Abstract
Umbraviruses are different from most other viruses in that they do not encode a conventional capsid protein (CP); therefore, no recognizable virus particles are formed in infected plants. Their lack of a CP is compensated for by the ORF3 protein, which fulfils functions that are provided by the CPs of other viruses, such as protection and long-distance movement of viral RNA. When the Groundnut rosette virus (GRV) ORF3 protein was expressed from Tobacco mosaic virus (TMV) in place of the TMV CP [ΤMV(ORF3)], in infected cells it interacted with the TMV RNA to form filamentous ribonucleoprotein (RNP) particles that had elements of helical structure but were not as uniform as classical virions. These RNP particles were observed in amorphous inclusions in the cytoplasm, where they were embedded within an electron-dense matrix material. The inclusions were detected in all types of cells and were abundant in phloem-associated cells, in particular companion cells and immature sieve elements. RNP-containing complexes similar in appearance to the inclusions were isolated from plants infected with ΤMV(ORF3) or with GRV itself. In vitro, the ORF3 protein formed oligomers and bound RNA in a manner consistent with its role in the formation of RNP complexes. It is suggested that the cytoplasmic RNP complexes formed by the ORF3 protein serve to protect viral RNA and may be the form in which it moves through the phloem. Thus, the RNP particles detected here represent a novel structure which may be used by umbraviruses as an alternative to classical virions.
One of the main characteristics of viruses is the formation of virus particles or virions, in which the viral genomic nucleic acid (RNA or DNA) is protected by encapsidation with one or more types of capsid (or coat) protein (CP). In virions, molecules of CP are packed into regular uniform structures with either helical or icosahedral symmetry. In plant viruses, the CP is also involved in transmission by biological vectors and often in the spread of viruses in infected plants. Some plant viruses, such as Cowpea mosaic virus (32), Potato virus X (PVX) (26), or Cucumber mosaic virus (CMV) (2), require the CP for movement both through plasmodesmata (cell-to-cell movement) and via the phloem (long-distance movement). Others, such as Tobacco mosaic virus (TMV), require the CP for long-distance movement but not for cell-to-cell movement (for a review, see reference 4).
Umbraviruses are unusual in that they do not encode a CP. Nevertheless, they accumulate and spread efficiently within and between plants. Moreover, unlike most single-stranded viral RNAs, umbraviral RNA in crude sap extracts remains infective for several hours and is resistant to degradation by RNase A (15, 16). These observations suggest that umbraviruses exploit alternative mechanisms for RNA protection and systemic spread, mechanisms different from those provided by a CP.
For aphid transmission between plants, umbraviruses depend on the assistance of a luteovirus, the CP of which forms transmissible nucleoprotein particles encapsidating umbraviral RNA (for a review, see reference 29). However, within infected plants, functions such as protection and movement of umbraviral RNA do not require the presence of the luteovirus and its CP (6). Therefore, some other umbravirus-specific product(s) must functionally replace the absent CP. The RNA genomes of umbraviruses contain four open reading frames (ORFs). Although ORF1 at the 5′ end of the RNA encodes a putative product of 31 to 37 kDa, this ORF together with ORF2 can also be expressed by a −1 frameshift to give a single protein that appears to be an RNA-dependent RNA polymerase (5, 10, 28). The other two ORFs overlap each other in different reading frames. ORF4 encodes a 27- to 29-kDa protein, which contains stretches of similarity with several plant virus-encoded movement proteins (MPs) that control virus movement from cell to cell (10, 28). In gene replacement experiments, the ORF4 protein encoded by one of the umbraviruses, Groundnut rosette virus (GRV), was shown to be able to replace functionally the MPs of PVX or CMV (20, 21), confirming that the GRV ORF4 protein is a cell-to-cell MP. The ORF4 protein enabled cell-to-cell movement of PVX and CMV regardless of the presence or absence of their CPs, but the CPs were still required for long-distance movement (20, 21).
Database searches with the sequences of the umbraviral 26- to 29-kDa ORF3 proteins revealed no significant similarity with any other recorded or predicted proteins (28). In infected cells, the GRV ORF3 protein expressed from a TMV vector as a fusion with green fluorescent protein (GFP) was located in cytoplasmic granules, some of which were associated with TMV-specific amorphous X-body inclusions, and in nuclei, preferentially targeting nucleoli (20). Subsequently, we showed that umbravirus ORF3 proteins could stabilize TMV RNA and facilitate its long-distance movement, replacing the TMV CP that is normally essential for both these functions (22, 23). These properties of the ORF3 proteins suggest that they can bind to viral RNA to protect it and transport it to and through the phloem. In support of this hypothesis, we show in this paper that the GRV-encoded ORF3 protein is able to interact with viral RNA to form filamentous ribonucleoprotein (RNP) particles, which have elements of regular helical structure but not the uniformity typical of virus particles. In the cytoplasm, the RNP particles are found in amorphous aggregates (cytoplasmic complexes) of various sizes, which may serve to protect the RNA, and which may be the form in which the viral genome is transported through the phloem.
MATERIALS AND METHODS
Chimeric TMV constructs, in vitro transcription, and inoculation of plants.
Chimeric TMV constructs were based on the TMV vector TMV(30B), kindly provided by W. O. Dawson (Citrus Research and Education Center, Lake Alfred, Fla.). TMV(ORF3) (the GRV ORF3 protein was expressed from TMV instead of the TMV CP) was as described by Ryabov et al. (22). For TMV(ORF3-His), the sequence coding for the GRV ORF3 protein with six histidine residues fused to the C terminus was amplified by using oligonucleotides 5′-GCATTTAATTAATGGACACCACCC-3 and 5′-GCATCTCGAGTCAATGGTGATGGTGATGGTGCCACTTATTGGCAGCGG-3′ as primers and pTMV(ORF3) as a template. The PCR fragment was cloned between the PacI and XhoI sites of pTMV(ΔCP) (22) to give plasmid pTMV(ORF3-His). Plasmids were linearized by digestion with KpnI, and in vitro transcripts were synthesized with T7 RNA polymerase by using the mMESSAGE mMACHINE T7 kit (Ambion, Inc., Austin, Tex.). The transcripts derived from 0.2 μg of plasmid template were inoculated directly to corundum-dusted leaves of Nicotiana benthamiana. This work was done under license no. GM126/2001 from the Scottish Executive Environment and Rural Affairs Department.
Generation of antibody against the GRV ORF3 protein.
The antibody against the GRV ORF3 protein was generated by injecting a rabbit with a synthetic oligopeptide (produced by Genosys, Pampisford, Cambridgeshire, United Kingdom) corresponding to part of the deduced sequence of the GRV ORF3 protein (amino acid residues 71 to 92 [28]), conjugated to keyhole limpet hemocyanin. The specificity of the antibody was confirmed by reactions with GRV- or TMV(ORF3)-infected material but not with healthy plants in Western blots. A rabbit polyclonal antibody against purified recombinant ORF3-His protein (see below) was also generated. Results for these two types of antibodies were essentially similar in all tests including immunogold labeling and Western blotting.
