/ET |
| E1202G+T1280I+V1897A | 84.6 ± 36.9 | |
| E1202G+T1280I+S2197P | 36.5 ± 17.2 | rep PI-luc/5.1c |
| E1202G+T1280I+G1304S+ S2197P | 5.2 ± 2.2 | rep PI-luc/5.1c+G1304S |
| E1202G+T1280I+2202delS | 86.6 ± 62.9 | |
| E1202G+T1280I+S2204I | 105.6 ± 64.0 | |
| E1202G+T1280I+S2204R | 49.1 ± 33.0 | |
| E1202G+T1280I+R2884G | 26.4 ± 7.3 | |
| E1202G+T1280I+Q2933R | 3.4 ± 0.8 | |
| K1846T+V1897A | 11.5 ± 4.7 | |
| 2202delS+S2204R | 1.0 ± 0.1 | |
| S2204R+S2197P | 0.2 ± 0.1 | |
| V1897A+S2204R | 8.0 ± 4.2 | |
| V1897A+2202delS | 3.0 ± 0.6 | |
| K1846T+S2204R | 7.7 ± 2.6 | |
| K1846T+S2204R | 7.1 ± 3.2 | |
| V1897A+R2884G | 1.8 ± 0.2 | |
| K1846T+R2884G | 4.1 ± 1.6 | |
| S2197P+R2884G | 0.3 ± 0.1 |
Ratio of RLU at 48 h and at 4 h after electroporation (percent). Mean values and standard deviations of at least three independent experiments are shown. For all experiments, Huh-7 cells with passage numbers above 90 were used.
Data were taken from Krieger et al. (28).
Note that clone 5.1 contains additional mutations that do not contribute to the adaptive phenotype (28).
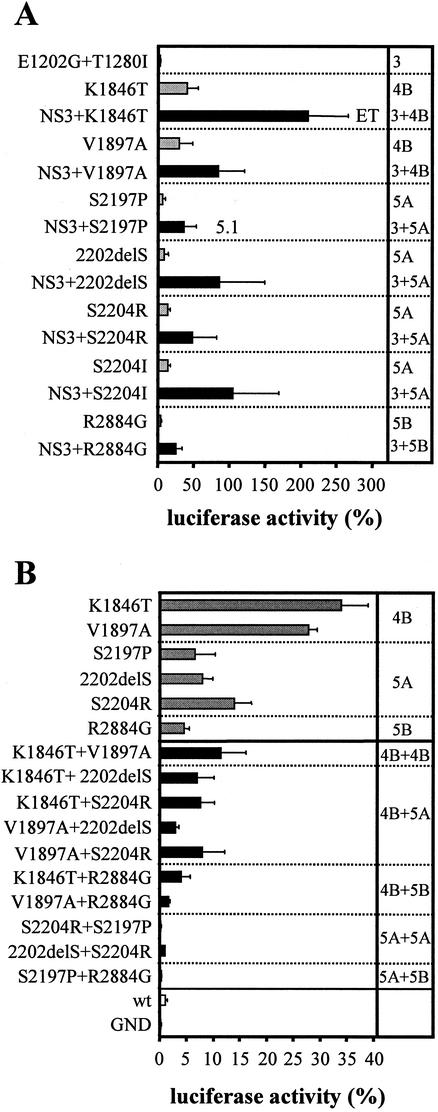
Cooperativity and incompatibility of adaptive mutations.
Since we have recently shown that the combination of two NS3 mutations (E1202G and T1280I) with one NS5A substitution (S2197P) led to a cooperative increase in RNA replication (28), we wanted to know whether the same was true for other combinations of adaptive mutations. Therefore, a series of highly adaptive mutations was combined with E1202G and T1280I in NS3 and analyzed for RNA replication in the transient assay (Fig. 4A and Table 2). In all cases but one, the combination was beneficial, and replication efficiency was enhanced 3- to 10-fold when these NS3 mutations were present. The exception was the NS5B mutation Q2933R which, when combined with the two substitutions in NS3 (Table 2) or other adaptive mutations (data not shown), did not enhance RNA replication cooperatively. The most efficient replicon we generated by this combinatorial approach carried one mutation in NS4B (K1846T) and the two mutations in NS3. Based on luciferase assays, replication of this replicon, designated rep PI-luc/ET, was ∼200-fold higher compared to the nonadapted RNA, and it was sixfold more efficient than rep PI-luc/5.1, which was the best-adapted replicon described thus far (28). An analogous result was found by Northern hybridization, corroborating that luciferase activities accurately reflect the level of RNA replication (Fig. 2C and D).
FIG. 4.
Cooperativity and incompatibility of adaptive mutations. Huh-7 cells were transfected with replicons containing the mutations specified in the left of each panel and harvested 4 and 48 h later. Luciferase activity is expressed as the percent RLU determined in lysates of cells at 48 h compared to those at 4 h after transfection. (A) NS3 mutations cooperatively enhance transient replication of replicons with NS4B, NS5A, and NS5B mutations. Pairs of replicons containing highly adaptive mutations without (gray bars) or with (black bars) two adaptive NS3 mutations are separated by dotted lines. The replication efficiency of the replicon carrying only the two NS3 mutations is given at the top. (B) Incompatibility of mutations in NS4B, NS5A, and NS5B. Replication efficiencies of replicons containing single highly adaptive mutations (gray bars) or combinations of these mutations (black bars) are compared. Replication levels of a wild-type replicon (wt) and a replication-deficient mutant (GND) are shown at the bottom of the panel (white bars). Values in both panels are means and standard deviations of at least three independent experiments. For all experiments, Huh-7 cells with passage numbers of >90 were used.
