Abstract
The C-terminal 500 amino acids of herpes simplex virus type 1 ICP4 are required for full activator function and viral growth and are known to participate in interactions consistent with the role of ICP4 as an activator of transcription. Oligonucleotide mutagenesis was used to target stretches of amino acids that are conserved with the ICP4 analogs of other alphaherpesviruses and were also predicted to be exposed on the surface of the molecule. Seven mutants were isolated that possessed one to three amino acid changes to the residue alanine in four regions between residues 1000 and 1200. The mutants generated were analyzed first in transfection assays and subsequently after introduction into the viral genome. A number of phenotypes representing different degrees of functional impairment were observed. In transient assays conducted at 37°C, mutant M2 was indistinguishable from wild-type ICP4. Mutants M6 and M7 were marginally impaired. M3, M4, and M5 were more significantly impaired but still able to activate transcription, and M1 was completely impaired. In the context of the viral genome, M1, M3, and M7 were found to be temperature sensitive for growth. All three overproduced immediate-early (IE) proteins at the nonpermissive temperature (NPT). M3 and M7 produced early but not late proteins, and M1 produced neither early nor late proteins, at the NPT. The ICP4 proteins synthesized by all of the mutants tested were able to bind to specific ICP4 binding sites in electrophoretic mobility shift experiments. However, the DNA-protein complexes formed with the ICP4 from M1, M3, or M7 produced at the NPT possessed altered mobility. These complexes were not supershifted by a monoclonal antibody that recognizes an epitope in the C terminus; however, they were supershifted by a monoclonal antibody that recognizes the N terminus. The results suggest that the mutant forms of ICP4, while able to bind to DNA, are conformationally altered at the NPT, thus impairing the ability of the protein to activate transcription to different extents. The complete lack of ICP4 function characteristic of the M1 protein, and the inability of all the mutants to attenuate IE gene expression, suggest that the mutations additionally affect functions of the N terminus to different extents.
The first five gene products of herpes simplex virus type 1 (HSV-1) to be expressed during infection of permissive cells are referred to as the immediate early, or IE, genes (27, 45, 49). The IE genes are transcriptionally activated by VP16 (6), which is delivered by the virion (3, 46) and binds as part of a complex (32, 37, 40) to specific sequence elements in the upstream regions of IE promoters (9, 35). The protein product of one IE gene, infected cell polypeptide 4 (ICP4), is required for the induced transcription of most of the remaining HSV genes (59). In the absence of ICP4, the early (E) genes are poorly expressed, and the remaining IE genes are overexpressed relative to those of wild-type (wt) virus (14, 18, 47). The overexpression of IE genes in the absence of ICP4 in part reflects the ability of ICP4 to repress transcription in certain contexts (15, 21, 26, 41). VP16 and ICP4 are the only known HSV proteins to function at the level of transcription initiation, acting sequentially in their contributions to the lytic cascade of viral gene expression.
ICP4 is a large and structurally complex molecule. Its mobility on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels corresponds to a molecular weight of 175 kDa (11), and it exists in cells as a ∼350-kDa dimer (38, 55). It has a Stokes radius of ∼90 Å, and its hydrodynamic properties suggest that it is a very elongated molecule (38, 55). This and the ability of ICP4 to bind to DNA (50) may enable it to function as an activator of transcription over long distances. Furthermore, ICP4 localizes in infected cell nuclei (11) in viral replication compartments (51), where it can be found with viral DNA, RNA polymerase II, and ICP22 (34).
HSV promoters are transcribed by cellular RNA polymerase II (1, 10). In reconstituted in vitro transcription reactions, ICP4 has been shown to activate transcription of a core RNA polymerase II promoter with a relatively simple set of polymerase II general transcription factors (7, 8, 24). ICP4 can form tripartite complexes with TFIID and TFIIB on DNA containing a TATA box and a properly positioned ICP4 binding site (33, 57). It can interact with TFIID in solution via its TAF250 subunit (8). ICP4 promotes the formation of transcription preinitiation complexes by enhancing TFIID binding to the promoter (22). These activities are consistent with the transcriptional regulatory activities and genetics of ICP4 (8, 22, 25).
HSV-1 ICP4 contains 1,298 amino acids (36). All the alphaherpesviruses have analogs of ICP4, and while there is very little nucleotide sequence homology among the analogs, there are easily recognizable blocks of amino acid similarity (36). These regions of amino acid similarity correspond to regions that are important for ICP4 function during infection (17, 42, 43, 54). While the amino-terminal 500 amino acids of ICP4 contain blocks of similarity, the carboxyl-terminal 500 amino acids contain the greatest degree of amino acid similarity among the analogs. ICP4 mutants consisting of the amino-terminal ∼800 amino acids are sufficient to activate transcription in transient assays (16), during virus infection (17), and in reconstituted in vitro transcription reactions (8). However, activation by such mutants is not as efficient as that by full-length ICP4, and viruses containing these mutations do not grow (17). Therefore, the large regions of amino acid similarity in the carboxyl terminus of ICP4 must specify functions that are important for activation and thus viral growth.
In an effort to dissect the functions of the carboxyl terminus, we used an oligonucleotide mutagenesis procedure to target conserved regions within the C terminus. The oligonucleotides were designed to change conserved residues to alanines and also to introduce new restriction sites in order to follow the mutations through plasmid and virus construction. Much of the conservation in the carboxyl-terminal region is in hydrophobic domains that are predicted to be in the interior of the molecule. Changes in such regions would most probably destabilize the structure of the entire molecule and provide little useful information. Therefore, we targeted hydrophilic regions in an effort to affect structures involved in interactions that directly affect the function of ICP4. The mutants generated were analyzed in transient assays for the ability to activate a reporter gene and the ability to complement the growth of an ICP4 deletion mutant. Five of the mutants were also recombined into the HSV genome at both ICP4 loci. The viral mutants were analyzed with respect to their growth characteristics, their ability to bind to DNA, and changes in the regulated cascade of HSV gene expression.
MATERIALS AND METHODS
Cells and viruses.
Monolayer cultures of Vero and E5 cells were maintained by standard procedures in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. E5 cells are transformed Vero cells that express complementing levels of wt ICP4 upon HSV-1 infection (14, 16). The HSV-1 strains KOS (wt) and Elmo were used as the parental viruses for generation of recombinant viruses. Elmo is a recombinant HSV that has the green fluorescent protein (GFP) driven by the human cytomegalovirus IE promoter inserted into both copies of ICP4 (M. Evans and N. DeLuca, unpublished data). The d120 mutant of HSV-1 has large deletions in both genes for ICP4 (14). For growth and burst size experiments, monolayers of 5 × 105 Vero cells in 35-mm-diameter culture dishes were exposed to 0.1 ml of virus (multiplicity of infection [MOI], 10 PFU/cell) for 1 h. After the 1-h adsorption period, the cells were washed twice with Tris-buffered saline (TBS) and incubated for the desired times at the indicated temperatures. Following incubation, the monolayers were scraped into the medium, subjected to two freeze-thaw cycles, sonicated, and titered by plaque assay on monolayers of E5 cells.
DNA isolation.
All plasmids were purified by the method described by Birnboim and Doly (4). To obtain infectious viral DNA, subconfluent monolayers of Vero or E5 cells were infected at an MOI of 0.1 PFU/cell with strain KOS or Elmo, respectively. After viral cytopathic effect (CPE) was generalized, the infected cells and the supernatant were concentrated by centrifugation, combined, and incubated at 37°C in TE (10 mM Tris-HCl-1 mM EDTA [pH 8.0]) containing 0.6% SDS and 0.4 mg of proteinase K/ml for at least 4 h. Following incubation, CsCl was added to give a final concentration of 57% (wt/wt). The precipitated SDS was removed by floating the precipitate by low-speed centrifugation. The remaining clarified solution was spun at 35,000 rpm in a Beckman 90 Ti rotor for 65 h at 25°C. Fractions were collected from the bottom, and the tube containing the viral DNA was inferred from the refractive index of 10 μl of each fraction and by an ethidium bromide (EtBr)-stained dot blot for DNA. Viral DNA was then dialyzed against TE and quantified. Viral DNA minipreps were prepared from culture tubes of 105 cells inoculated with a single plaque. When CPE was generalized, 500 μl of the virus-cell suspension was pelleted for 10 min at 12,000 × g. The pellets were washed with the same volume of ice-cold TBS and resuspended in 450 μl of TE. SDS (0.6%) and proteinase K (0.4 mg/ml) were added, and the samples were incubated for 6 h at 37°C. The mixture was then extracted twice with a mixture of phenol-chloroform-isoamyl alcohol (25:24:1) and once with chloroform-isoamyl alcohol (24:1) and was then precipitated with ethanol at −20°C for 30 min. The precipitate was spun for 15 min at 12,500 × g, washed with 70% ethanol, air dried, and resuspended in 100 μl of TE buffer.
Introduction of mutants into the ICP4 gene.