Electron microscopy, immunogold labeling, and in situ hybridization.
Pieces of leaf material systemically infected with TMV(ORF3) were sampled 12 to 15 days postinoculation, fixed, and embedded essentially as described before (18). In addition to conventional embedding in Araldite resin, some samples were embedded in either L.R. white resin or a 1:1 mixture of Araldite-L.R. white for in situ hybridization studies. All samples included areas of leaf blade and some of the major (class III) veins, and sections were cut using a Reichert Ultracut ultramicrotome. Ultrathin sections were mounted on pyroxylin-coated nickel grids for conventional studies and for immunogold labeling or on pyroxylin-coated gold grids for in situ hybridization. After the sections were labeled, they were stained by the isobutanol staining technique (19) and examined with a Philips CM 10 electron microscope.
To examine the structure of selected particles in more detail, linear displacement was done in Adobe Photoshop version 6.0. Images of particles that showed apparent helical symmetry (see Fig. 1D, left insert) were superimposed and axially disturbed relative to one another to detect repeat structures. This was done using either two copies of the same particle image or two different images. Positions at which a repeat structure was reinforced were observed, and relative axial translation was measured.
FIG.1.
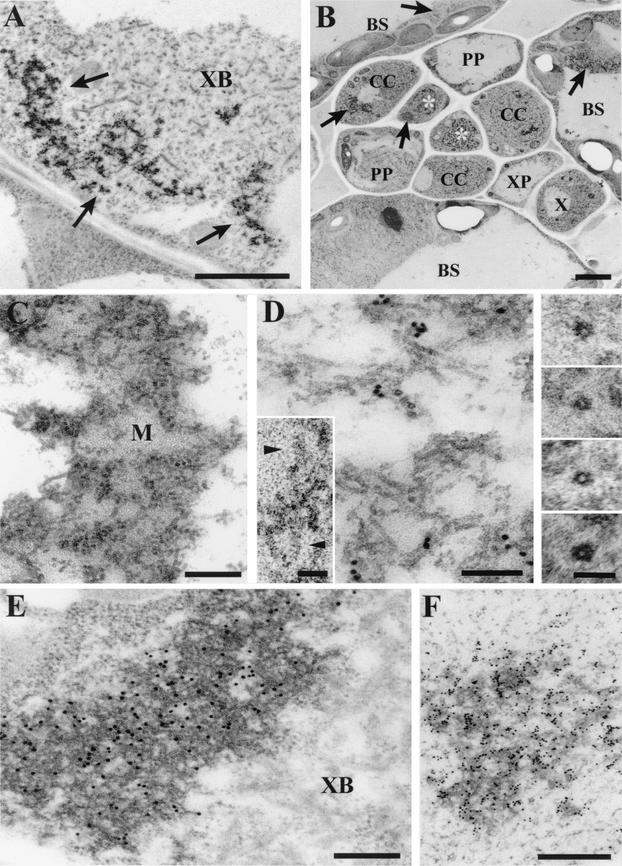
Electron micrographs showing the localization in TMV(ORF3)-infected N. benthamiana cells of RNP particles containing the ORF3 protein. (A) Section of an infected companion cell showing typical ORF3 protein-related cytoplasmic inclusions (indicated by arrows), associated with an X body (XB). Bar, 2 μm. (B) Typical section of a class V vein showing the presence of the ORF3 protein-related inclusions (indicated by arrows) in bundle sheath (BS) cells, companion cells (CC), and sieve elements (indicated by asterisks). In other sections, inclusions were also detected in all other types of cells, including phloem parenchyma (PP) and xylem parenchyma (XP) cells. X, xylem vessel. Bar, 2 μm. (C) Section showing an inclusion composed of a complex of filamentous structures embedded in an electron-dense matrix (M). Bar, 200 nm. (D) Section showing filamentous particles at higher magnification. The section was labeled by in situ hybridization with an RNA probe specific for TMV(ORF3) positive-strand RNA. Bar, 100 nm. Inserts show selected transverse sections of the filamentous particles, showing the electron-lucent central hole and parts of two particles which show helical structure (arrowheads). Bars in inserts, 50 nm. (E) Immunogold labeling of a section containing an ORF3 protein-related inclusion, using rabbit antibody against the ORF3 protein. XB, X body. Bar, 200 nm. (F) In situ hybridization, as in panel D, of a section containing an ORF3 protein-specific inclusion. Bar, 500 nm.
Immunogold labeling was done as described by Roberts (18). Mounted sections were incubated with antibody specific for GRV ORF3 protein at various dilutions for 18 h at room temperature, followed by washes with buffer and incubation with the goat anti-rabbit 15-nm-diameter gold probe for 5 h. After successive washes with buffer and distilled water, the sections were stained as described above.
For in situ hybridization, sections of material that had been embedded in L.R. white resin or a 1:1 mixture of Araldite-L.R. white were processed essentially as described by Leitch et al. (13). The sections were labeled using TMV(ORF3)-specific digoxigenin-labeled RNA probes (GRV ORF3 positive-strand-specific probe corresponding to nucleotides 2641 to 3396 of GRV RNA [ORF3] and TMV replicase gene negative-strand-specific probe corresponding to nucleotides 270 to 2675 of TMV RNA). Hybridization mixtures contained 50% formamide and 25% dextran sulfate. The probes were visualized by using antibody against digoxigenin conjugated to 10-nm-diameter gold particles. Some sections were treated with an immunogold silver staining kit (IntenSEM; Amersham International, Little Chalfont, Buckinghamshire, United Kingdom) to make the gold probe more easily visible and to allow more direct comparisons with the conventional immunogold labeling results.
The cytoplasmic RNP complexes isolated from extracts were negatively stained with 2% uranyl acetate or 2% sodium phosphotungstate, examined, and photographed in a Philips CM 10 electron microscope.
Fractionation of leaf extracts and isolation of the cytoplasmic RNP complexes.
Leaf extracts were made by grinding systemically infected N. benthamiana leaf tissues in 0.2 M Tris-HCl buffer (pH 9.0) containing 0.2 M KCl, 30 mM MgCl2, 0.2 M sucrose, and 10 mM 2-mercaptoethanol. The extracts were separated into enriched subcellular fractions by the method of Donald et al. (7). Thus, after filtering, the extract was centrifuged for 10 min at 1,000 × g to yield a low-speed pellet fraction (P1). The resulting supernatant was then centrifuged at 30,000 × g for 30 min to produce a high-speed pellet fraction (P30) and a supernatant fraction (S30). For further isolation and purification of the cytoplasmic ORF3 protein-containing RNP complexes, fraction P30 was centrifuged through a 30% sucrose cushion for 180 min at 90,000 × g. The resulting pellet was resuspended and fractionated by centrifugation for 20 h at 33,000 rpm in a Spinco L2-65B centrifuge in a Cs2SO4 density gradient generated from layers of 30 and 36% Cs2SO4 in a Beckman SW41 tube. Fractions obtained after centrifugation were dialyzed against 20 mM Tris-HCl buffer (pH 7.5) containing 10% glycerol to eliminate Cs2SO4 and analyzed by electron microscopy, Western blotting, dot blot hybridization, and infectivity assays.