In an attempt to increase RNA replication even further, we introduced an additional NS3 mutation (G1304S) into rep PI-luc/5.1. Surprisingly, this substitution decreased replication efficiency by 85% (Table 2), although G1304S itself had no significant effect on transient replication (Fig. 3). All combinations of highly adaptive mutations in NS4B, NS5A, and NS5B also showed a significant decrease in replication efficiency, albeit to different extents (Fig. 4B and Table 2): the combinations of NS4B mutations with others in NS4B, NS5A, or NS5B reduced replication to the level of single NS5A or NS5B adapted mutants. In contrast, combinations of NS5A mutations with each other or with R2884G were either completely inactive or replicated less efficiently than the wild-type replicon. This result was consistent with our previous observation showing that the combination of a highly adaptive mutation in NS5A (E2163G) with R2884G led to a completely inactive replicon (31). Thus, the incompatibility of highly adaptive mutations appears to be a general rule.
Influence of Huh-7 passage number on the efficiency of transient HCV replication.
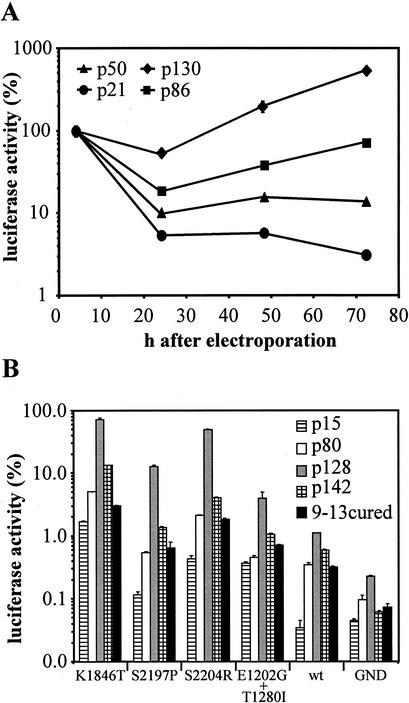
Since Huh-7 is the only permissive cell line in the replicon system, it seemed likely that host cell factor(s) contribute substantially to the efficiency of HCV RNA replication. Moreover, we observed a high consistency of the results within a given experiment, whereas the variation of replication efficiency of a replicon was high between independent experiments. After we had excluded technical problems, we assumed that variations in the quality of the host cells were responsible for this phenomenon. To address this question, four different passages of the Huh-7 cell line we had in the laboratory (p21, p50, p86, and p130) were each transfected with 1 μg of rep PI-luc/ET RNA, and replication was measured as described above (Fig. 5A). Transfection efficiency was similar, yielding ∼106 RLU in each tested passage 4 h after transfection, whereas at later time points dramatic differences between the passages were found. For Huh-7 cells of passage 21, the RLU count permanently dropped during the observation period. In contrast, a strong increase in luciferase activity was observed for Huh-7 cells of passage 130 within the same interval, resulting in a nearly 200-fold difference between these two passages 72 h after electroporation. This corresponded to ∼5,000,000 RLU for passage 130 versus 30,000 RLU for passage 21 at this time point. It is important to note that the replication efficiency of the highest adapted replicon (rep PI-luc/ET) and the wild-type replicon differed ∼200-fold (Table 2), and comparable differences were found for a given replicon transfected into a highly and a low permissive Huh-7 passage. In consequence, the permissiveness of Huh-7 cells used for transfection was as important for the efficiency of HCV replication as the adaptive mutations.
FIG. 5.
Influence of Huh-7 passage number on transient HCV replication. (A) Replicon PI-luc/ET (1 μg) was transfected into different passages of Huh-7 cells that were harvested after 4, 24, 48, and 72 h. Luciferase activity measured in the 4-h lysate was set as 100% for every passage tested. Data are from a representative experiment. Note the logarithmic scale of the ordinate. (B) Replicons carrying different mutations specified at the bottom were transfected into different passages of Huh-7 cells or cell clone 9-13 that was cured from the replicon by treatment with IFN-α (9-13cured). Luciferase activity is expressed as the percent RLU determined at 48 h compared to that determined at 4 h after transfection. Note the logarithmic scale of the ordinate.
To further substantiate this observation, we tested a panel of adapted replicons and the wild-type replicon for their ability to replicate in different Huh-7 passages. As shown in Fig. 5B, replication of the wild-type replicon could not be detected in p15 and replication efficiency in the remaining passages varied by only threefold (0.34% in p80 versus 1.1% in p128). In case of the NS3 mutant (E1202G+T1280I), the ratio between the highest and the least permissive Huh-7 passage was 10-fold but 50-fold for the NS4B mutant (K1846T) and 100-fold for the NS5A mutants S2197P and S2204R, indicating that the fluctuations in the permissiveness of different Huh-7 passages had a greater impact on highly adaptive mutations in NS5A and NS4B than on NS3 mutations and the wild-type sequence. Another conclusion that can be drawn from the experiment shown in Fig. 5B is that a replicon with the wild-type sequence replicated more efficiently in Huh-7 cells of passage 128 compared to replicons harboring the highly adaptive mutations S2197P and S2204R in passage 15. This clearly emphasizes the important role of the host cell environment for HCV replication and opens the possibility that, under particular selection conditions, by using a highly permissive Huh-7 population and a low selective pressure, it should be possible to generate cell clones persistently replicating the unmodified wild-type genome.