Construction of ICP4 mutants was based on subcloning of a 903-bp MscI fragment of the ICP4 gene (amino acids [aa] 969 to 1270) from pK1-2 into pUC-18 at the SmaI restriction site in the polylinker. pK1-2 contains the promoter and entire ICP4 coding sequence from the KOS strain of HSV-1 cloned into pUC-8 (16). The desired mutations were introduced into the plasmid containing the 903-bp MscI fragment by using the Gene Editor In Vitro Site-Directed Mutagenesis system (Promega) and the 5′-phosphorylated mutagenic oligonucleotides PC-1 (GCGCGCCTGCGCATGGCCCGCCGCGGCGCCCGCGGTGTCG), PC-2 (CGTGCAGTGCGCAGTGGCCTGGCCGGCGGC), PC-3 (GCCATGGCGCCCGCGGCGCCGGCCTTCTGCGAGG), PC-4 (GACTTCTGCGAGGCGGAGGCCGCCTCGGCCGCCTGCGCGCG), PC-5 (GCCGCCTGCGCGGCCTGGGGCCTGGCGGCGCCGCTGCGG), PC-6 (CATGTCCCCGGCCGAGTACCGCCG), and PC-7 (CTTCGGCCCGGACGCACCGGTGCCCATG). The changed nucleotides (relative to the wt sequence) are boldfaced. The newly introduced restriction sites (underlined) are as follows: FspI (PC-1, PC-2), SacII (PC-3), SfoI (PC-5), BstZI (PC-4, PC-6), and PinAI (PC-7). Incorporation of these nucleotide sequences into the 903-bp fragment was verified by restriction enzyme digestion with the enzymes listed above and subsequently by DNA sequencing. The mutant 903-bp inserts were subcloned back into pK1-2 by using the BsaAI and AflIII restriction sites, replacing the preexisting wt sequence.
Transfection assays and marker transfer.
All transfection and marker rescue experiments were carried out in 35-mm-diameter culture dishes by using LipofectAMINE PLUS reagent (Gibco BRL) according to the manufacturer's instructions. For complementation, in vitro expression, and primer extension experiments, 1 μg of total plasmid DNA was used to transfect Vero cells. In complementation experiments the transfected cells were infected with the d120 virus (MOI, 1 PFU/cell) 24 h following transfection. Eighteen hours later, the cells were scraped into the medium, disrupted by one freeze-thaw cycle and a 45-s sonication, and then spun for 10 min at 1,000 × g in a tabletop centrifuge. The clarified supernatants were then titered by a plaque assay on E5 cells. Cells were also transfected with different ICP4 alleles in order to observe the expression of the ICP4 protein from the plasmids. These transfections were performed in the same way as those for the complementation experiments. Following incubation, the cells were scraped into 200 μl of Laemmli sample buffer (Bio-Rad) with 0.72 M β-mercaptoethanol, boiled for 3 min, and analyzed by Western blotting. To determine the ability of the mutant ICP4 proteins expressed from plasmids to activate the thymidine kinase gene (tk) promoter, the mutant plasmids or wt pK1-2 was cotransfected together with plasmid pLSWT into monolayers of Vero cells as described above. pLSWT contains the entire promoter and coding sequence of tk from HSV-1 (30). Total RNA was isolated 40 h after transfection by using the Ultraspec RNA isolation system (BioTecx). Primer extension was performed as described previously (29). For marker rescue, a mixture of 0.5 μg of infectious virus DNA (KOS for M1; Elmo for M2, M3, M6, and M7) and 0.5 μg of gel-eluted 4,771-bp EcoRI fragments from the mutant pK1-2 plasmids was used to transfect E5 cells.
Isolation of mutant viruses.
To isolate a mutant virus containing the M1 mutations, the progeny from the transfection with pM1 and KOS DNA were diluted and plated on monolayers of E5 cells to produce isolated plaques. A number of plaques were picked and used to inoculate culture tubes containing 105 E5 cells. When CPEs were generalized, viral DNA minipreps were prepared from one-half of the culture as described above. The ICP4 genotypes were determined by restriction enzyme analysis of PCR products. Isolates containing the correct PCR products were plaque purified three times and then rechecked by PCR and DNA sequencing. To isolate the M2, M3, M6, and M7 viruses, the progeny from the transfections of the corresponding plasmid fragments and Elmo were diluted and plated onto monolayers of E5 cells to produce isolated plaques. Elmo produces plaques that fluoresce when viewed with a fluorescent microscope due to the insertion of the GFP genes in both copies of ICP4. Therefore, the progeny plaques were screened for the GFP-negative phenotype by using a fluorescent microscope. These plaques were picked, amplified, and analyzed as described above for M1. The primers for the PCRs used in this analysis were as follows: forward, GCGGCTCATCGTGGTCAA, corresponding to aa 1024 to 1030 of ICP4, and reverse, GTTGCCGTTTATTGCGTCTT, which is complementary to nucleotides 36 to 55 3′ to the translation termination codon of ICP4. These were chosen to amplify an 884-bp fragment flanking the loci of the introduced mutations. Approximately 1 μl of viral DNA miniprep (∼10 ng) was combined on ice with 3 μl of the forward primer and 2 μl of the reverse primer in 24 μl of distilled H2O. To this 30 μl of JumpStart REDTaq ReadyMix (Sigma) was added. The reaction mixture was subjected to initial denaturation (at 97.5°C for 3 min) followed by 31 cycles of denaturation (95.5°C for 1 min), annealing (58.8°C for 1 min), and elongation (72°C for 2 min with 3 s of extension per cycle). All PCRs were performed in a Perkin-Elmer Automatic Thermal Cycler. A 15-μl volume of final PCR products was combined with the appropriate buffers and restriction enzymes that recognize the sites introduced during mutagenesis, and then the mixture was incubated for 2 h at the appropriate temperatures. The final products were resolved in 3% agarose gels, which were subsequently stained with EtBr.
Infected cell polypeptide labeling.
Confluent monolayers of Vero cells in 35-mm-diameter cell culture dishes were infected for 1 h with wt virus or mutant viruses (MOI, 10 PFU/ml). l-Methionine- and l-cysteine-free Dulbecco's modified Eagle medium (Sigma) was added to the cultures, which were subsequently incubated at the indicated temperatures. Infected cultures were radioactively labeled at 6 or 12 h postinfection by replacing the medium with 50.0 μCi of l-[35S]methionine in 1 ml of TBS buffer equilibrated to the appropriate temperature and then continuing incubation for 1 h longer. Following the labeling period, the monolayers were washed with cold TBS containing 0.1 mM Nα-p-tosyl-l-lysine chloromethyl ketone (TLCK) and were subsequently solubilized in 200 μl of Laemmli sample buffer with 0.72 M β-mercaptoethanol. The samples were then boiled for 3 min, and the SDS peptides were electrophoretically resolved by SDS-7.5% PAGE. The gels were then fixed in H2O-methanol-acetic acid (6:3:1), dried, and exposed to Hyperfilm(Amersham Pharmacia Biotech).
Western blot analysis.
Protein samples generated from transfected or infected cells were heat denatured for 5 min at 95°C, resolved by SDS-6% PAGE, and electrophoretically transferred to polyvinylidene difluoride membranes (Hybond). The ICP4 was then detected with ICP4-specific antibodies. Rabbit polyclonal serum N15 (53) and monoclonal antibodies 58S (56) and 1101 (Goodwin Institute) were used at dilutions of 1:500. The desired protein-antibody complexes were visualized by using the appropriate secondary immunoglobulin G (whole molecule)-peroxidase conjugate and the ECL+Plus Western blotting chemiluminescence detection system (Amersham Pharmacia Biotech) as described by the manufacturer.
EMSA.
Electrophoretic mobility shift analysis (EMSA) was performed as previously described (17). The 32P end-labeled 135-bp EcoRI-BamHI fragment used in the EMSA reactions contains sequences from the ICP4 promoter from −108 to +27 relative to the transcription start site. This fragment contains a strong ICP4-binding site centered at +2 relative to the ICP4 mRNA start site. Protein extracts from infected Vero cells were prepared as described previously (17). Vero cell monolayers (106 cells) were infected with wt (KOS) or mutant viruses (MOI, 10 PFU/cell) at the indicated temperatures. Six hours after infection, the cells were washed twice with ice-cold TBS plus 0.1 mM TLCK and were lysed for 30 min on ice in 100 μl of 50 mM Tris-HCl (pH 8.0)-500 mM KCl-4% NP-40-0.1 mM TLCK. The extracts were then centrifuged at 13,000 × g for 15 min, and the supernatants were subsequently used for EMSA experiments. One microliter of the extract or purified protein was combined in a 20-μl DNA binding reaction mixture as specified elsewhere (31, 57). After 40 min the DNA-protein complexes were resolved by 4% native PAGE at 200 V for 2.5 to 3 h. The gels were then dried and exposed to Hyperfilm. Where indicated, monoclonal antibody 58S (1:25) or 1101(1:20) was added to the binding reaction mixture 10 min before it was loaded onto the gel. Some of the EMSA reactions utilized purified protein as a source of ICP4. ICP4 molecules were purified from nuclei of Vero cells that had been infected as previously described (31, 53).
RESULTS
Targeted mutagenesis of the C terminus of ICP4.
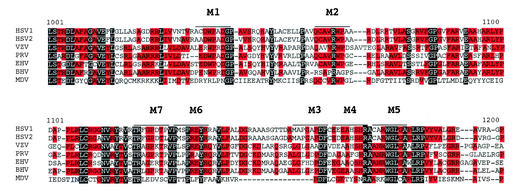
Alphaherpesviruses all possess ICP4 analogs, and except for HSV-1 and HSV-2, there is very little nucleotide sequence homology between the genes. However, there are regions within the proteins that possess discernible amino acid similarity. This was first described by McGeoch and colleagues in their comparison of the varicella-zoster virus (VZV) and HSV ICP4 genes (36). An amino acid sequence alignment of the ICP4 proteins from HSV-1, HSV-2, VZV, pseudorabies virus, equine herpesvirus (EHV), bovine herpesvirus, and Marek's disease virus demonstrates that while the primary sequences of these proteins range from 1,298 residues for HSV-1 to 1,487 for EHV, the alignment of these genes spans 1,817 residues (data not shown). The expanded consensus alignment is due to the fact that blocks of amino sequence similarity are often separated by nonconserved regions that are highly variable in length. One highly conserved region of ICP4 that is relatively void of gaps in the alignment is the region between aa 900 and 1200. Within this region one can identify stretches of primary sequence that are both conserved and relatively hydrophilic. Functions for this region of the protein have not been described. Therefore, oligonucleotide mutagenesis was used to target specific areas of this region.