Viral protein and RNA analyses.
For viral protein analysis, samples of plant tissues were ground in liquid nitrogen, resuspended in dissociation buffer (50 mM Tris-HCl [pH 6.8] containing 2 M urea, 3% sodium dodecyl sulfate [SDS], and 1.5% 2-mercaptoethanol) and incubated at 95°C for 5 min. Samples were then separated by electrophoresis in SDS-10% polyacrylamide gels using the Tris-Tricine buffer system (27). Gels were stained with Coomassie blue. For Western blot analysis, proteins were transferred to nitrocellulose membrane Protran BA85 (Schleicher & Schuell, Dietzenbach, Germany), and blots were treated with the ORF3 protein antibodies. Immunoreactions were detected using the ECL Western blotting system (Amersham Pharmacia Biotech, Uppsala, Sweden).
Total RNA was isolated from infected N. benthamiana leaf tissues using TRI reagent (Sigma, St. Louis, Mo.) according to the manufacturer's protocol. After the isopropanol precipitation, the RNA pellet was washed with 75% ethanol and dissolved in water. For dot blot hybridization analysis, samples of RNA were spotted onto Hybond N nylon membranes. Hybridization was done by the method of Sambrook et al. (25) with 32P-labeled cDNA probes corresponding to the GRV ORF3 nucleotide sequence.
Infectivity assay.
Infectivity of fractions containing TMV(ORF3) was assayed by counting necrotic lesions in inoculated leaves of Nicotiana tabacum Samsun NN. Infectivity of fractions from GRV-infected plants was assayed by counting local lesions in inoculated leaves of Chenopodium amaranticolor.
Purification and analysis of GRV ORF3-His protein.
The ORF3 protein containing a C-terminal six-histidine tag (ORF3-His) was overexpressed in N. benthamiana plants from TMV and purified by Ni2+ affinity chromatography. The purification involved the following steps: solubilization of the P1 and P30 fractions in buffer containing 8 M urea, 0.1 M NaH2PO4, and 0.01 M Tris-HCl (pH 8.0); binding to Ni2+-nitrilotriacetic acid-agarose; washing with buffer containing 8 M urea, 0.1 M NaH2PO4, and 0.01 M Tris-HCl (pH 6.3); and elution with buffers containing 8 M urea, 0.1 M NaH2PO4, and 0.01 M Tris-HCl (pH 5.9 and pH 4.5). To allow protein refolding, the solubilized protein was dialyzed sequentially against buffer consisting of 10 mM Tris-HCl (pH 7.0), 50 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, and 10% glycerol containing 4 or 1 M urea or no urea.
To analyze the ability of the ORF3 protein to form oligomers, the purified ORF3-His protein was sedimented in Beckman SW41 tubes through a 10 to 30% sucrose gradient for 20 h at 150,000 × g. Fractions obtained were analyzed by Western blotting.
For RNA-binding assays, GRV cDNA clone grpol corresponding to the 5′-terminal 2,556 nucleotides of the GRV genome (28) was transcribed in the presence of 5 μCi of [32P]UTP (Amersham Life Science, Ltd., Little Chalfont, Buckinghamshire, United Kingdom) using a mMESSAGE mMACHINE T7 kit (Ambion, Inc.). The labeled transcripts were mixed with purified ORF3-His protein in 15 μl of binding buffer A (50 mM Tris-HCl [pH 7.0], 1 mM EDTA, 50 mM NaCl, 1 mM dithiothreitol, 1 mg of bovine serum albumin [BSA] per ml, and 10% glycerol). After incubation on ice for 30 min, the mixture was subjected to electrophoresis in 1% agarose in TAE (Tris-acetate-EDTA) buffer by the method of Li and Palukaitis (14). North-Western assay was performed by the method of Kalinina et al. (12). Purified protein (1.5 μg) was subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and electroblotted onto nitrocellulose membranes. Membrane-bound protein was denatured in a mixture of 6 M urea and 0.1% Tween 20 and renatured in a solution containing 20 mM Tris-HCl (pH 7.5), 0.2 g of BSA per liter, 0.2 g of Ficoll per liter, and 0.2 g of polyvinylpyrrolidone per liter. Membranes were incubated in the same buffer with 32P-labeled RNA transcripts at room temperature and then were washed, dried, and autoradiographed. Nitrocellulose membrane filter-binding assays were performed by the method of Li and Palukaitis (14). To analyze the stability of the ORF3 protein-RNA complex, buffer A contained different concentrations of NaCl. In competition binding assays, different amounts (250 and 1,000 ng) of competitor RNA or DNA (CMV single-stranded RNA [ssRNA], TMV ssRNA, bacteriophage M13 ssDNA, simian rotavirus SA11 double-stranded RNA [dsRNA], plasmid dsDNA fragment [SmaI-digested pUC18 DNA], tRNA or rRNA hydrolyzed by NaOH [about 50 nucleotides in length]), were added to the mixtures. The mixtures were filtered through a 45-μm-pore-size nitrocellulose membrane (Schleicher and Schuell). The membranes were washed, dried, and either used for determining radioactivity by liquid scintillation counting or autoradiographed. Quantitative analysis of RNA binding was done by densitometry of the autoradiographic images with an Intelligent Quantifier, version 2.5.0 (Bio Image, Ann Arbor, Mich.).
RESULTS
Structural features and localization in infected cells of RNP particles containing the ORF3 protein.
Electron microscopy of ultrathin sections of N. benthamiana leaf tissue systemically infected with a TMV(30B) vector expressing the GRV ORF3 protein in place of its own CP [ΤMV(ORF3)] revealed characteristic cytoplasmic inclusions (Fig. 1A) that were not found in tissue infected with the empty TMV(30B) vector or with wild-type TMV. These inclusions were found in all cell types. They were present in epidermal cells and in the spongy and palisade mesophyll, as well as in the bundle sheath cells surrounding the veins. Within the vascular bundles of minor veins, the inclusions were found in immature sieve elements, phloem parenchyma and companion cells, and also in some xylem tissues (Fig. 1B). Inclusions were not found in mature sieve elements, indicating that the virus may have invaded the immature sieve element-companion cell complex while it was still symplastically connected to surrounding cells.
The GRV ORF3-related inclusions varied in size from 300 nm to 4 μm, and as many as 10 separate inclusions could be found in a single cell section. The larger inclusions were mainly found associated with the X bodies or viroplasms typical of TMV infection (Fig. 1A), which contain rod-like structures that have been described previously and that are structurally and serologically different from TMV particles (8, 11). The smaller inclusions were generally distributed elsewhere in the cytoplasm. Close examination showed the inclusions to be composed of a complex of thin filamentous structures embedded in an electron-dense matrix (Fig. 1C and D) that appeared quite different from the ground cytoplasm. The filaments were flexuous, not rigid, structures of indeterminate length up to about 200 nm (Fig. 1D). In transverse section, they had a diameter of 13 to 14 nm and appeared to have an electron-lucent central hole about 4 nm in diameter (Fig. 1D, see inserts). Center-to-center spacing of adjacent filaments (in both cross and longitudinal section) was about 20 nm. Occasionally, a helical repeat, which never exceeded four turns, could be seen on some filaments (Fig. 1D, see inserts). Linear displacement of these images showed reinforcement consistent with a helical pitch estimated to be about 6 nm. In cross sections, a single helix turn could be seen to comprise five or six subunits (Fig. 1D, see inserts). No similar filaments were found in the cytoplasm outside the inclusions or in nuclei.