Furthermore, we analyzed the Huh-7 replicon cell clone 9-13 (31, 32), after removal of the replicon by IFN-α treatment for 2 weeks (Fig. 5B). These cells were included because we assumed that the selective pressure used to generate a replicon cell clone would not only select for cell culture-adaptive mutations but also for more permissive Huh-7 cells (32). In contrast to our assumption, the efficiency of the cured replicon cell clone 9-13 to support transient HCV replication was comparable to naive Huh-7 cells of passage 80 (Fig. 5B). We repeated this experiment with other cured replicon cell clones and found significant variations in the level of permissiveness. From seven cured replicon cell clones tested the best was about as efficient as the most permissive naive Huh-7 cells (data not shown). We also did not find a higher replication efficiency of replicons that carried the same adaptive mutations than those that were present in the selectable replicons originally replicating in the cells (data not shown), suggesting that various adapted mutants do not differ in their host cell requirements.
Huh-7 cells with higher passage numbers tended to be more efficient than earlier passages (Fig. 5A), but this phenotype was not stable. As shown in Fig. 5B, Huh-7 cells from p142, which were direct descendants of the cells from p128, were less permissive. Within 14 passages, the efficiency of replication decreased by up to 1 order of magnitude. Thus, the ability of Huh-7 cells to support replication of HCV was subject to a variability that we were unable to control. Nevertheless, in our hands passage numbers higher than 120 were generally more permissive, and this phenotype was conserved even after storage in liquid nitrogen (data not shown).
No difference of IRES-dependent translation or RNA stability between various Huh-7 passages.
Several possibilities could explain the differences in HCV replication observed with the various Huh-7 passages. Potential explanations were differences of IRES activity or RNA stability, variations in the amount of a cellular factor or factors required for replication, differences in tolerance against cytopathic effects that were caused by HCV RNAs or proteins, or variations in the level of an antiviral state induced by replicating HCV RNAs. To analyze translation efficiencies of the replicons in different Huh-7 passages, we had to test the EMCV IRES that directed expression of the HCV NS proteins and the PV IRES required for translation of the reporter gene (Fig. 2A). EMCV IRES activity was examined by transfection of an in vitro transcript depicted in Fig. 6A into various Huh-7 passages. Luciferase activity measured 2 h after transfection was normalized for transfection efficiency by using β-galactosidase activity expressed from a cotransfected capped RNA carrying the lacZ gene. No evidence for variations in the efficiency of translation from the EMCV IRES between the Huh-7 passages tested was found (Fig. 6A). The activity of the PV IRES was assayed in the same way except that a replication-deficient replicon was used to quantify PV IRES activity, and β-galactosidase expression was controlled by an EMCV IRES present in the cotransfected control RNA (Fig. 6B). No significant variations of PV IRES activity were found with the different Huh-7 passages.
FIG. 6.
Comparable translation from the EMCV and the PV IRES in different Huh-7 passages. Replication-deficient RNAs containing a luciferase gene under the translational control of an EMVC (A) or a PV IRES (B) as indicated at the top of each panel were electroporated into different passages of Huh-7 cells. In vitro transcripts coding for β-galactosidase were cotransfected to normalize for transfection efficiency. Cells were harvested 2 h after electroporation, and the luciferase and β-galactosidase activities were determined. The given values are corrected for transfection efficiency. Passage numbers of the Huh-7 cells used for transfection are indicated under each column.
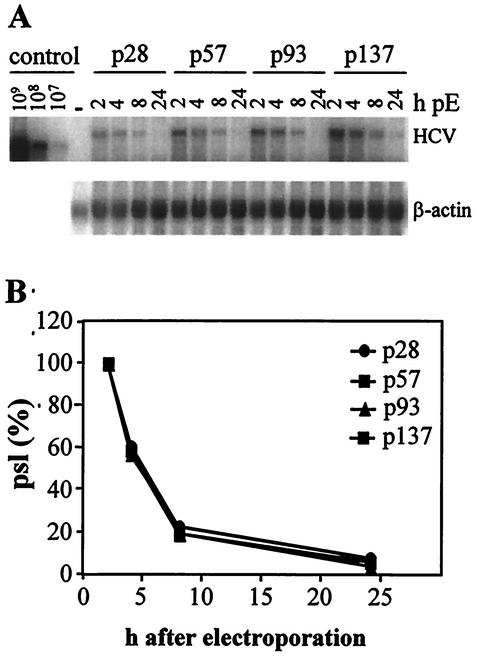
The stability of the transfected replicon RNA also was a factor that could contribute to differences in replication efficiency. Therefore, the RNA half-lives in various Huh-7 passages were determined by transfection of a replication-deficient replicon and Northern blot analysis of total RNA that was isolated from cells 2, 4, 8, and 24 h after transfection (Fig. 7A). The HCV-specific RNA was quantified by phosphorimaging as described in Materials and Methods, and values obtained at 2 h posttransfection were set to 100% (22,778 photostimulated light units [PSL] with passage 28 cells, 43,149, 44,226, and 77,349 PSL with cells of passage 57, 93, and 137, respectively) (Fig. 7B). Transfected RNAs decayed with a half-life of 2.5 to 3 h, and no significant differences were observed between the various Huh-7 passages. The generally reduced signal intensity for samples of p28 was due to a lower transfection efficiency in this particular experiment.