Figure 1 shows the portion of the entire ICP4 amino acid alignment for seven alphaherpesviruses that corresponds to the region of HSV-1 ICP4 between aa 1001 and 1200. Oligonucleotides were designed for mutagenesis by taking several parameters into account. Regions of similarity were targeted, because these regions are most likely important for the function of ICP4. Relatively hydrophilic regions were chosen because these would most likely be near the surface of the molecule and may engage in interactions important for ICP4 function, and also in order to avoid global structural changes that may result from changes in the interior of the protein. In addition, all substitutions were made to change existing residues to alanine, resulting in relatively conservative substitutions. Lastly, the nucleotide changes were designed to also insert a new restriction enzyme cleavage site in order to facilitate tracking of the mutations through plasmid and virus construction. The targeted residues were between aa 1032 and 1050, 1065 and 1070, 1110 and 1135, and 1158 and 1180. The precise loci of the mutations were constrained by the need to fulfill all these criteria, as well as by the details of subsequent construction steps. This resulted in our inability to isolate mutants in an apparently important region between aa 1110 and 1120. In total, seven site-directed mutants, designated M1 through M7, were isolated (Fig. 1). The wt residues for each mutant locus are listed in Table 1 along with the new sequence that was generated by the oligonucleotides described in Materials and Methods. Three mutants have a single alanine substitution, two have two substitutions, one has three, and the last mutant has three alanine substitutions and a deletion of an arginine residue. The mutations listed in Fig. 1 and Table 1 were ultimately inserted into pK1-2, which contains the entire ICP4 gene under the control of its own promoter (16). The pK1-2 derivatives of M1 (pM1) through M7 (pM7) were identified by the presence of the new restriction enzyme cleavage sites and DNA sequence analysis (data not shown).
FIG. 1.
Sequence alignment of seven alphaherpesviruses corresponding to aa 1001 to 1200 of HSV-1 ICP4 and mutant loci. The entire amino acid sequences of the ICP4 analogs from HSV-1 (36), HSV-2 (19), VZV (12), pseudorabies virus (58), EHV (23), bovine herpesvirus (61), and Marek's disease virus (2) were aligned by using the Vector-NTI alignment program. Shown is the region of the alignment corresponding to the region of HSV-1 ICP4 between residues 1001 and 1200. Residues shaded in black are conserved in all seven proteins, while those shaded in red are conserved in at least four of the seven proteins. Also shown are the loci (lines above the sequence) for the M1 through M7 mutants. The line above the HSV sequence for each mutant corresponds to the sequences described in Table 1.
TABLE 1.
Amino acid changes in the mutants constructed in this study
| Mutant | Oligonucleotidea | wt sequenceb | Mutant sequencec |
|---|---|---|---|
| M1 | PC-1 | CDWPADGP | CAWPAAAP |
| M2 | PC-2 | CAVRWPAA | CAVAWPAA |
| M3 | PC-3 | APGAPDFC | APAAPAFC |
| M4 | PC-4 | EEAHSHRA | AEAASAΔA |
| M5 | PC-5 | ARWGLGAP | AAWGLAAP |
| M6 | PC-6 | MSPREYRR | MSPAEYRR |
| M7 | PC-7 | RFGPDTPV | RFGPDAPV |
Oligonucleotides, used to create the indicated mutants, are described in Materials and Methods.
The wt ICP4 sequence at the locus of the indicated mutant depicted in Fig. 1.
Mutant sequence relative to the indicated wt ICP4 sequence. The changed nucleotides are boldfaced.
Activities of ICP4 expressed from mutated plasmids.
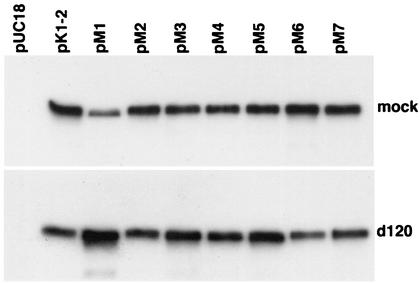
pK1-2 and pM1 through pM7 were first used to transfect Vero cells in order to verify the expression of full-length ICP4 from each of the mutant plasmids. Figure 2 shows a Western blot for ICP4 in samples derived from Vero cells transfected with pK1-2 and the mutant plasmids, with and without superinfection by the ICP4 deletion mutant d120. d120 has most of the ICP4 gene deleted, is replication defective, and does not result in the induction of early and late genes during infection (14). Figure 2 shows that all the plasmids express stable full-length ICP4 in the presence and absence of superinfection by d120. The abundance of ICP4 expressed from pM1 was somewhat lower in the sample without superinfection, suggesting that this protein may be less stable in the absence of HSV infection.
FIG. 2.
Expression of ICP4 from mutant plasmids. Vero cells were transfected with the indicated plasmids and were subsequently either mock infected or infected with d120 as described in Materials and Methods. Following incubation, SDS samples prepared from the cultures were subjected to SDS-PAGE, followed by Western blot analysis as described in Materials and Methods. The membranes containing the transferred SDS peptides were probed with a rabbit polyclonal antiserum raised against ICP4.
As an initial assessment of the effects of the mutations on ICP4 activity, each of the mutants was compared to pK1-2 in transient transfection assays in order to determine the abilities of the mutants to activate the tk gene and to provide complementing activity for the ICP4 deletion mutant d120. These two assays provide a preliminary assessment of the activities of the mutants and allow selection of mutants with interesting phenotypes for transfer to the viral genome.
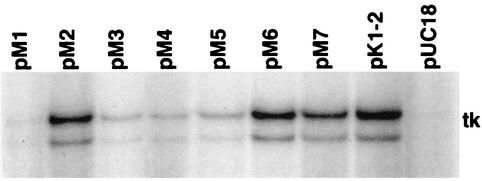
The ability of the transiently expressed ICP4 proteins to activate the tk gene in cotransfection assays was measured by primer extension analysis for tk mRNA (Fig. 3). This assay measures the abundance of tk mRNA expressed from the transfected tk gene as a function of the cotransfected ICP4 plasmids. The abundance of tk mRNA in the absence of ICP4 was too low to quantify accurately; however, it is clear from Fig. 3 that the different ICP4 alleles fall into three groups with respect to ability to activate the tk gene. The level of tk RNA resulting from the pM1 cotransfection could not be unambiguously differentiated from that resulting from transfection without an ICP4 plasmid. Therefore, this mutant specifies a very low, if any, ICP4 activation function. The levels of tk RNA resulting from the pM2, pM6, and pM7 transfections could not be unambiguously differentiated from those resulting from transfection with pK1-2. Therefore, these mutants specify nearly wt levels of ICP4 activation function. pM3, pM4, and pM5, all of which cluster between aa 1158 and 1180, showed significantly reduced ICP4 activation function.
FIG. 3.
Activation of tk gene expression as a function of wt and mutant ICP4. Shown are results of a primer extension analysis for tk mRNA isolated from cells cotransfected with plasmid pLSWT and the indicated plasmid. pLSWT encodes the entire tk gene of HSV-1. The double band corresponds to the primer extension typically seen with the tk promoter/start site (29, 30).
The ability of the transiently expressed ICP4 proteins to complement d120 for viral growth was measured by infecting with d120 cells that had previously been transfected with a wt or mutant ICP4 plasmid and subsequently measuring viral yield by a plaque assay on E5 cells (16). This assay for ICP4 function is more sensitive and more quantitative than the cotransfection assay used above. Therefore, it is possible to reproducibly observe more subtle differences by using this assay. The results shown in Table 2 divide the mutants into four groups. pM1 provided very little to no complementing activity as inferred by comparison to the negative control. pM3, pM4, and pM5 provided 4.0 to 6.2% of the complementing activity of pK1-2. It should be noted that this represents a 20- to 30-fold increase in d120 yield over that of the negative control. pM6 and pM7 provided 38 and 55% of the complementing activity of pK1-2, respectively. pM2 was indistinguishable from pK1-2. Therefore, the results of the complementation analysis (Table 2), which measures the abilities of the mutants to support viral growth, are qualitatively similar to the results of the assay that measures the abilities of the mutants to activate the tk promoter (Fig. 3).
TABLE 2.
Complementation of d120 by plasmids expressing mutant ICP4
| Plasmida | Yieldb | Complementationc | % of wtd | Avge |
|---|---|---|---|---|
| None | 3.92 | 1.0 | 0.2 | 0.4 |
| pK1-2 | 6.60 | 482 | 100 | 100 |
| pM1 | 4.28 | 2.3 | 0.5 | 0.8 |
| pM2 | 6.58 | 458 | 95 | 93 |
| pM3 | 5.38 | 29 | 6.0 | 4.5 |
| pM4 | 5.65 | 19 | 4.0 | 3.5 |
| pM5 | 5.40 | 30 | 6.2 | 7.0 |
| pM6 | 6.34 | 265 | 55 | 50 |
| pM7 | 6.18 | 181 | 38 | 32 |
The indicated plasmid was used to transfect Vero cells. Eighteen hours later, the cultures were infected with d120 at an MOI of 1 PFU/cell. Viral lysates were prepared 24 h following infection.