Immunogold labeling of sections with antibodies against the GRV ORF3 protein showed a high specificity of labeling over the inclusions (Fig. 1E), confirming that they contain ORF3 protein. No gold label was found anywhere else in the cytoplasm, and the background level of label was very low. Close examination of many micrographs indicated that the gold particles over the inclusions were associated with the filamentous structures and were rarely found over the electron-dense matrix (Fig. 1E). Bearing in mind that the antibody-antibody-gold complexes may be >35 nm in size and therefore, that the location of the gold particles may be some distance from the antigen site, the filamentous particles with elements of helical structure may be built from ORF3 protein molecules. Neither the virus (TMV)-encoded 30-kDa MP nor the 126-kDa RNA-dependent RNA polymerase was detected in the inclusions by immunogold labeling (data not shown).
In in situ hybridization of the sections, using a probe complementary to the TMV(ORF3) positive-strand RNA, the gold label specifically targeted the ORF3 protein-associated inclusions and particularly the filaments but not the surrounding matrix material (Fig. 1D and F), indicating that TMV(ORF3) RNA molecules are incorporated in the filamentous particles together with the ORF3 protein. No in situ hybridization signal was found over the ORF3 protein-associated inclusions when a probe specific for TMV negative-strand RNA was used (data not shown).
Isolation and characterization of the ORF3 protein-containing cytoplasmic complexes.
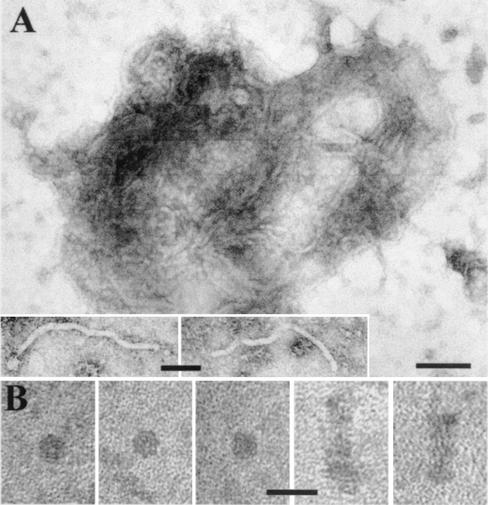
Negatively stained extracts of N. benthamiana leaf tissues systemically infected with TMV(ORF3) contained complexes of filamentous particles embedded in an electron-dense matrix (similar to those shown in Fig. 2A), which resembled the ORF3 protein-associated cytoplasmic inclusions detected in ultrathin sections and described above. The sizes of the complexes and the diameters of the individual filaments (Fig. 2A, inserts) were similar to those found in the ultrathin sections. Extracts obtained from uninfected plants or from plants inoculated with the empty vector TMV(30B) did not contain such complexes.
FIG. 2.
Electron micrographs of cytoplasmic RNP complexes isolated from N. benthamiana leaves systemically infected with TMV(ORF3). (A) Typical RNP complexes with buoyant densities in Cs2SO4 of 1.34 to 1.45 g/ml. Similar complexes were isolated from N. benthamiana plants infected with GRV. Bar, 100 nm. Inserts show individual filamentous particles within complexes. Insert bar, 50 nm. (B) Small rods and disks with buoyant densities of 1.22 to 1.29 g/ml isolated from TMV(ORF3)-infected plants. Bar, 25 nm.
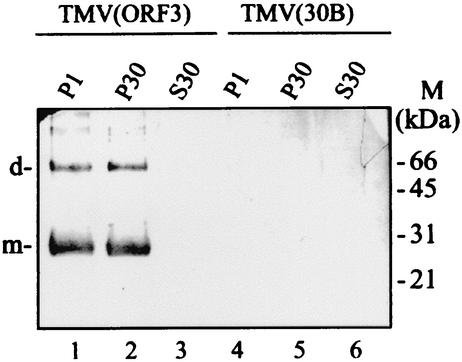
The extracts of infected plants were fractionated by differential centrifugation. Electron microscopy showed that the complexes were present in low-speed (P1) and high-speed (P30) pellet fractions, but none were detectable in the 30,000 × g supernatant fraction (S30). Western blot analyses of the fractions showed that the ORF3 protein (represented by monomers and dimers [see below]) was equally distributed between fractions P1 and P30 (Fig. 3, lanes 1 and 2), but it was not detected in fraction S30 (Fig. 3, lane 3), indicating that it is not present in the cytoplasm in a soluble nonaggregated form.
FIG. 3.
Western blot analysis of fractionated extracts from N. benthamiana plants infected with TMV(ORF3) or TMV(30B). Fractions were enriched for nuclei and chloroplasts (P1), membranes and mitochondria (P30), or soluble proteins (S30), and 20 μl of each fraction was loaded. The blot was probed with antibodies against a synthetic oligopeptide corresponding to part of the deduced sequence of the GRV ORF3 protein (see Materials and Methods). The positions of the molecular mass markers (M) and of the dimers (d) and monomers (m) of the ORF3 protein are indicated to the right and left of the blot, respectively.
Fraction P30 was centrifuged again through a 30% sucrose cushion, and the resuspended pellet was further fractionated by centrifugation in a Cs2SO4 density gradient. Electron microscopy demonstrated that the ORF3-specific complexes, such as those shown in Fig. 2A, were present in all fractions with a density between 1.27 and 1.49 g/ml but that most were found in fractions with a density between 1.34 to 1.45 g/ml. Western blotting, dot blot hybridization, and infectivity assays showed that the bulk of the ORF3 protein, TMV RNA, and infectivity cosedimented with the complexes in fractions with densities between 1.34 to 1.45 g/ml (Fig. 4A, B, and D). The appearance and composition of the complexes observed in extracts suggest that they correspond to the inclusions observed in ultrathin sections and that the filamentous particles observed in both complexes and inclusions contain both the ORF3 protein and TMV RNA. The RNA in the complexes seems to be protected against RNase attack because incubation of plant sap for 30 min at room temperature before isolation of complexes did not eliminate infectivity: the average number of local necrotic lesions in N. tabacum cv. Samsun NN leaves decreased only from 54 to 49. Treatment of the complexes with organic solvents (such as chloroform) or detergents (such as Triton X-100) decreased the infectivity (for example, for treatment with chloroform, from 49 lesions to 3), suggesting that the complexes may contain lipid. However, no membrane-like material was observed by electron microscopy associated either with the isolated complexes or with the inclusions in ultrathin sections. In some fractions with lower density (1.22 to 1.29 g/ml), small rods and disks with a diameter of 13 to 14 nm could sometimes be observed (Fig. 2B and 4A) and may represent products of disintegration of the cytoplasmic complexes.