FIG. 7.
Comparable stabilities of replicon RNAs in different Huh-7 passages. Cells specified in the top were transfected with 15 μg of the replication-defective replicon PI-luc/GND and harvested at different time points postelectroporation (pE) as indicated above each lane. (A) Total RNA was analyzed by Northern hybridization in parallel to known amounts of in vitro transcripts (control) and total RNA from naive Huh-7 cells (−). The positions of replicon RNA (HCV) and β-actin mRNA are given on the right. (B) Quantification of the Northern hybridization shown in panel A by phosphorimaging. Values were corrected for differences in loading by using the β-actin signal and are expressed as the percent PSL at a given time point relative to that observed 2 h after transfection.
Evidence for host cell factors or conditions determining HCV RNA replication.
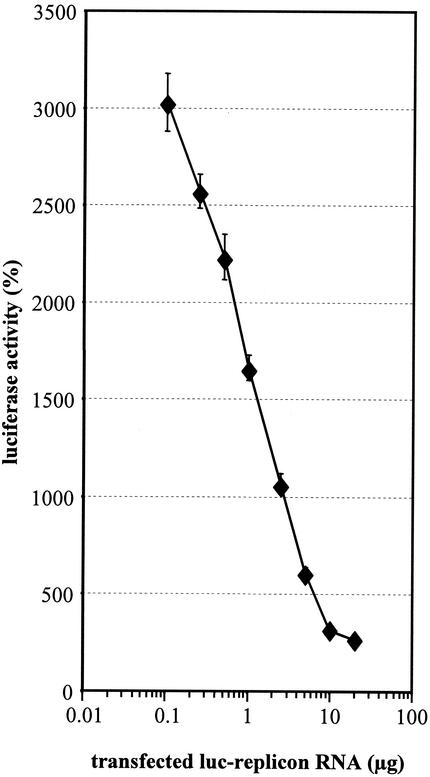
Another possibility explaining the different levels of permissiveness of Huh-7 passages was variability in the abundance or activity of cellular factor(s) affecting directly or indirectly HCV replication. This factor(s) could be either an inhibitor or an activator. In the case of an inhibitor, replication should increase with increasing amounts of transfected RNA. In contrast, a cellular activator should become limiting for RNA replication with increasing concentrations of replicon RNA. Consequently, replication efficiency should decrease with increasing amounts of transfected RNA. To differentiate between these alternatives, titration experiments were performed in which 0.1 to 20 μg of rep PI-luc/ET in vitro transcripts were transfected into Huh-7 cells. To avoid effects of various RNA concentrations on transfection efficiency, we adjusted the amount of transfected RNA to 20 μg for all samples by addition of yeast tRNA. Replication efficiency was expressed as the percent RLU, with 100% representing the luciferase activity measured 4 h after transfection. This normalization was necessary because the absolute luciferase values are determined by the amount of replicon RNA used for transfection. However, by using this correction, we were able to calculate the extent of HCV RNA amplification relative to the amount of replicon RNA introduced into cells. The results in Fig. 8 show that the replication efficiency was inversely correlated with the amount of transfected replicon RNA. Transfection of 0.1 μg of rep PI-luc/ET yielded 6.5 × 104 RLU (100%) and 2 × 106 (3,025%) 4 and 72 h after transfection, respectively, corresponding to a >30-fold amplification. In contrast, transfection of 20 μg of the same RNA resulted in 1.27 × 107 RLU (100%) at 4 h posttransfection, but only 3.37 × 107 RLU (266%) at 72 h after transfection, corresponding to only a 2.6-fold amplification. This negative correlation between the amount of replicon RNA present in a cell and its replication efficiency indicated that a host factor(s) or condition(s) limited HCV replication in Huh-7 cells.
FIG. 8.
Inverse correlation between the amount of transfected replicon RNA and the replication efficiency. Increasing amounts of rep luc/ET RNA were used for electroporation of Huh-7 cells. The amount of total transfected RNA in every sample was adjusted to 20 μg by the addition of yeast tRNA. Cells were harvested 4 and 72 h after electroporation and assayed for luciferase activity. The replication efficiency is expressed as the percent RLU determined at 72 h compared to that determined at 4 h after transfection. Note the logarithmic scale of the abscissa.