Viral lysates were assayed for infectivity by plaque assay on E5 cells. Shown are log10 titers.
Expressed as the ratio of the titer for a culture transfected with a given plasmid to the titer of the culture that received no plasmid.
Percent complementation relative to that with pK1-2.
Average percent complementation for three experiments, including the data given in footnotes b to d.
Characterization of viruses expressing mutant ICP4 molecules.
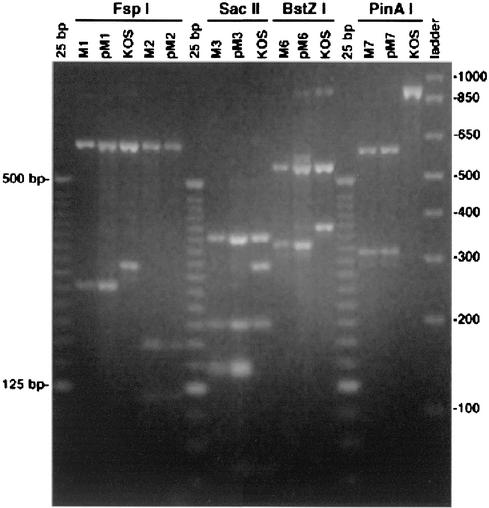
While transient assays can provide some information relevant to the activity of a transfected effector molecule, additional and more physiologically relevant information can be obtained by observing the activity of the effector in its natural context. Therefore, mutants M1, M2, M3, M6, and M7 were recombined into the HSV genome at the native ICP4 loci. Progeny plaque isolates resulting from the cotransfections of the mutant plasmids with HSV DNA were screened as described in Materials and Methods, and ultimately by PCR and restriction enzyme digestion, for incorporation of the mutant ICP4 alleles at both ICP4 loci. Figure 4 shows a comparison of the PCR-amplified 903-bp MscI fragments from the mutant viruses, wt (strain KOS) virus, and the plasmids used to generate the mutant viruses. Following PCR and prior to electrophoretic separation, the amplified products were digested with the restriction enzymes whose cognate sites were generated by the oligonucleotides used to create each of the mutations. Since the sites for these restriction enzymes are relatively frequent in the HSV genome, some of these sites are also present within the wt 903-bp MscI fragment. PinAI, the site specified by the oligonucleotide used to create the M7 mutant, does not cleave the wt 903-bp MscI fragment (Fig. 4). However, a PinAI site is present in the 903-bp MscI fragment from the M7 virus and the plasmid used to generate it, pM7. The electrophoretic profile of pM7 is identical to that of M7, and there is no evidence of the KOS profile in either. Therefore, the M7 virus has the same ICP4 allele as the pM7 plasmid. Similarly, the electrophoretic profile of each of the mutants is the same as that of the corresponding plasmid used to generate the mutant, and these profiles are distinct from that of KOS. Therefore, all mutants possess the intended allele and do not contain the gene for wt ICP4.
FIG. 4.
Restriction enzyme analysis of PCR-amplified products of mutant viruses and the plasmids used to generate them. DNA was isolated from cells infected with the M1, M2, M3, M6, M7, or KOS virus. The region containing the mutational loci in the DNA of the infected cell as well as that in the plasmids used to generate the mutant viruses was PCR amplified as described in Materials and Methods. The amplified products were digested with the indicated restriction enzyme and electrophoresed on agarose gels. The enzymes used to digest a mutant virus-plasmid pair were those whose sites were engineered into the plasmid by oligonucleotide mutagenesis. Shown are the EtBr-stained gel of the restriction digests and size markers for reference.
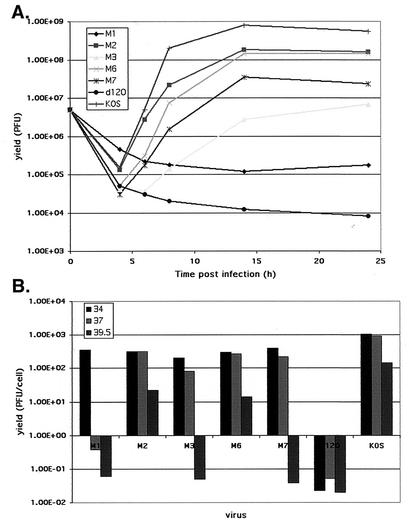
To determine the effects of the alanine mutations on viral growth, single-step growth experiments for each of these mutants were carried out using Vero cells, and results for these mutants were compared to those for d120 and KOS. Initially, replicate cultures of Vero cells were infected at an MOI of 10 PFU/cell as described in Materials and Methods and were incubated at 37°C. At different times postinfection, virus suspensions were prepared from the cultures and yields were determined by plaque assays on E5 cells (Fig. 5A). M2 and M6 had yields only marginally reduced relative to that of wt virus. The yields of M7 and M3 were reduced by approximately 1 and 2 orders of magnitude, respectively, relative to that of wt virus. While new progeny were created in the M3 infection, the total yield was very similar to the input used to initiate the infection. M1, like d120, showed no evidence of viral growth in the course of the experiment.
FIG. 5.
Single-step growth of mutant viruses in Vero cells. (A) Mutant viruses M1, M2, M3, M6, M7, and d120 and wt virus (KOS) were compared in a single-step growth experiment conducted at 37°C. Shown are total yields (PFU) per infected Vero cell culture as a function of time postinfection. (B) Burst sizes of mutant viruses M1, M2, M3, M6, M7, and d120 and wt virus (KOS) were compared in a single-step growth experiment conducted at 34, 37, and 39.5°C. Shown are the yields (PFU) per Vero cell for each of the viruses after 24 h of incubation at the indicated temperatures (degrees Celsius).
Thus far, all analyses of mutants were conducted at 37°C. Alanine substitutions often result in the generation of temperature-sensitive (ts) mutants (60). To explore this possibility, single-step growth experiments were performed at three temperatures: 34, 37, and 39.5°C. Infected cultures were harvested at a single time postinfection (24 h), and the viral suspensions prepared subsequently were analyzed for infectivity by a plaque assay on E5 cells at 37°C (Fig. 5B). All of the mutants, except d120, produced more than 100 PFU per infected Vero cell at 34°C. M1 produced less than 1 PFU/cell at both 37 and 39.5°C, indicating that M1 was temperature sensitive at both 37 and 39.5°C. M3 and M7 were temperature sensitive at 39.5°C. M2 and M6 possess some ts character in that both produced only 10 to 20 PFU per cell at 39.5°C, while KOS produced 100 to 200 PFU/cell at this temperature. Therefore, three out of the five alanine substitution mutants that were analyzed in the context of viral infection were temperature sensitive.
Lastly, we investigated the plaquing efficiency of the mutants as a function of temperature and cell type. Table 3 shows the plaquing characteristics of stocks of the alanine mutants, KOS, and d120 at different temperatures, with and without complementation provided by the E5 cell line. Consistent with the growth properties depicted in Fig. 5, M1, M3, and M7 were temperature sensitive for plaque formation on Vero cells at 39.5°C, as indicated by the ratios of the log10 titers at 39.5°C to those at 34°C (−3.8, −4.9, and −4.62, respectively). M1 was as temperature sensitive for plaquing at 37°C as it was at 39.5°C. d120 does not plaque at any temperature on Vero cells. The ts plaquing phenotype of M1, M3 and M7 on Vero cells was abolished by ICP4 supplied in E5 cells, as demonstrated by the ratios of the log10 titers at 39.5°C to those at 34°C (0.05, −0.02, and −0.24, respectively). This supports the conclusion that the ts phenotypes of M1, M3, and M7 were due to mutations in ICP4 and are not functions of mutations outside of ICP4. The ratios of the log10 titers on Vero cells to those on E5 cells at each temperature additionally support the conclusions that M1 is strongly temperature sensitive for plaque-forming ability at 37 and 39.5°C, M3 and M7 are temperature sensitive for plaque-forming ability at 39.5°C, and these phenotypes are due to mutations in ICP4.
TABLE 3.
Plating characteristics of mutants as a function of cell type and temperature
| Virus | Log10 titera on the following culture at the indicated temp:
|
Log10 titer on Vero cell culture − log10 titer on E5 cell cultureb at:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| E5
|
Vero
|
||||||||
| 34°C | 37°C | 39.5°C | 34°C | 37°C | 39.5°C | 34°C | 37°C | 39.5°C | |
| M1 | 8.56 | 8.64 | 8.61 | 8.4 | 4.7 | 4.6 | −0.16 | −3.94 | −4.01 |
| M2 | 9.04 | 9.04 | 8.93 | 9.08 | 9.08 | 8.93 | 0.04 | 0.04 | 0.0 |
| M3 | 8.95 | 9.02 | 7.76 | 8.97 | 7.89 | 4.0 | 0.02 | −0.13 | −3.76 |
| M6 | 8.46 | 8.48 | 7.82 | 8.62 | 8.56 | 8.18 | 0.16 | 0.08 | 0.36 |
| M7 | 8.56 | 8.61 | 8.32 | 8.62 | 8.52 | 4.0 | 0.06 | −0.09 | −4.3 |
| d120 | 7.51 | 7.23 | 7.32 | <3 | <3 | <3 | <−4.5 | <−4.2 | <−4.3 |
| KOS | 9.78 | 9.84 | 9.75 | 9.78 | 9.78 | 9.76 | 0.0 | −0.06 | 0.01 |
Titers of representative virus stocks were determined by standard plaque assays on monolayer cultures of E5 cells or Vero cells.