FIG. 4.
Fractionation of the cytoplasmic RNP complexes isolated from plants infected with TMV(ORF3) by Cs2SO4 density gradient centrifugation. (A) Diagram showing density (ρ) gradient in the fractions obtained after centrifugation. The positions of the rods and disks (rods) and cytoplasmic RNP complexes (RNP-C) determined by electron microscopy and of fractions possessing infectivity are indicated. (B) Western blot analysis of the fractions, using antibody against the GRV ORF3 protein. The positions of the dimers (d) and monomers (m) of the ORF3 protein are indicated. Fraction b (bottom) is a sample of the resuspended pellet from the gradient. (C) Electrophoretic protein analysis (SDS-PAGE) of a partially purified preparation of the cytoplasmic RNP complexes (pooled fractions 11 to 14). Gel was stained with Coomassie blue (lane 1) or analyzed by Western blotting as in panel B (lane 2). The positions of the molecular mass markers (M) and of the dimers (d) and monomers (m) of the ORF3 protein are indicated to the right and left of the gel, respectively. (D) Dot blot hybridization analysis of TMV(ORF3) RNA contained in combined fractions 3 to 6, 7 to 10, and 11 to 14. Hybridization was with a probe specific to the ORF3 nucleotide sequence.
Although cytoplasmic inclusions containing filamentous RNP particles were not observed in ultrathin sections of N. benthamiana plants infected with GRV, complexes consisting of filamentous particles embedded in a matrix similar to those shown in Fig. 2A were detected in extracts of these plants (data not shown). Like the complexes in extracts from TMV(ORF3)-infected plants, those from GRV-infected plants had a buoyant density of about 1.34 to 1.45 g/ml, and fractions containing them also contained ORF3 protein and virus (GRV) RNA and were infective. However, the complexes were present in much smaller amounts in extracts from GRV-infected plants, presumably because much less protein is produced from ORF3 in its natural context than when it is coupled to the TMV CP promoter.
The nature of the matrix in the complexes remains obscure. SDS-PAGE of partially purified preparations of the complexes from TMV(ORF3)-infected plants (fractions with a density 1.34 to 1.45) revealed several protein bands in addition to the ORF3-specific bands (representing monomers and dimers [see below]) (Fig. 4C).
Properties of the ORF3 protein in vitro.
The chimeric virus TMV(ORF3-His) is identical to TMV(ORF3) except that it expresses the GRV ORF3 protein tagged with six histidine residues (ORF3-His). In preliminary experiments, it was shown that this construct was able to mediate protection and long-distance movement of the viral RNA in the same way as TMV(ORF3), indicating that the main functional properties of the ORF3 protein are not affected by addition of the histidine residues. Preparations of the ORF3-His protein from N. benthamiana plants infected with TMV(ORF3-His) gave two major bands (about 28 and 56 kDa) on SDS-polyacrylamide gels. Western blot analysis revealed that both of these bands reacted with an antibody against the ORF3 protein, suggesting that they represent monomer (28-kDa) and dimer (56-kDa) forms of the ORF3-His protein (Fig. 5A). In some preparations of the ORF3-His protein, a trimer (84 kDa) was also detected (data not shown). Two bands, corresponding to the monomer (27-kDa) and dimer (54-kDa) forms of the native (nontagged) ORF3 protein, were also detected by Western blotting in extracts obtained from plants infected with GRV or ΤMV(ORF3) (Fig. 3 and 4B), indicating that oligomerization is not a consequence of tagging the protein with histidine residues or of the purification procedure. In nondenaturing conditions in a 10 to 30% sucrose gradient, the purified ORF3-His protein sedimented in positions expected for dimers, trimers, tetramers, pentamers, and oligomers consisting of more than 20 ORF3-His protein molecules (Fig. 5B), confirming its strong tendency to aggregate. All forms of the ORF3-His protein separated by sucrose gradient centrifugation behaved as a mixture of monomers and dimers in subsequent SDS-PAGE (Fig. 5B). Moreover, reelectrophoresis of either monomer or dimer bands isolated from a gel gave rise to both monomers and dimers in SDS-PAGE (data not shown), indicating that the formation of oligomers of ORF3-His protein at least partly involves noncovalent bonds between subunits.
FIG. 5.
Oligomerization of the ORF3 protein in vitro. (A) Western blot analysis, using antibody against the GRV ORF3 protein, of the recombinant ORF3-His protein isolated from TMV(ORF3-His)-infected plants and fractionated by SDS-PAGE (lane 1). Control sample isolated from TMV(30B)-infected plants was loaded in lane 2. The positions of the molecular mass markers (M) and of the dimers (d) and monomers (m) of the ORF3 protein are indicated to the right and left of the blot, respectively. (B) Sedimentation analysis of the ORF3-His protein oligomers in a 10 to 30% sucrose density gradient. Fractions obtained after centrifugation were subjected to Western blot analysis as in panel A. The positions in the gradient of the molecular mass markers (M) (carbonic anhydrase [29 kDa], BSA [66 kDa], alcohol dehydrogenase [150 kDa], β-amylase [200 kDa], and apoferritin [443 kDa]) are shown above the blot. The positions of dimers (d) and monomers (m) of the ORF3-His protein are indicated to the right of the blot.
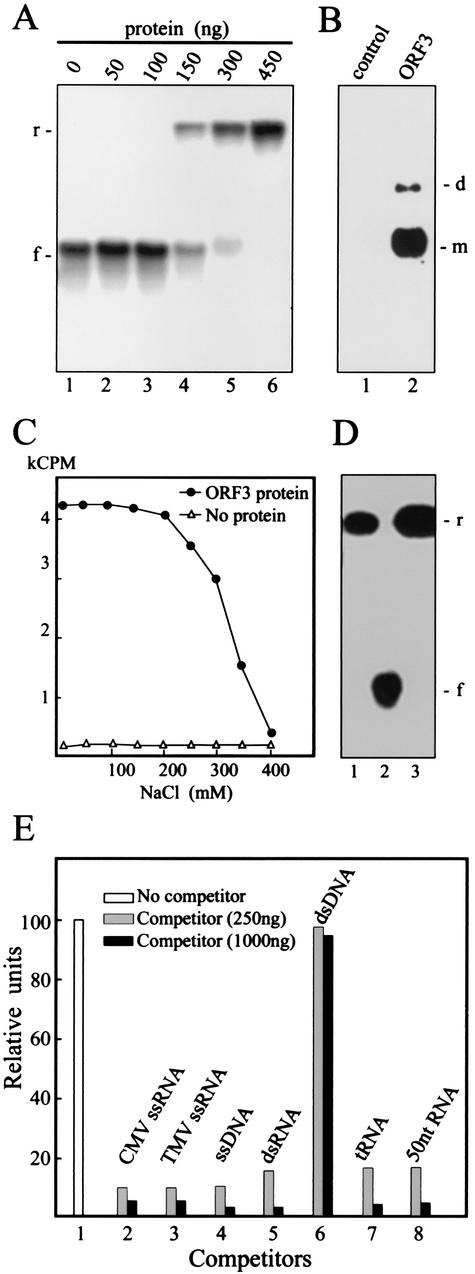
Binding of the ORF3-His protein to ssRNA was detected by electrophoretic retardation in nondenaturing agarose gels. Figure 6A illustrates the binding of the protein to 32P-labeled ssRNA corresponding to the 5′-terminal 2,556 nucleotides of the GRV genome. At low ORF3-His protein/RNA ratios, most of the labeled GRV RNA migrated as protein-free RNA (Fig. 6A), but when the amount of protein was increased (150 to 300 ng or more), the RNA barely entered the gel (Fig. 6A), indicating that it had formed large complexes with the ORF3-His protein. Moreover, the rapid shift of mobility with increasing protein concentration and the absence of labeled RNA with a mobility intermediate between these two extremes (Fig. 6A) showed that binding to the labeled RNA probe occurred in a highly cooperative fashion. If one or a few molecules of ORF3-His protein are bound to a labeled RNA molecule, this RNA molecule appears to become a preferential substrate for rapid binding of additional protein molecules. Although the cooperative binding was observed only at comparatively high protein/RNA ratios (75:1, wt/wt), this may not be a true reflection of the relative concentrations of the reacting species. As shown above, ORF3-His protein exists in solution as a mixture of monomers and oligomers, and not all of these forms may be capable of binding to RNA.