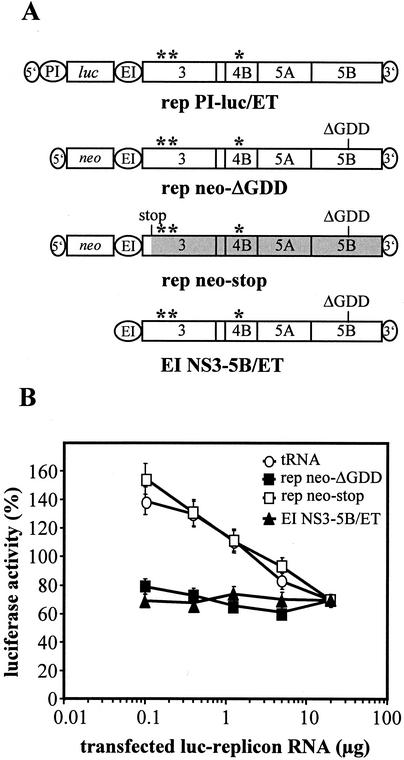
Such factors could either alter cellular conditions in a way to generate a more favorable environment or modulate the replicase complex, e.g., via direct binding. In the latter case one would expect that replication efficiency of a replicon RNA is reduced by increasing amounts of replicase complexes competing for binding of the cellular factor. Therefore, cotransfection experiments were performed in which increasing amounts of replicon PI-luc/ET (0.1, 0.4, 1.25, 5, and 20 μg) were adjusted with decreasing amounts of each of the following RNAs to a total of 20 μg of RNA per transfection: (i) tRNA, (ii) a replicon with an inactive NS5B RdRp (rep neo-ΔGDD), (iii) a replicon carrying a stop codon about 200 nt downstream of the NS3 start codon (rep neo-stop), or (iv) a nonreplicating RNA encoding only the NS3 to NS5B polyprotein (EI NS3-5B/ET; Fig. 9A). Cells were harvested 4 and 24 h after transfection, and the replication efficiency was calculated by determining the ratio of luciferase activities measured at these time points. As shown in Fig. 9B, cotransfection of the parental replicon PI-luc/ET with in vitro transcripts directing the expression of the HCV polyprotein (rep neo-ΔGDD or EI NS3-5B/ET) led to a constantly low replication. In contrast, when yeast tRNA or the RNA carrying the stop codon in NS3 were cotransfected, no interference with RNA replication was observed (Fig. 9B). The difference of the RLU observed with the various competitor RNAs was not due to different transfection efficiencies. The 4-h values that only measure translation of the luciferase reporter gene from the replicon RNA were comparable in the different transfections (e.g., 252,591 RLU or 218,419 RLU, in the case of 0.4 μg of replicon RNA and 19.6 μg of cotransfected rep neo-stop or rep neo-ΔGDD competitor, respectively).
FIG. 9.
Coexpression of the HCV NS proteins, not the cotransfected RNA itself, affects RNA replication. (A) Schematic structure of RNAs used for cotransfection experiments with rep PI-luc/ET. Adaptive mutations are indicated by asterisks. “ΔGDD” refers to the position of the 10-amino-acid deletion spanning the GDD motif in NS5B, and “stop” refers to an engineered stop codon that inhibits translation ∼70 amino acids downstream of the NS3 inititation codon. For further details, see the legend to Fig. 2A. (B) Increasing amounts of rep PI-luc/ET-RNA (0.1, 0.4, 1.25, 5, and 20 μg) were cotransfected with different competitors specified at the top. In each case the total amount of transfected RNA was adjusted to 20 μg by the addition of decreasing amounts of competitor RNA (19.9, 19.6, 18.75, 15, and 0 μg). Huh-7 cells were electroporated with this mixture, seeded into aliquots, and harvested at 4 and 24 h after transfection. Replication efficiency is expressed as the percent RLU determined 24 h compared to 4 h after transfection. Note the logarithmic scale of the abscissa.
Although this result was well concordant with our assumption that a cellular factor or factors interacted with the HCV replicase complex, we could not exclude the possibility that high amounts of the NS proteins might have caused cytotoxic effects, leading to an apparent reduction of HCV replication. To address this question, we cotransfected 20 μg of different replication deficient in vitro transcripts (Fig. 9A) with a plasmid coding for luciferase. Since we did not observe an effect of the viral proteins on luciferase expression (data not shown), it seemed unlikely that cytopathic effects or a host cell shutoff could account for the inhibition of HCV replication observed with RNAs encoding NS3 to NS5B. We also did not observe a correlation between high levels of transient HCV replication and apoptosis in a double immunofluorescence analysis in which we stained for HCV NS5B and caspase-3 (data not shown). In summary, these data provide evidence for a host cell factor(s) or conditions that modulate the HCV replicase complex and limit HCV replication in Huh-7 cells.
DISCUSSION
In the present study, we quantified the effect of cell culture-adaptive mutations on HCV RNA replication. By combining a highly adaptive mutation in NS4B (K1846T) with two NS3 mutations (E1202G+T1280I [28]), we obtained a replicon, designated rep PI-luc/ET, that replicated six times more efficiently than the best replicon we had described so far (rep5.1 [28]) and 15 times more efficiently than the best replicon described by another group (7) using the mutation S2204I. Under optimal conditions, luciferase activity obtained with rep PI-luc/ET is up to 10,000-fold above background as determined with a replication-deficient control. Another advantage of this replicon is due to the fact that cell culture adaptation is achieved by an NS4B mutation that is more compatible with other adaptive mutations than those in NS5A (Fig. 4B). Although replication efficiency was reduced upon combination of this NS4B mutation (K1846T) with other highly adaptive mutations, all combinations involving this particular substitution were viable, whereas some combinations involving highly adaptive mutations in NS5A (e.g., S2204R+S2197P) resulted in complete replication deficiency. This is especially important when mutational analyses in NS5A and NS5B are performed because mutations leading by chance to cell culture adaptation would inactivate the replicon when combined with an adaptive NS5A mutation but would be viable when combined with the adaptive NS4B mutations described here.