Boldfaced ratios indicate temperature sensitivity.
Biochemical characterization of the ICP4 mutants.
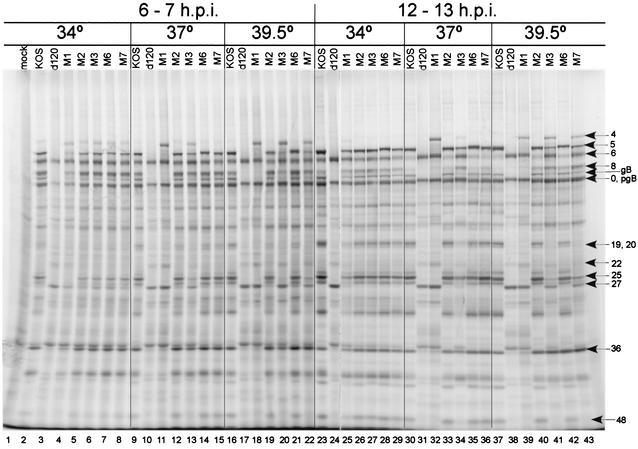
While ICP4 functions at the level of transcription to regulate gene expression (21, 59), a convenient way to assess the gene expression phenotype of ICP4 mutants is to observe the profile of viral polypeptides synthesized during infection (13, 18, 44, 47, 54). Accordingly, Vero cells were infected with the indicated viruses, at the indicated temperature, and labeled with [35S]methionine either from 6 to 7 h postinfection or from 12 to 13 h postinfection. Cultures were harvested immediately following the labeling interval and were analyzed by SDS-PAGE. Since d120 has large deletions in the genes encoding ICP4, it does not express ICP4. As a consequence, later viral proteins were not detectable at this level of resolution. However, large quantities of the IE proteins ICP0, ICP22, and ICP27, as well as ICP6, were synthesized in d120-infected cells (Fig. 6, lanes 3, 10, 17, 24, 31, and 38). This phenotype is consistent with the findings of previous studies (14). The polypeptide profiles of M1 through M7 at 34°C were similar to that of KOS (Fig. 6; compare lane 2 with lanes 4 to 8 and lane 23 with lanes 25 to 29). The phenotypes of M1 at 37°C (Fig. 6, lanes 11 and 32) and 39.5°C (lanes 18 and 39) were similar to that of d120 at both time points (lanes 10, 17, 31, and 38), except for the observation that M1 overexpressed the mutant forms of ICP4, ICP22, and ICP27, and the mutant forms of ICP4 were also overexpressed from the M3 and M7 genomes during infection at 39.5°C (Fig. 6, lanes 20 and 22, respectively, and lanes 41 and 43, respectively). However, in contrast to the M1 phenotype, representative early proteins such as ICP8, pgB, and ICP36, as well as some “leaky-late” (βγ) proteins (ICP5 and ICP25), were also expressed during M3 and M7 infection at 39.5°C. Representative late genes (γ), such as ICP19, -20, and -48, were not detected (Fig. 6, lanes 41 and 43). M3 also displayed a partial ts phenotype at 37°C (Fig. 6, lanes 13 and 34). Consistent with their growth properties, the gel profiles for M2 and M6 were similar to those for KOS at the times and temperatures sampled. However, M2 and M6 may possess some ts character. At 39.5°C their burst sizes were reduced more than that of KOS (Fig. 5B), and it appears that ICP8 was not shut off as completely at late times as in KOS-infected cells (Fig. 6, lanes 37, 40, and 42).
FIG. 6.
Synthesis of viral polypeptides in Vero cells infected with wt and ICP4-mutant viruses. Vero cells were infected with 10 PFU of the indicated viruses/cell and were incubated at the indicated temperatures. Cultures were labeled with l-[35S]methionine from 6 to 7 h and from 12 to 13 h postinfection as described in Materials and Methods. Following the labeling period, monolayers were harvested and analyzed by SDS-PAGE as described in Materials and Methods. Shown are autoradiographic images of infected cell polypeptides. Positions of representative infected cell polypeptides are indicated by arrows and numbers on the right (for example, “4” stands for ICP4).
An activity that is crucial for the ability of ICP4 to regulate transcription is its ability to bind to DNA (20, 39, 50). This activity is specified by a region of ICP4 between residues 262 and 490 (62). The region of ICP4 containing the mutations in M1, M3, and M7 can be excluded from the ICP4 protein without greatly affecting the ability of ICP4 to bind to DNA (17). However, it is possible that the mutations specifying the M1, M3, and M7 mutants may affect the conformation of the protein such that the DNA binding domain is affected. Cell extracts were prepared from cells infected with the mutants at 34, 37, and 39.5°C in order to determine the ability of the ICP4 protein to bind to a specific ICP4 binding site in EMSA assays. Two monoclonal antibodies were used to supershift potential ICP4-DNA complexes: 1101, which recognizes an epitope near the amino terminus of ICP4 (28), and 58S (56), which recognizes an epitope contained in the carboxyl-terminal 270 aa of ICP4 (17). The region containing the 58S epitope also contains the loci for all the mutants generated in this study.
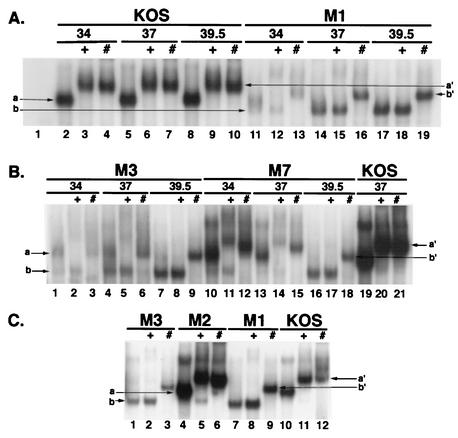
Figure 7 shows the regions of the gels containing the DNA-protein complexes from three EMSA experiments. The specific ICP4-DNA complexes formed with extracts from cells infected at 34, 37, and 39.5°C with M1 and KOS are shown in Fig. 7A. The KOS ICP4-DNA complexes formed with proteins synthesized at the three temperatures were very similar (mobility a) (Fig. 7A, lanes 2, 5, and 8). These complexes were supershifted by antibodies 58S and 1101 to mobility a′ (Fig. 7A, lanes 3, 4, 6, 7, 9, and 10). The DNA-protein complexes formed with extracts prepared from M1-infected cells at 34°C were somewhat heterogeneous (Fig. 7A, lane 11). A complex similar to the “a” complex was seen as well as one with a greater mobility (mobility b). The diffuse “a” complex from lane 11 either disappeared or was supershifted by 58S (Fig. 7A, lane 12) and 1101 (Fig. 7A, lane 13). The novel “b” complex was not supershifted by 58S but was supershifted by 1101 to b′. The novel mobility form of the M1 ICP4-DNA complex and its lack of reactivity with 58S is more clearly seen with the M1 extracts prepared from proteins synthesized at 37 and 39.5°C (Fig. 7A, lanes 14 to 19).
FIG. 7.
DNA binding of mutant ICP4 proteins. EMSA was used to observe the abilities of the mutant ICP4 proteins to bind to a specific ICP4 binding site. Details of the EMSA are provided in Materials and Methods. In some reactions the ICP4-specific monoclonal antibody 58S (+) or 1101 (#) was used to supershift complexes formed by ICP4 protein. Designations: a, mobility of ICP4-DNA complexes formed with extracts from KOS-infected cells; a′, mobility to which these complexes were supershifted by antibodies 58S and 1101; b, mobility (greater than a) of some ICP4-DNA complexes formed with extracts from M1-infected cells; b′, mobility to which “b” complexes were supershifted by 1101. (A) Comparison of DNA-protein complexes formed with KOS and M1 extracts prepared from cells infected at 34, 37, and 39.5°C. Shown are autoradiographic images of the region of the gel containing the ICP4-DNA protein complexes. Lane 1 represents the DNA-protein complexes in this region of the gel formed by using extracts from d120-infected cells. (B) Comparison of DNA-protein complexes formed with M3, M7, and KOS extracts prepared from cells infected at 34, 37, and 39.5°C. For KOS, only the 37°C samples are shown. (C) Comparison of DNA-protein complexes formed with M3, M2, M1, and KOS ICP4s purified from cells infected at 37°C.
The specific ICP4-DNA complexes formed with extracts from cells infected at 34, 37, and 39.5°C with M3, M7, or KOS are shown in Fig. 7B. When the 39.5°C extracts were used, the ICP4-DNA complexes formed with both M3 (Fig. 7B, lanes 7 to 9) and M7 (Fig. 7B, lanes 16 to 18) were very similar to those formed with the 39.5°C M1 extracts (Fig. 7A, lanes 15 and 16) in that the ICP4-DNA complexes had greater mobilities than the KOS complex and were supershifted only by monoclonal antibody 1101. The complexes formed with the 34 and 37°C M7 extracts (Fig. 7B, lanes 10 to 15) resembled those formed by KOS extracts (Fig. 7B, lanes 19 to 21) with respect to mobility and the ability of 58S and 1101 to supershift the complexes. The complexes formed with the 34 and 37°C M3 extracts (Fig. 7B, lanes 1 to 6) were a mixture of low- and high-mobility complexes (mobilities a and b, respectively).