FIG. 6.
Analysis of nucleic acid binding by the ORF3-His protein. (A) Gel retardation electrophoresis assay for RNA binding by the ORF3 protein. Increasing amounts of the ORF3 protein were incubated in 15 μl of binding buffer A (see Materials and Methods) with 2ng of 32P-labeled GRV RNA transcript, and the mixtures were electrophoresed in 1% nondenaturing agarose gel. The amount of the ORF3 protein used in the assay is indicated above the lanes. The positions of free (f) and retarded (r) RNAs are shown to the left of the gel. (B) North-Western blot analysis of RNA binding by the ORF3 protein. After SDS-PAGE, the ORF3 protein was transferred onto a nitrocellulose membrane and renatured. The membrane was treated with 32P-labeled GRV RNA transcript. The ORF3 protein was substituted by BSA mixed with an equivalent protein preparation from plants infected with the empty TMV(30B) vector as a control. The positions of dimers (d) and monomers (m) are indicated to the right of the blot. (C) Salt stability of RNA-ORF3 protein complex. The ORF3 protein (450 ng) was incubated with labeled GRV RNA (4 ng) in the presence of the indicated concentrations of NaCl. After incubation, the mixtures were analyzed by nitrocellulose membrane filter-binding assay. RNA binding was quantified by determining the radioactivity of the membrane by liquid scintillation counting. kCPM, 1,000 cpm. (D) Gel retardation electrophoresis assay for RNA binding by the ORF3 protein (400 ng) as in panel A but in the presence of 350 mM (lane 1) or 200 mM (lane 3) NaCl. Lane 2 contained RNA without the ORF3 protein, preincubated in 350 mM NaCl. The positions of free (f) and retarded (r) RNAs are shown to the right of the gel. (E) Competition binding assay of the ORF3 protein to GRV ssRNA. The protein (450 ng in 15 μl) was incubated with labeled GRV RNA (4 ng) either in the absence (column 1) or in the presence of the following unlabeled competitors: CMV ssRNA (column 2), TMV ssRNA (column 3), bacteriophage M13 ssDNA (column 4), simian rotavirus SA11 dsRNA (column 5), plasmid dsDNA fragment (SmaI-digested pUC18 DNA) (column 6), tRNA (column 7), and rRNA hydrolyzed by NaOH (about 50 nucleotides in length) (column 8). The amounts of the competitors are indicated. After incubation, the mixtures were analyzed by the nitrocellulose membrane filter-binding assay. RNA binding was quantified by densitometry of autoradiographic images in relative units (100 units corresponds to RNA binding by the ORF3 protein without competitor).
To verify the ability of the ORF3-His protein (and not just undetected contaminating plant proteins) to bind ssRNA, North-Western blot experiments were done. After SDS-PAGE, the purified protein was transferred onto a nitrocellulose membrane and renatured. The membrane was then incubated with a 32P-labeled RNA probe, and binding was analyzed by autoradiography. As shown in Fig. 6B, two bands could be seen in the positions expected for the ORF3-His protein monomers and dimers. In control experiments using an equivalent protein preparation from plants infected with the empty TMV(30B) vector together with BSA to give an equal protein concentration, no RNA binding was observed (Fig. 6B). As the recombinant ORF3 protein tested in these experiments contained a His tag, it was possible that RNA-binding activity was due to a nonspecific effect of the additional His residues. However, the North-Western blot assay results showed that the ORF3 protein expressed in plants from TMV(ORF3) without the His tag and present in the cytoplasmic RNP complexes also bound to the RNA probe (data not shown), suggesting that RNA-binding activity of the ORF3 protein is independent of the presence or absence of a His tag.
The stability of protein-RNA complexes with respect to salt concentration is often used as a criterion to measure the strength of protein-RNA association. 32P-labeled GRV RNA and the ORF3-His protein were incubated at different salt concentrations, and the incubation mixtures were filtered through membranes and washed to remove unbound RNA. As shown in Fig. 6C, maximal complex formation was observed at NaCl concentrations of up to 200 mM; at higher salt concentrations, the activity was inhibited, disappearing completely at 400 mM. The ability of the ORF3-His protein to bind RNA at high NaCl concentrations (200 or 350 mM) was also shown by using the electrophoretic retardation assay (Fig. 6D).
To determine whether the binding of ORF3-His protein to nucleic acid is sequence specific, competition binding assays were performed by incubating the ORF3-His protein with 32P-labeled GRV RNA transcript and unlabeled competitor nucleic acids of various types and sizes and analyzing the incubation mixture using nitrocellulose membrane-binding assays (Fig. 6E). Full-length single-stranded nucleic acids (such as TMV RNA or CMV RNA) and short RNAs (such as tRNA) or fragments of rRNA hydrolyzed by NaOH (about 50 nucleotides in length) were able to compete efficiently with the labeled transcript for binding to the ORF3-His protein. Bacteriophage M13 ssDNA or simian rotavirus SA11 dsRNA were also able to compete, whereas dsDNA (SmaI-digested pUC18 plasmid DNA) was not (Fig. 6E). Thus, the ORF3-His protein apparently binds to ssRNA, ssDNA, and dsRNA without sequence specificity.