From 26 independent replicon cell clones, 16 had a highly adaptive mutation in a region spanning 27 amino acids that is presumed to be important for phosphorylation and hyperphosphorylation of NS5A (42). The same region was also found to be the main site for adaptation by Blight et al. (7). Although they demonstrate a lack of correlation between hyperphosphorylation and adaptation, and this was also found by our group (N. Krieger and R. Bartenschlager, unpublished observations), the adaptive mutations in this region show some striking similarities, suggesting that changes in phosphorylation play a role in adaptation: (i) most changes in this region affect serine residues either by substitution or deletion; (ii) at one position (D2177) a negative charge that may mimic a phosphorylated serine residue is removed; and (iii) alanine at position 2199 was replaced by threonine or glutamic acid (Fig. 1) or serine (7). Since a negative charge (glutamic acid) at this position could functionally replace serine or threonine, this observation suggests that serine or threonine are regularly phosphorylated at this site. It seems plausible that subtle changes in the net charge or phosphorylation pattern of this region undetectable by conventional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis confer the adaptive phenotype. Therefore, more detailed analyses of the phosphorylation status of NS5A and its possible influence on RNA replication are required.
A second set of highly adaptive mutations was found in NS4B. In a total of six cell clones we found changes in only two different positions (K1846 and V1897). Mutations at these sites had the strongest impact on HCV replication. Interestingly, Guo et al. (18) also identified one of these mutations (K1846T) in replicons isolated from two independent Huh-7 clones after G418 selection. These results suggest an important role of NS4B for HCV replication. It was shown that NS4B induces the formation of a cytoplasmic membrane structure that appears to be the HCV replication complex (9). It is tempting to speculate that formation or activity of this structure in some way is affected by cell culture-adaptive mutations. Further studies in this direction will be required.
Interestingly, most mutations conferring cell culture adaptation, in particular the highly adaptive ones, have not been found in natural isolates of HCV. This observation suggests that these mutations do not increase replication efficiency per se but rather represent an adaptation that is specific for Huh-7 cells. The mechanism by which this adaptation is achieved currently is not known. The fact that mutations in NS4B, NS5A, and NS5B led to a decrease of replication when combined with each other suggests that these mutations mediate cell culture adaptation via the same or a very similar mechanism. For instance, some of the adaptive mutations may directly modify the interaction with a cellular protein, whereas others may act indirectly via conformational changes within the replicase complex. Alternatively, the mutations found to be highly adaptive in the present study, although in different regions of the polyprotein, could be in close proximity in the replicase complex and simultanously or sequentially interact with this cellular factor. Finally, some of the adaptive mutations may cause changes in the viral RNA sequence that lead to a loss of inhibitory interactions between HCV sequences and the heterologous sequences. However, we have recently shown that the introduction of cell culture-adaptive mutations into authentic HCV full-length genomes that lack any heterologous sequences leads to a similar increase of RNA replication, making this possibility unlikely (35).
In contrast to highly adaptive mutations, we also found several alterations exhibiting only little or no effect on HCV replication efficiency. One cluster of these mutations was found in domain 1 of the NS3 helicase (for the three-dimensional structure, see reference 25; for the location of some adaptive mutations, see reference 31). Because of their cooperative increase of replication efficiency when combined with highly adaptive mutations, it seems likely that the mechanism of cell culture adaptation achieved by mutations in NS3 is different from the one mediated by mutations in NS4B, NS5A, and NS5B. This assumption fits to the incompatibility observed when mutations within NS3 or within the NS4B to NS5B region were combined.
The observation that mutations in NS3 were always found in conjuction with highly adaptive ones indicates that the capability of NS3 mutations to increase RNA replication was very low. This finding implies a threshold value for HCV RNA replication that had to be exceeded to confer G418 resistance to the cell. Under the selection conditions used here (1 mg of G418/ml), this threshold value seemed to be about four- to fivefold above the replication level of the wild-type replicon. Mutations analyzed in the present study that supported replication to a level below this threshold were only found in cooperation with other adaptive mutations in selected cell clones, whereas mutations that supported a higher replication level could be found as single conserved mutations. The selective pressure used in the present study to generate the replicon cell clones was very high. Therefore, it should be possible to obtain cell clones carrying replicons with only moderate or even no adaptive mutations when the G418 concentration was reduced during selection.
The mutation located at the very C terminus of the protease, E1202G, was remarkable for several reasons. (i) It was the only one in NS3 that was found several times in independent replicon cell clones. (ii) It was compatible with highly adaptive mutations, as well as with NS3-helicase mutations. (iii) This mutation was found in >5% of the published NS3 sequences available in the database and in a cDNA clone that was infectious in chimpanzees (46). These results suggest that E1202G leads to cell culture adaptation via a mechanism that is independent from all other mutations.