The EMSA for which results are shown in Fig. 7A and B utilized whole-cell extracts. Therefore, it is possible that the mobility difference between the novel “b” complexes formed with M1, M3, and M7 extracts and the wt “a” complex was due to other proteins in the extract differentially participating in the two complexes. To address this issue, the M1 and M3 ICP4 molecules were purified as described in Materials and Methods and compared to purified M2 and KOS ICP4 molecules by EMSA (Fig. 7C). M2 is similar to KOS with respect to growth parameters (Fig. 5B; Table 3) and gene expression profile (Fig. 6). Preliminary EMSA experiments with cell extracts also revealed that the M2 ICP4-DNA complexes are similar to the KOS complexes (data not shown). The DNA-protein complexes formed with purified M2 ICP4 (Fig. 7C, lanes 4 to 6) are also similar to those formed with purified KOS ICP4 (Fig. 7C, lanes 10 to 12) and are shifted with both monoclonal antibodies. The lower-mobility complexes seen with purified KOS and M2 ICP4s are probably due to multimerization of ICP4 on the DNA. The purified M1 (Fig. 7C, lanes 1 to 3) and M3 (Fig. 7C, lanes 7 to 9) proteins both formed the higher-mobility “b” complex and were supershifted only by antibody 1101. Therefore, we conclude that the difference in mobility between the “a” and “b” complexes is a consequence of the altered conformation of the mutant ICP4 molecule and that this altered conformation is also indicated by the differential reactivity to antibody 58S.
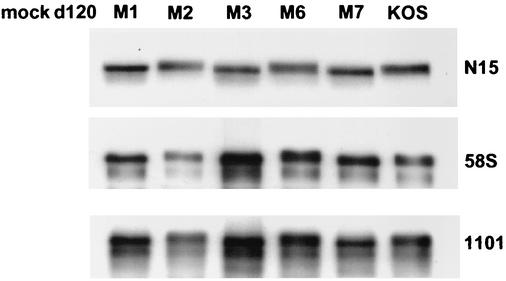
It is significant that antibody 1101 reacts with the amino-terminal region of ICP4, while 58S reacts with an epitope in the same region of the protein where the mutations in M1, M3, and M7 were created. The ability of 58S to react with these proteins synthesized at 34°C in EMSAs suggests that the mutations do not directly affect the epitope. To further address this issue, extracts from Vero cells infected at 39.5°C with d120, M1, M2, M3, M6, M7, or KOS were analyzed by Western blot analysis for ICP4 by probing with N15, 58S, and 1101 (Fig. 8). N15 is a rabbit polyclonal antibody raised against a recombinant ICP4 polypeptide (54). As expected, N15 and 1101 reacted with all the ICP4 proteins synthesized from M1 to M7 and KOS. In contrast to the EMSA experimental results (Fig. 7), 58S reacted just as well with M1, M3, and M7 ICP4s synthesized at 39.5°C as it did with M2, M6, and KOS ICP4s. Therefore, we conclude that the lack of reactivity of 58S in the EMSA experiments is a consequence of the altered conformation of the region of ICP4 where 58S binds.
FIG. 8.
Western blot analysis of mutant ICP4 proteins using different antibody preparations. Three-microliter portions of the protein extracts that were also used in the EMSA experiments (Fig. 7) were combined with 3 μl of Laemmli sample buffer, boiled for 3 min, and subsequently resolved by SDS-PAGE (6% polyacrylamide). The SDS-peptides were transferred to polyvinylidene difluoride membranes for Western blot analysis as described in Materials and Methods. Three different antibodies were used to probe the membranes: N15 (rabbit polyclonal), 58S (mouse monoclonal), and 1101 (mouse monoclonal).
DISCUSSION
ICP4 is a large and structurally complex protein consisting of many regions that collectively contribute to its role in the regulation of HSV gene transcription and in viral growth (5, 17, 42, 54). In the absence of the C-terminal 524 aa, the remaining peptide composed of 774 aa can bind to specific ICP4 binding sites (17), form tripartite complexes on promoters with TFIID and TATA-binding protein (57), facilitate the formation of preinitiation complexes on TATA boxes (22), repress transcription of promoters containing ICP4 binding sites near their mRNA start sites (17, 25), and activate transcription of many β and βγ genes during viral infection (17). Proteins lacking the C-terminal region are also partially active in transient transfection assays (16) and in reconstituted in vitro transcription (8) assays. However, viruses expressing ICP4 proteins lacking the C-terminal region are defective with regard to full activation of β and βγ genes and viral DNA synthesis, and γ genes are not expressed (17). As a consequence, viral growth is severely impaired. While the amino-terminal two-thirds of the ICP4 has many of the activities that are ascribed to wt ICP4, the C-terminal region of ICP4 is clearly important for full activation and viral growth. With the exception of the DNA binding domain (approximate position, aa 300 to 500), the bulk of amino acid conservation between HSV ICP4 and the ICP4 homologues of other alphaherpesviruses resides in the C-terminal one-third of ICP4. In an effort to understand how this region contributes to the activity and structure of ICP4, several site-directed mutants were constructed in regions within the C terminus that are both similar to other ICP4 homologues and relatively hydrophilic.
Seven site directed mutants between aa 1000 and aa 1200 fell into three general categories with respect to ability to support the growth of d120 and activate the tk gene in transient assays: those that were similar to wt ICP4 (M2, M6, and M7), those with partial activity (M3, M4, and M5), and one with undetectable activity (M1) (Table 2 and Fig. 3). Similar results were obtained when the plasmid mutant alleles for M1, M2, M3, M6, and M7 were analyzed in the context of viral infection (Fig. 5A). In addition, it was discovered that M1, M3, and M7 were temperature sensitive for growth. M3 and M7 were temperature sensitive for growth at 39.5°C, with M3 being partially impaired at 37°C, and M1 was temperature sensitive for growth at 37 and 39.5°C (Fig. 5 and Table 3). The ts phenotypes were due to mutations in ICP4, as evidenced by the fact that the ICP4 supplied by E5 cells completely complemented the ts growth defect (Table 3). All of the mutants also contained the new restriction site introduced as a consequence of the engineered mutation and did not contain an ICP4 gene lacking the introduced restriction site (Fig. 4). Importantly, the yields (Fig. 5B) and plaquing efficiencies (Table 3) of M1, M3, and M7 at 39.5°C were similar. Despite this, while all three overproduced ICP4, ICP22, and ICP27 at 39.5°C, significant levels of β and βγ gene expression were observed in M3- and M7-infected cells. β and βγ gene expression were as defective in M1 as in d120 (Fig. 6).
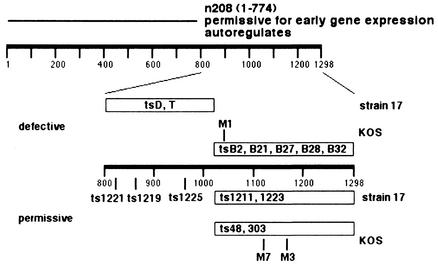
ts mutants displaying these two phenotypes have been described previously (13, 18, 44, 48), and their map locations within the ICP4 protein are shown in Fig. 9. All the previously mapped mutants either are the products of generalized mutagenesis or arose spontaneously. The base changes in M1, M7, and M3 are centered at aa 1038, 1125, and 1160, respectively (Fig. 1), and are shown in Fig. 9 relative to the other mutants. The previously described strain KOS ts mutants in which early gene expression is completely defective were mapped between residues 1030 and 1298 (18), as were those permissive for early gene expression (13) (Fig. 9). The mutations in the strain 17 mutants, tsD and tsT, in which early gene expression is relatively defective, were localized between aa 868 and 1030 (48). Strain 17 ts mutants permissive for early gene expression possess mutations throughout the region from aa 800 to aa 1298 (44). At present there is not enough mutant sequence information to ascribe the two phenotypes to specific domains of the protein. However, it is noteworthy that the base changes in plasmid mutants M3, M4, and M5 are all located in the same conserved stretch between aa 1155 and 1180 (Fig. 1.) and that these mutants behave similarly in that they partially complement d120 (Table 2) and activate tk (Fig. 3) in cotransfection experiments at 37°C.
FIG. 9.
Loci for known ts mutants in the C terminus of ICP4. (Top) The region of ICP4 expressed from mutant n208 (17) and its phenotype with respect to activation of early genes and autoregulation are shown above a scale representing the entire 1,298 aa of HSV-1 ICP4. (Bottom) The loci of known ts mutants of strains KOS and 17 that are defective and permissive for early gene expression are shown above and below a scale representing aa 800 to 1298, respectively. Loci for defective strain 17 mutants tsD and tsT (48), defective KOS mutants tsB2, tsB21, tsB27, tsB28, and tsB32 (18), permissive strain 17 mutants ts1221, ts1219, ts1225, ts1211, and ts1223 (44), and permissive KOS mutants ts48 and ts303 (13) are represented as described previously. Those for M1, M3, and M7 are from this study.