DISCUSSION
It has been shown that despite the lack of virus particles, umbraviral RNA remains infective in crude sap extracts for several hours (15, 16). In contrast, the stability of deproteinized umbraviral RNA is no greater than that of any other ssRNA. Recently, using a gene replacement approach, we have demonstrated that the ORF3 protein encoded by umbraviruses can stabilize TMV RNA in the absence of the TMV CP (23). The ORF3 protein was also able to facilitate rapid long-distance spread of TMV RNA, presumably through the phloem, when it replaced CP in the hybrid virus TMV(ORF3) (22, 23). In this paper, direct experimental evidence is presented that during systemic infection by TMV(ORF3), both ORF3 protein and viral RNA are detected in all types of phloem-associated cells, including phloem parenchyma, companion cells, and sieve elements, which is consistent with the idea that the ORF3 protein facilitates long-distance RNA spread through the phloem. Within the phloem of minor veins, nucleoprotein complexes were present in immature sieve elements, indicating an early invasion of the phloem in sink leaves (17).
Confocal laser scanning microscopy showed that in epidermal and mesophyll cells, GRV ORF3 protein fused with GFP and expressed from a TMV vector was localized in cytoplasmic granules that were often associated with TMV-specific amorphous X bodies but were also distributed elsewhere in the cytoplasm in association with membrane structures, possibly sheets of the endoplasmic reticulum (reference 20 and our unpublished results). These cytoplasmic granules presumably correspond to the ORF3 protein-containing cytoplasmic inclusions observed in ultrathin sections and to the ORF3-associated complexes found in extracts. Although in this study we were unable to confirm directly the association of these complexes with membranes, possibly because of the procedure used to prepare the samples for electron microscopy, the observation that infectivity of extracts containing the cytoplasmic complexes was significantly decreased by treatment with organic solvents or detergents strongly suggests the association of some membrane-like material with them. Reddy et al. (16) also found that GRV infectivity in leaf extracts was abolished by treatment with organic solvents, as did Murant et al. (15) for Carrot mottle virus, another umbravirus.
The cytoplasmic complexes detected in this study were composed of filamentous RNP particles consisting of the ORF3 protein and viral RNA, embedded in an electron-dense matrix. This is the first report of visualization in vivo of RNP particles other than virions formed by plant viruses. The length of the RNP particles in ultrathin sections varied widely but rarely exceeded 200 nm. However, infectivity of the isolated complexes indicated that full-length TMV(ORF3) RNA molecules were present in the sections. This suggests that the RNP particles are quite flexible and never lie completely in the plane of a section. The filamentous RNP particles had elements of helical structure which never exceeded four turns. Thus, these particles resemble other virus-specific filamentous particles, such as the virions of potexviruses, but are not as uniform as them. These RNP particles are a novel form which may be used by umbraviruses as an alternative to classical virions. In vitro experimental results showed that binding of ORF3 protein to RNA is a highly cooperative process, but it seems to lead to a less well-ordered structure than does the assembly of CP and RNA into conventional virions. Moreover, it is still unclear if the RNA molecule is located inside or outside the protein skeleton of the RNP particles. Despite the apparent imperfection of the RNP structures, the RNA associated with them is protected from degradation. However, the RNP particles were always found within the cytoplasmic inclusions that were stable in tissue extracts, and the structure of the whole complex, including the matrix in which the RNP particles are embedded, may provide the RNA molecules with an additional level of protection against RNases.
The cytoplasmic RNP complexes are the only form of interaction between viral RNA and ORF3 protein that has been identified; therefore, it is tempting to suggest that they are involved in protection or stabilization of the RNA in all infected tissues and possibly in movement of the RNA through the phloem. The ORF3 protein is not required for movement through plasmodesmata between mesophyll cells; that function is provided by the ORF4 protein. Therefore, the ORF3 protein probably functions in protecting viral RNA during transit in the phloem. It is possible that the filamentous particles comprising RNA and the ORF3 protein are the form in which the viral genome passes through plasmodesmata modified by the ORF4 protein to permit movement, although ORF3 protein itself was not detected in transit within plasmodesmata by immunogold labeling. Significantly, the filamentous particles were never observed outside inclusions in the ultrathin sections and very rarely in the preparations of the RNP complexes; therefore, it seems that after passage of the viral genome through plasmodesmata, the complexes are immediately reassembled.
The ability to extract from GRV-infected tissue infective cytoplasmic ORF3 protein-containing RNP complexes with a structure similar to those from TMV(ORF3)-infected tissue suggests that they are not mere artifacts of ORF3 overexpression and that these complexes protect and traffic TMV and GRV RNAs by a common mechanism. The properties of the ORF3 protein in vitro, such as cooperative binding to RNA and oligomerization, are consistent with its direct role in the formation of the filamentous particles in vivo. Detection of the ORF3 protein as dimers (and sometimes trimers) in denaturing conditions (i.e., in the presence of SDS) suggested that its tendency to aggregate is strong. There was no indication that this aggregation involved covalent bonding, and other examples of noncovalent, SDS-resistant protein-protein interactions have been reported (1, 9).
Formation of the cytoplasmic RNP complexes may also be involved in protecting viral RNA from the plant's defensive RNA silencing response, which is based on a sequence-specific degradation of foreign, and in particular, viral RNA molecules. A key feature of the RNA silencing pathway is the generation of dsRNA that corresponds in sequence to the target (virus) RNA. This dsRNA is cleaved into short interfering RNAs 21 to 25 nucleotides in length and these are thought to mediate the target specificity for RNA degradation (for recent reviews, see references 3, 30, and 31). To combat host defense RNA silencing, some plant viruses encode silencing suppressor proteins that also block posttranscriptional gene silencing in transgenic plants. No suppressor activity has been associated with umbraviruses, and in particular the ORF3 protein does not suppress posttranscriptional gene silencing (24), suggesting that umbraviruses may possess another mechanism to escape from RNA silencing. One possible mechanism could be based on sequestration of viral RNA incorporated into the ORF3 cytoplasmic complexes, making it invisible to the silencing machinery.
GFP-labeled ORF3 protein was also found in nuclei, preferentially targeting nucleoli (20), but no ORF3 protein-associated RNP structures were identified in ultrathin sections of nuclei. Thus, the ORF3 protein in nuclei is probably in a different form from that in the cytoplasm, and the role of its nuclear or nucleolar localization is still unclear.
Acknowledgments
This work was supported in part by a grant-in-aid from the Scottish Executive Environment and Rural Affairs Department (SEERAD), by the Royal Society (Programme for Joint Projects with FSU and Ex-quota visit fellowship to N.K.), and by a Department of Biotechnology Fellowship (from the Ministry of Science and Technology of the Government of India to S.K.R.).