The distribution of highly adaptive mutations in the HCV polyprotein described here may provide a general map of the regions important for adaptation to Huh-7 cells that is applicable also to other HCV isolates. This assumption is supported by data obtained with a second genotype 1b-derived replicon (18, 23). The original genome used in that study, designated HCV-N, contains a unique four-amino-acid insertion in the IFN sensitivity-determining region of NS5A and was shown to be infectious in chimpanzees (5), albeit with very low efficiency. Colony formation efficiency of a subgenomic, selectable HCV-N replicon was much higher compared to our parental HCV Con1 replicon (18, 23) and could be further enhanced by introducing adaptive mutations found in our isolate. Removal of the four-amino-acid insertion from the HCV-N replicon dramatically reduced colony formation efficiency that could be increased again by using adaptive mutations identified with our isolate (23). Thus, the mutations described here appear to be applicable to HCV replicons of other isolates and most likely also other genotypes.
Apart from cell culture-adaptive mutations, we demonstrate here that the host cell plays a very important role for RNA replication, as we had already speculated earlier (32). In fact, in the most permissive Huh-7 passage the unmodified wild-type replicon replicated more efficiently than even highly adapted replicons in an Huh-7 passage that was poorly permissive. This difference between the Huh-7 passages was found with cells originating from one cell stock in the same laboratory, and we can expect that it will be even higher between Huh-7 stocks passaged in different laboratories. These fluctuations that we were unable to control may explain the quantitative variations of the results different groups observed even when working with the same HCV replicons and they will complicate the direct comparison of these results.
The different levels of permissiveness observed with individual Huh-7 passages may be due to variations in the abundance or activity of cellular factor(s) that enhance HCV RNA replication, e.g., via direct binding to the replicase complex. In agreement with this assumption, we found that cotransfected RNAs encoding the complete NS3 to NS5B region reduced RNA replication, whereas no reduction was found with RNAs encoding individual HCV proteins (Fig. 9 and unpublished data). These results suggest that binding of host cell protein(s) requires a complete NS3 to NS5B polyprotein complex, which will make the identification of such cellular protein(s) by biochemical interaction assays or in yeast two-hybrid screens very difficult.
Currently, it is not known whether the different capabilities of Huh-7 passages to support HCV replication are due to different numbers of cells that permit high-level replication or to different levels of permissiveness of all cells of a given passage. In agreement with the first assumption, immunofluorescence analysis of different Huh-7 passages revealed that far fewer than 1% of the cells of passage 20 expressed detectable amounts of HCV protein 72 h after transfection but that 10 to 50% of the cells of passage 140 did express HCV protein (V. Lohmann, unpublished data). Moreover, RNA titration experiments analogous to the one presented in Fig. 8 show that the reduction of RNA replication when the amount of transfected RNA was increased is similar with different Huh-7 passages, but the overall levels of RNA replication are higher in more-permissive Huh-7 cells (V. Lohmann, unpublished data). Thus, the permissiveness of a given Huh-7 passage is primarily determined by the number of permissive cells in this culture.
Huh-7 clones harboring a persistent replicon should provide an optimal environment for HCV RNA replication. If this property is stable, removal of the replicon should create a perfect host cell for transient replication assays. The cured replicon cell clones analyzed in the present study did not support replication to a level higher than the one obtained with the most permissive passages of naive Huh-7 cells. However, the permissiveness of naive Huh-7 cells varies tremendously and whether or not cured cells support RNA replication more efficiently primarily depends on which naive cells are used as a reference. Moreover, owing to this variation of permissiveness, a direct comparison of cured cells and naive Huh-7 originally used for transfection with the corresponding selectable replicon is very difficult.
In summary, we found that efficient replication of HCV in Huh-7 cells very much depends on cell culture-adaptive mutations and host cell conditions or factors. Taking into account the tight coupling between HCV replication and cell growth (36), the interactions between HCV and the host cell appear to be very complex. It will be an interesting and challenging task to identify the cellular factors involved and their contribution in RNA replication.
Acknowledgments
We thank Thomas Pietschmann, Nicole Krieger, Sandra Sparacio, and Michael Frese for critically reading the manuscript and for helpful discussions.
This work was supported in part by the SFB 490 (Teilprojekt A2) and the EU (QLK2-1999-00356).