All of the ts mutants shown in Fig. 9 map to a region of ICP4 that is not contained in the mutant protein expressed from the n208 virus (17). The pattern of β and βγ gene expression from the n208 genome (17), as well as the behavior of the plasmid mutant used to create n208 in transient complementation and cotransfection assays (16), is similar to that of M3 and M7 at elevated temperatures. Therefore, it is possible that these mutations may inactivate a function of the protein that is specific for the C terminus without affecting the ability of the amino-terminal part of the protein to activate transcription to the extent seen in n208-infected cells. However, unlike the n208 molecule, which is functional with respect to autoregulation (17), M3 and M7 ICP4s show defective autoregulation at the nonpermissive temperature (Fig. 6). This suggests that the mutations in these molecules affect this activity, which is specified by the N-terminal region of ICP4. Additionally, M3 and M7 retain the ability to bind to the specific ICP4 binding site located at the start site of the ICP4 promoter. Given that this activity is required for repression of the ICP4 promoter (52), the mutations in M3 and M7 must affect the conformation of the proteins such that ICP4 cannot participate in other interactions involved in autoregulation. These interactions may include the ability to participate in tripartite complexes with TFIID and TFIIB on repressed promoters (25, 33, 57) or additional unknown interactions that are also involved in repression. An additional possibility is that the conformationally altered ICP4 may activate rather than repress its own and other IE promoters at the elevated temperature. This has been observed previously in transient assays using cloned ICP4 genes from the tsL14 and tsB32 viruses (15). Additional studies are needed to investigate these possibilities.
Like some other ts mutants that map in the C terminus (Fig. 9), M1 also shows defective expression of early genes (Fig. 3 and 6). Considering that the truncated peptide expressed from the n208 virus allows for expression of early genes, either the M1 protein synthesized at the nonpermissive temperature must be globally misfolded or the mutation in M1 is in the region of ICP4 that interacts with structures specified by the amino terminus. The conformation of the M1 protein synthesized at the nonpermissive temperature was clearly altered, as demonstrated by the increased mobility of the protein-DNA complexes formed with a segment from the ICP4 promoter (Fig. 7A and C). Importantly, the mutation in M1 did not eliminate the ability of the protein to bind to DNA. The M1-DNA complexes were supershifted by the addition of monoclonal antibody 1101 (H943), which has been mapped to an epitope near aa 125 (28). However, antibody 58S, which maps to an epitope C-terminal to aa 1030 (17), a region containing all the mutations in this study, did not supershift DNA-protein complexes formed with the M1 ICP4 synthesized at the nonpermissive temperature. Antibody 58S did react with the SDS peptide of the M1 protein produced at the nonpermissive temperature, suggesting that the epitope conferring reactivity with 58S is intact in the M1 protein and that it is masked in the native structure by a temperature-induced conformational change. The same mobility changes of the protein-DNA complexes and antibody reactivity patterns were also seen with the M3 and M7 proteins produced at the nonpermissive temperature (Fig. 7B and C). Thus, while there is evidence for conformational changes in the C termini of the M1, M3, and M7 proteins produced at the nonpermissive temperature, there is no evidence of changes in the conformation of the N-terminal region as judged by the ability to bind DNA and react with antibody 1101. Clearly, these observations do not rule out changes in the N terminus.
The M1, M3, and M7 proteins produced at the nonpermissive temperature all bind to DNA. While specific DNA binding sites may not be important for activation of transcription by ICP4, the DNA binding activity appears to be involved, in that the ability of ICP4 mutants to bind to specific ICP4 binding sites correlates with activation (17, 43, 54). Therefore, the temperature-induced conformational change in the M1, M3, and M7 proteins inferred from the altered mobility of the protein-DNA complexes, as well as from the loss of reactivity to antibody 58S, may affect the ability of this region of the protein to interact productively with binding partners involved in activation. The observations of a previous study examining the activities of a transdominant-negative mutant of ICP4 (X25) are consistent with this hypothesis (55). The ICP4 mutant protein X25, which has a deletion between aa 32 and 275 as well as between aa 775 and 1298, forms dimers and binds to DNA but does not modulate transcription (55). When X25 is expressed in wt virus-infected cells, virus production and late gene expression are impaired. A large proportion of the wt ICP4 is sequestered into heterodimers consisting of wt and X25 polypeptides. The heterodimers bind to DNA, and while they contain one copy of the 58S epitope, their ability to react with the 58S epitope in supershift experiments is markedly reduced (55). It was suggested that the reactivity of 58S with the C terminus served as a paradigm or marker for the wt conformation of this region, which is necessary for interactions with other proteins involved in transcription activation. These may include TAFII250, as previously described (8), or some known protein(s) involved in activation. Additional studies to address this issue are under way.
Past studies have shown that most of the activities ascribed to ICP4 can be demonstrated with a fragment of ICP4 comprised of the N-terminal 774 aa. It is clear, however, that this is not sufficient to supply all the functionality needed to allow viral growth. The C terminus is a large, complex region of the protein that is required for full function. Additionally, changes in the C terminus can have effects on some activities inherent in the N terminus. Given these observations, and the large size and unusual shape of ICP4, it may be that regions engaging in protein-protein interactions that result in transcription regulation are not discrete stretches of primary sequence, as with other activators, but are composed of discontinuous regions of the protein interacting with each other to form interfaces for protein-protein interactions. This further illustrates the utility of analysis of ICP4 conducted in the context of viral infection, as well as the need for continued genetic analysis.
Acknowledgments
This work was supported by NIH grant AI27431.
We are grateful to Mary Evans for constructing the Elmo virus used for the marker transfer in these studies. We are also grateful to Susan Zabeirowski, Sara Jackson, and Padmavathi Sampath for discussions and for comments on the manuscript.
REFERENCES
- 1.Alwine, J. C., W. L. Steinhart, and C. W. Hill. 1974. Transcription of herpes simplex type 1 DNA in nuclei isolated from infected HEp-2 and KB cells. Virology 60:302-307. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, A. S., A. Francesconi, and R. W. Morgan. 1992. Complete nucleotide sequence of the Marek's disease virus ICP4 gene. Virology 189:657-667. [DOI] [PubMed] [Google Scholar]
- 3.Batterson, W., and B. Roizman. 1983. Characterization of the herpes simplex virion-associated factor responsible for the induction of alpha genes. J. Virol. 46:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruce, J. W., and K. W. Wilcox. 2002. Identification of a motif in the C terminus of herpes simplex virus regulatory protein ICP4 that contributes to activation of transcription. J. Virol. 76:195-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, M. E., J. W. Palfreyman, and C. M. Preston. 1984. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J. Mol. Biol. 180:1-19. [DOI] [PubMed] [Google Scholar]
- 7.Carrozza, M. J., and N. DeLuca. 1998. The high mobility group protein 1 is a coactivator of herpes simplex virus ICP4 in vitro. J. Virol. 72:6752-6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrozza, M. J., and N. A. DeLuca. 1996. Interaction of the viral activator protein ICP4 with TFIID through TAF250. Mol. Cell. Biol. 16:3085-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordingley, M. G., M. E. Campbell, and C. M. Preston. 1983. Functional analysis of a herpes simplex virus type 1 promoter: identification of far-upstream regulatory sequences. Nucleic Acids Res. 11:2347-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costanzo, F., G. Campadelli-Fiume, L. Foa-Tomasi, and E. Cassai. 1977. Evidence that herpes simplex virus DNA is transcribed by cellular RNA polymerase B. J. Virol. 21:996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Courtney, R. J., and M. Benyesh-Melnick. 1974. Isolation and characterization of a large molecular-weight polypeptide of herpes simplex virus type 1. Virology 62:539-551. [DOI] [PubMed] [Google Scholar]
- 12.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759-1816. [DOI] [PubMed] [Google Scholar]
- 13.DeLuca, N. A., M. A. Courtney, and P. A. Schaffer. 1984. Temperature-sensitive mutants in herpes simplex virus type 1 ICP4 permissive for early gene expression. J. Virol. 52:767-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeLuca, N. A., A. M. McCarthy, and P. A. Schaffer. 1985. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J. Virol. 56:558-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeLuca, N. A., and P. A. Schaffer. 1985. Activation of immediate-early, early, and late promoters by temperature-sensitive and wild-type forms of herpes simplex virus type 1 protein ICP4. Mol. Cell. Biol. 5:1997-2008. [DOI] [PMC free article] [PubMed]
- 16.DeLuca, N. A., and P. A. Schaffer. 1987. Activities of herpes simplex virus type 1 (HSV-1) ICP4 genes specifying nonsense peptides. Nucleic Acids Res. 15:4491-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeLuca, N. A., and P. A. Schaffer. 1988. Physical and functional domains of the herpes simplex virus transcriptional regulatory protein ICP4. J. Virol. 62:732-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dixon, R. A., and P. A. Schaffer. 1980. Fine-structure mapping and functional analysis of temperature-sensitive mutants in the gene encoding the herpes simplex virus type 1 immediate early protein VP175. J. Virol. 36:189-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dolan, A., F. E. Jamieson, C. Cunningham, B. C. Barnett, and D. J. McGeoch. 1998. The genome sequence of herpes simplex virus type 2. J. Virol. 72:2010-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faber, S. W., and K. W. Wilcox. 1986. Association of the herpes simplex virus regulatory protein ICP4 with specific nucleotide sequences in DNA. Nucleic Acids Res. 14:6067-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Godowski, P. J., and D. M. Knipe. 1986. Transcriptional control of herpesvirus gene expression: gene functions required for positive and negative regulation. Proc. Natl. Acad. Sci. USA 83:256-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grondin, B., and N. DeLuca. 2000. Herpes simplex virus type 1 ICP4 promotes transcription preinitiation complex formation by enhancing the binding of TFIID to DNA. J. Virol. 74:11504-11510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grundy, F. J., R. P. Baumann, and D. J. O'Callaghan. 1989. DNA sequence and comparative analyses of the equine herpesvirus type 1 immediate early gene. Virology 172:223-236. [DOI] [PubMed] [Google Scholar]