REFERENCES
- 1.Bentley, M., M. J. Ladu, C. Rajan, G. S. Getz, and C. A. Reardon. 2002. Apolipoprotein E structural requirements for the formation of SDS-stable complexes with beta-amiloid-(1-40): the role of salt bridges. Biochem. J. 366:273-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Canto, T., D. A. M. Prior, K.-H. Hellwald, K. J. Oparka, and P. Palukaitis. 1997. Characterization of cucumber mosaic virus. IV. Movement protein and coat protein are both essential for cell-to-cell movement of cucumber mosaic virus. Virology 237:237-248. [DOI] [PubMed] [Google Scholar]
- 3.Carrington, J. C. 2000. RNA silencing: moving targets. Nature 408:150-151. [DOI] [PubMed] [Google Scholar]
- 4.Carrington, J. C., K. D. Kasschau, S. K. Mahajan, and M. C. Schaad. 1996. Cell-to-cell and long-distance transport of viruses in plants. Plant Cell 8:1669-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demler, S. A., D. G. Rucker, and G. A. de Zoeten. 1993. The chimeric nature of the genome of pea enation mosaic virus: the independent replication of RNA-2. J. Gen. Virol. 74:1-14. [DOI] [PubMed] [Google Scholar]
- 6.Demler, S. A., O. N. Borkhsenious, D. G. Rucker, and G. A. de Zoeten. 1994. Assessment of the autonomy of replicative and structural functions encoded by the luteo-phase of pea enation mosaic virus. J. Gen. Virol. 75:997-1007. [DOI] [PubMed] [Google Scholar]
- 7.Donald, R. G. K., H. Zhou, and A. O. Jackson. 1993. Serological analysis of barley stripe mosaic virus-encoded proteins in infected barley. Virology 195:659-668. [DOI] [PubMed] [Google Scholar]
- 8.Esau, K., and J. Cronshaw. 1967. Relation of tobacco mosaic virus with host cells. J. Cell Biol. 33:665-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gentile, F., P. Amodeo, F. Febbraio, F. Picaro, A. Motta, S. Formisano, and R. Nucci. 2002. SDS-resistant active and thermostable dimers are obtained from the dissociation of homotetrameric β-glycosidase from hyperthermophilic Sulfolobus solfataricus in SDS. Stabilizing role of the A-C intermonomeric interface. J. Biol. Chem. 277:44050-44060. [DOI] [PubMed] [Google Scholar]
- 10.Gibbs, M. G., J. I. Cooper, and P. M. Waterhouse. 1996. The genome organization and affinities of an Australian isolate of carrot mottle umbravirus. Virology 224:310-313. [DOI] [PubMed] [Google Scholar]
- 11.Hull, R. 2002. Matthews' plant virology. Academic Press, Ltd., London, United Kingdom.
- 12.Kalinina, N. O., O. N. Fedorkin, O. V. Samuilova, E. Maiss, T. Korpela, S. Y. Morozov, and J. G. Atabekov. 1996. Expression and biochemical analyses of the recombinant potato virus X 25K movement protein. FEBS Lett. 397:75-78. [DOI] [PubMed] [Google Scholar]
- 13.Leitch, A. R., T. Schwargachas, D. Jackson, and I. I. Leitch. 1994. In situ hybridisation. RMS Microscopy handbook. BIOS Scientific Publishers, Oxford, United Kingdom.
- 14.Li, Q., and P. Palukaitis. 1996. Comparison of the nucleic acid- and NTP-binding properties of the movement protein of cucumber mosaic cucumovirus and tobacco mosaic tobamovirus. Virology 216:71-79. [DOI] [PubMed] [Google Scholar]
- 15.Murant, A. F., R. A. Goold, I. M. Roberts, and J. Cathro. 1969. Carrot mottle-a persistent aphid-borne virus with unusual properties and particles. J. Gen. Virol. 4:329-341. [Google Scholar]
- 16.Reddy, D. V. R., A. F. Murant, J. H. Raschké, M. A. Mayo, and O. A. Ansa. 1985. Properties and partial purification of infective material from plants containing groundnut rosette virus. Ann. Appl. Biol. 107:65-78. [Google Scholar]
- 17.Roberts, A. G., S. Santa Cruz, I. M. Roberts, D. A. M. Prior, R. Turgeon, and K. J. Oparka. 1997. Phloem unloading in sink leaves of Nicotiana benthamiana: comparison of a fluorescent solute with a fluorescent virus. Plant Cell 9:1381-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts, I. M. 1994. Factors affecting the immunogold labelling of plant virus antigens in thin sections. J. Virol. Methods 50:155-166. [DOI] [PubMed] [Google Scholar]
- 19.Roberts, I. M. 2002. Iso-butanol saturated water: a simple procedure for increasing staining intensity of resin sections for light and electron microscopy. J. Microsc. 207:97-107. [DOI] [PubMed] [Google Scholar]
- 20.Ryabov, E. V., K. J. Oparka, S. Santa Cruz, D. J. Robinson, and M. E. Taliansky. 1998. Intracellular location of two groundnut rosette umbravirus proteins delivered by PVX and TMV vectors. Virology 242:303-313. [DOI] [PubMed] [Google Scholar]
- 21.Ryabov, E. V., I. M. Roberts, P. Palukaitis, and M. Taliansky. 1999. Host-specific cell-to-cell and long-distance movements of cucumber mosaic virus are facilitated by the movement protein of groundnut rosette virus. Virology 260:98-108. [DOI] [PubMed] [Google Scholar]
- 22.Ryabov, E. V., D. J. Robinson, and M. E. Taliansky. 1999. A plant virus-encoded protein facilitates long-distance movement of heterologous viral RNA. Proc. Natl. Acad. Sci. USA 96:1212-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryabov, E. V., D. J. Robinson, and M. E. Taliansky. 2001. Umbravirus-encoded proteins both stabilize heterologous viral RNA and mediate its systemic movement in some plant species. Virology 288:391-400. [DOI] [PubMed] [Google Scholar]
- 24.Ryabov, E. V., G. Fraser, M. A. Mayo, H. Barker, and M. E. Taliansky. 2001. Umbravirus gene expression helps Potato leafroll virus to invade mesophyll tissues and to be transmitted mechanically between plants. Virology 286:363-372. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Santa Cruz, S., A. G. Roberts, D. A. M. Prior, S. Chapman, and K. J. Oparka. 1998. Cell-to-cell and phloem-mediated transport of potato virus X: the role of virions. Plant Cell 10:495-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schagger, H., and G. von Jagow. 1987. Tricine sodium dodecyl sulfate polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 kDa to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 28.Taliansky, M. E., D. J. Robinson, and A. F. Murant. 1996. Complete nucleotide sequence and organization of the RNA genome of groundnut rosette umbravirus. J. Gen. Virol. 77:2335-2345. [DOI] [PubMed] [Google Scholar]
- 29.Taliansky, M. E., D. J. Robinson, and A. F. Murant. 2000. Groundnut rosette disease virus complex: biology and molecular biology. Adv. Virus Res. 55:357-400. [DOI] [PubMed] [Google Scholar]
- 30.Vance, V. B., and H. Vaucheret. 2001. RNA silencing in plants: defense and counterdefense. Science 292:2277-2280. [DOI] [PubMed] [Google Scholar]
- 31.Voinnet, O. 2001. RNA silencing as a plant immune system against viruses. Trends Genet. 17:449-459. [DOI] [PubMed] [Google Scholar]
- 32.Wellink, J., and A. van Kammen. 1989. Cell-to-cell transport of cowpea mosaic virus requires both the 58K/48K proteins and the capsid proteins. J. Gen. Virol. 70:2279-2286. [Google Scholar]