REFERENCES
- 1.Bartenschlager, R., L. L. Ahlborn, J. Mous, and H. Jacobsen. 1993. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J. Virol. 67:3835-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 3.Bartenschlager, R., and V. Lohmann. 2001. Novel cell culture systems for the hepatitis C virus. Antiviral Res. 52:1-17. [DOI] [PubMed] [Google Scholar]
- 4.Bartenschlager, R., V. Lohmann, T. Wilkinson, and J. O. Koch. 1995. Complex formation between the NS3 serine-type proteinase of the hepatitis C virus and NS4A and its importance for polyprotein maturation. J. Virol. 69:7519-7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beard, M. R., G. Abell, M. Honda, A. Carroll, M. Gartland, B. Clarke, K. Suzuki, R. Lanford, D. V. Sangar, and S. M. Lemon. 1999. An infectious molecular clone of a Japanese genotype 1b hepatitis C virus. Hepatology 30:316-324. [DOI] [PubMed] [Google Scholar]
- 6.Behrens, S. E., L. Tomei, and R. DeFrancesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 7.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 8.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 9.Egger, D., B. Wölk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed]
- 10.Failla, C., L. Tomei, and R. DeFrancesco. 1995. An amino-terminal domain of the hepatitis C virus NS3 protease is essential for interaction with NS4A. J. Virol. 69:1769-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friebe, P., and R. Bartenschlager. 2002. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 76:5326-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friebe, P., V. Lohmann, N. Krieger, and R. Bartenschlager. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gale, M. J., S. M. Blakely, B. Kwieciszewski, S. L. Tan, M. Dossett, N. M. Tang, M. J. Korth, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1998. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanism of kinase regulation. Mol. Cell. Biol. 18:5208-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gale, M. J., M. J. Korth, N. M. Tang, S. L. Tan, D. A. Hopkins, T. E. Dever, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1997. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 230:217-227. [DOI] [PubMed] [Google Scholar]
- 16.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. A second hepatitis C virus-encoded proteinase. Proc. Natl. Acad. Sci. USA 90:10583-10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J. Virol. 67:2832-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hijikata, M., N. Kato, Y. Ootsuyama, M. Nakagawa, and K. Shimotohno. 1991. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc. Natl. Acad. Sci. USA 88:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hijikata, M., H. Mizushima, T. Akagi, S. Mori, N. Kakiuchi, N. Kato, T. Tanaka, K. Kimura, and K. Shimotohno. 1993. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J. Virol. 67:4665-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 22.Hugle, T., F. Fehrmann, E. Bieck, M. Kohara, H. G. Krausslich, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology 284:70-81. [DOI] [PubMed] [Google Scholar]
- 23.Ikeda, M., M. Yi, K. Li, and S. M. Lemon. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 76:2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, D. W., Y. Gwack, J. H. Han, and J. Choe. 1995. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem. Biophys. Res. Commun. 215:160-166. [DOI] [PubMed] [Google Scholar]
- 25.Kim, J. L., K. A. Morgenstern, J. P. Griffith, M. D. Dwyer, J. A. Thomson, M. A. Murcko, C. Lin, and P. R. Caron. 1998. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure 6:89-100. [DOI] [PubMed] [Google Scholar]
- 26.Koch, J. O., and R. Bartenschlager. 1999. Modulation of hepatitis C virus NS5A hyperphosphorylation by nonstructural proteins NS3, NS4A, and NS4B. J. Virol. 73:7138-7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, C., J. A. Thomson, and C. M. Rice. 1995. A central region in the hepatitis C virus NS4A protein allows formation of an active NS3-NS4A serine proteinase complex in vivo and in vitro. J. Virol. 69:4373-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lohmann, V., F. Körner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lohmann, V., F. Körner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lohmann, V., F. Körner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 33.Murphy, F. A., C. M. Fauquet, D. H. L. Bishop, S. A. Ghabrial, A. W. Jarvis, G. P. Martelli, M. A. Mayo, and M. D. Summers (ed.). 1995. Classification and nomenclature of viruses: sixth report of the international committee on taxonomy of viruses, p. 424-426. Springer-Verlag, Vienna, Austria.
- 34.Nakabayashi, H., K. Taketa, K. Miyano, T. Yamane, and J. Sato. 1982. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 42:3858-3863. [PubMed] [Google Scholar]
- 35.Pietschmann, T., V. Lohmann, A. Kaul, N. Krieger, G. Rinck, G. Rutter, D. Strand, and R. Bartenschlager. 2002. Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J. Virol. 76:4008-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pietschmann, T., V. Lohmann, G. Rutter, K. Kurpanek, and R. Bartenschlager. 2001. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J. Virol. 75:1252-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poynard, T., V. Ratziu, Y. Benhamou, P. Opolon, P. Cacoub, and P. Bedossa. 2000. Natural history of HCV infection. Best Practice Res. Clin. Gastroenterol. 14:211-228. [DOI] [PubMed] [Google Scholar]
- 38.Reed, K. E., and C. M. Rice. 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242:55-84. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Suzich, J. A., J. K. Tamura, H. F. Palmer, P. Warrener, A. Grakoui, C. M. Rice, S. M. Feinstone, and M. S. Collett. 1993. Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J. Virol. 67:6152-6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanji, Y., M. Hijikata, S. Satoh, T. Kaneko, and K. Shimotohno. 1995. Hepatitis C virus-encoded nonstructural protein NS4A has versatile functions in viral protein processing. J. Virol. 69:1575-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanji, Y., T. Kaneko, S. Satoh, and K. Shimotohno. 1995. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J. Virol. 69:3980-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsukiyama, K. K., N. Iizuka, M. Kohara, and A. Nomoto. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van den Hoff, M. J., A. F. Moorman, and W. H. Lamers. 1992. Electroporation in “intracellular” buffer increases cell survival. Nucleic Acids Res. 20:2902.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, C., P. Sarnow, and A. Siddiqui. 1993. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J. Virol. 67:3338-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1997. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl. Acad. Sci. USA 94:8738-8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yanagi, M., M. St. Claire, S. U. Emerson, R. H. Purcell, and J. Bukh. 1999. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc. Natl. Acad. Sci. USA 96:2291-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]