- 24.Gu, B., and N. DeLuca. 1994. Requirements for activation of the herpes simplex virus glycoprotein C promoter in vitro by the viral regulatory protein ICP4. J. Virol. 68:7953-7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu, B., R. Kuddus, and N. A. DeLuca. 1995. Repression of activator-mediated transcription by herpes simplex virus ICP4 via a mechanism involving interactions with the basal transcription factors TATA-binding protein and TFIIB. Mol. Cell. Biol. 15:3618-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu, B., R. Rivera-Gonzalez, C. A. Smith, and N. A. DeLuca. 1993. Herpes simplex virus infected cell polypeptide 4 preferentially represses Sp1-activated over basal transcription from its own promoter. Proc. Natl. Acad. Sci. USA 90:9528-9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Honess, R. W., and B. Roizman. 1974. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol. 14:8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hubenthal-Voss, J., R. A. Houghten, L. Pereira, and B. Roizman. 1988. Mapping of functional and antigenic domains of the α4 protein of herpes simplex virus 1. J. Virol. 62:454-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imbalzano, A. N., D. M. Coen, and N. A. DeLuca. 1991. Herpes simplex virus transactivator ICP4 operationally substitutes for the cellular transcription factor Sp1 for efficient expression of the viral thymidine kinase gene. J. Virol. 65:565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imbalzano, A. N., and N. A. DeLuca. 1992. Substitution of a TATA box from a herpes simplex virus late gene in the viral thymidine kinase promoter alters ICP4 inducibility but not temporal expression. J. Virol. 66:5453-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imbalzano, A. N., A. A. Shepard, and N. A. DeLuca. 1990. Functional relevance of specific interactions between herpes simplex virus type 1 ICP4 and sequences from the promoter-regulatory domain of the viral thymidine kinase gene. J. Virol. 64:2620-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kristie, T. M., and P. A. Sharp. 1993. Purification of the cellular C1 factor required for the stable recognition of the Oct-1 homeodomain by the herpes simplex virus alpha-trans-induction factor (VP16). J. Biol. Chem. 268:6525-6534. [PubMed] [Google Scholar]
- 33.Kuddus, R., B. Gu, and N. A. DeLuca. 1995. Relationship between TATA-binding protein and herpes simplex virus type 1 ICP4 DNA-binding sites in complex formation and repression of transcription. J. Virol. 69:5568-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leopardi, R., P. L. Ward, W. O. Ogle, and B. Roizman. 1997. Association of herpes simplex virus regulatory protein ICP22 with transcriptional complexes containing EAP, ICP4, RNA polymerase II, and viral DNA requires posttranslational modification by the UL13 protein kinase. J. Virol. 71:1133-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mackem, S., and B. Roizman. 1982. Structural features of the herpes simplex virus alpha gene 4, 0, and 27 promoter-regulatory sequences which confer alpha regulation on chimeric thymidine kinase genes. J. Virol. 44:939-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGeoch, D. J., A. Dolan, S. Donald, and D. H. Brauer. 1986. Complete DNA sequence of the short repeat region in the genome of herpes simplex virus type 1. Nucleic Acids Res. 14:1727-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKnight, J. L., T. M. Kristie, and B. Roizman. 1987. Binding of the virion protein mediating alpha gene induction in herpes simplex virus 1-infected cells to its cis site requires cellular proteins. Proc. Natl. Acad. Sci. USA 84:7061-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Metzler, D. W., and K. W. Wilcox. 1985. Isolation of herpes simplex virus regulatory protein ICP4 as a homodimeric complex. J. Virol. 55:329-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muller, M. T. 1987. Binding of the herpes simplex virus immediate-early gene product ICP4 to its own transcription start site. J. Virol. 61:858-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Hare, P., C. R. Goding, and A. Haigh. 1988. Direct combinatorial interaction between a herpes simplex virus regulatory protein and a cellular octamer-binding factor mediates specific induction of virus immediate-early gene expression. EMBO J. 7:4231-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Hare, P., and G. S. Hayward. 1985. Three trans-acting regulatory proteins of herpes simplex virus modulate immediate-early gene expression in a pathway involving positive and negative feedback regulation. J. Virol. 56:723-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paterson, T., and R. D. Everett. 1988. Mutational dissection of the HSV-1 immediate-early protein Vmw175 involved in transcriptional transactivation and repression. Virology 166:186-196. [DOI] [PubMed] [Google Scholar]
- 43.Paterson, T., and R. D. Everett. 1988. The regions of the herpes simplex virus type 1 immediate early protein Vmw175 required for site specific DNA binding closely correspond to those involved in transcriptional regulation. Nucleic Acids Res. 16:11005-11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paterson, T., V. G. Preston, and R. D. Everett. 1990. A mutant of herpes simplex virus type 1 immediate early polypeptide Vmw175 binds to the cap site of its own promoter in vitro but fails to autoregulate in vivo. J. Gen. Virol. 71:851-861. [DOI] [PubMed] [Google Scholar]
- 45.Pereira, L., M. H. Wolff, M. Fenwick, and B. Roizman. 1977. Regulation of herpesvirus macromolecular synthesis. V. Properties of alpha polypeptides made in HSV-1 and HSV-2 infected cells. Virology 77:733-749. [DOI] [PubMed] [Google Scholar]
- 46.Post, L. E., S. Mackem, and B. Roizman. 1981. Regulation of alpha genes of herpes simplex virus: expression of chimeric genes produced by fusion of thymidine kinase with alpha gene promoters. Cell 24:555-565. [DOI] [PubMed] [Google Scholar]
- 47.Preston, C. M. 1979. Control of herpes simplex virus type 1 mRNA synthesis in cells infected with wild-type virus or the temperature-sensitive mutant tsK. J. Virol. 29:275-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Preston, V. G. 1981. Fine-structure mapping of herpes simplex virus type 1 temperature-sensitive mutations within the short repeat region of the genome. J. Virol. 39:150-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Preston, V. G., A. J. Davison, H. S. Marsden, M. C. Timbury, J. H. Subak-Sharpe, and N. M. Wilkie. 1978. Recombinants between herpes simplex virus types 1 and 2: analyses of genome structures and expression of immediate early polypeptides. J. Virol. 28:499-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Purifoy, D. J., and K. L. Powell. 1976. DNA-binding proteins induced by herpes simplex virus type 2 in HEp-2 cells. J. Virol. 19:717-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quinlan, M. P., L. B. Chen, and D. M. Knipe. 1984. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36:857-868. [DOI] [PubMed] [Google Scholar]
- 52.Roberts, M. S., A. Boundy, P. O'Hare, M. C. Pizzorno, D. M. Ciufo, and G. S. Hayward. 1988. Direct correlation between a negative autoregulatory response element at the cap site of the herpes simplex virus type 1 IE175 (alpha 4) promoter and a specific binding site for the IE175 (ICP4) protein. J. Virol. 62:4307-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shepard, A. A., and N. A. DeLuca. 1991. Activities of heterodimers composed of DNA-binding- and transactivation-deficient subunits of the herpes simplex virus regulatory protein ICP4. J. Virol. 65:299-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shepard, A. A., A. N. Imbalzano, and N. A. DeLuca. 1989. Separation of primary structural components conferring autoregulation, transactivation, and DNA-binding properties to the herpes simplex virus transcriptional regulatory protein ICP4. J. Virol. 63:3714-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shepard, A. A., P. Tolentino, and N. A. DeLuca. 1990. trans-dominant inhibition of herpes simplex virus transcriptional regulatory protein ICP4 by heterodimer formation. J. Virol. 64:3916-3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Showalter, S. D., M. Zweig, and B. Hampar. 1981. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP4. Infect. Immun. 34:684-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith, C. A., P. Bates, R. Rivera-Gonzalez, B. Gu, and N. A. DeLuca. 1993. ICP4, the major transcriptional regulatory protein of herpes simplex virus type 1, forms a tripartite complex with TATA-binding protein and TFIIB. J. Virol. 67:4676-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vlcek, C., Z. Kozmik, V. Paces, S. Schirm, and M. Schwyzer. 1990. Pseudorabies virus immediate-early gene overlaps with an oppositely oriented open reading frame: characterization of their promoter and enhancer regions. Virology 179:365-377. [DOI] [PubMed] [Google Scholar]
- 59.Watson, R. J., and J. B. Clements. 1978. Characterization of transcription-deficient temperature-sensitive mutants of herpes simplex virus type 1. Virology 91:364-379. [DOI] [PubMed] [Google Scholar]
- 60.Wertman, K. F., D. G. Drubin, and D. Botstein. 1992. Systematic mutational analysis of the yeast ACT1 gene. Genetics 132:337-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wirth, U. V., C. Fraefel, B. Vogt, C. Vlcek, V. Paces, and M. Schwyzer. 1992. Immediate-early RNA 2.9 and early RNA 2.6 of bovine herpesvirus 1 are 3′ coterminal and encode a putative zinc finger transactivator protein. J. Virol. 66:2763-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu, C. L., and K. W. Wilcox. 1990. Codons 262 to 490 from the herpes simplex virus ICP4 gene are sufficient to encode a sequence-specific DNA binding protein. Nucleic Acids Res. 18:531-538. [DOI] [PMC free article] [PubMed] [Google Scholar]