Abstract
Sequences of the untranslated regions at the 5′ and 3′ ends (5′UTR and 3′UTR) of the hepatitis C virus (HCV) RNA genome are highly conserved and contain cis-acting RNA elements for HCV RNA replication. The HCV 5′UTR consists of two distinct RNA elements, a short 5′-proximal stem-loop RNA element (nucleotides 1 to 43) and a longer element of internal ribosome entry site. To determine the sequence and structural requirements of the 5′-proximal stem-loop RNA element in HCV RNA replication and translation, a mutagenesis analysis was preformed by nucleotide deletions and substitutions. Effects of mutations in the 5′-proximal stem-loop RNA element on HCV RNA replication were determined by using a cell-based HCV replicon replication system. Deletion of the first 20 nucleotides from the 5′ end resulted in elimination of cell colony formation. Likewise, disruption of the 5′-proximal stem-loop by nucleotide substitutions abolished the ability of HCV RNA to induce cell colony formation. However, restoration of the 5′-proximal stem-loop by compensatory mutations with different nucleotides rescued the ability of the subgenomic HCV RNA to replicate in Huh7 cells. In addition, deletion and nucleotide substitutions of the 5′-proximal stem-loop structure, including the restored stem-loop by compensatory mutations, all resulted in reduction of translation by two- to fivefold, suggesting that the 5′-proximal stem-loop RNA element also modulates HCV RNA translation. These findings demonstrate that the 5′-proximal stem-loop of the HCV RNA is a cis-acting RNA element that regulates HCV RNA replication and translation.
Hepatitis C virus (HCV) is an enveloped RNA virus containing a single-stranded, positive-sense RNA genome approximately 9.6 kb in length (11, 35). The viral RNA genome is composed of a single long open reading frame, flanked by untranslated regions at the 5′ and 3′ ends (5′UTR and 3′UTR) (7, 9, 11, 23, 39, 47). Sequence analysis and comparison studies revealed that both the 5′ and 3′ UTRs are the most-conserved regions of the viral RNA genome among different HCV genotypes and isolates (9, 23, 39, 47). The conservation of the 5′ and 3′ UTR sequences suggests that they contain cis-acting RNA elements required for HCV RNA expression, replication, and regulation. A number of studies have demonstrated that the 5′UTR harbors an internal ribosome entry site (IRES) that mediates initiation of translation of the viral polyprotein (6, 7, 36, 40, 42-44). Biochemical and functional studies have revealed that the HCV 5′UTR folds into a highly ordered complex structure with multiple stem-loops (27, 36). These highly folded secondary and tertiary RNA elements function as cis-signals for interaction with the 40S ribosome subunit and/or eukaryotic translation initiation factors (24, 32, 38). Recent studies have revealed that two RNA stem-loops, domains IIId and IIIe of the IRES, directly interact with the 40S ribosome subunit and eukaryotic translation initiation factor 3 binds the upstream stem-loop III of the IRES (27, 38). Genetic studies using chimeric poliovirus (PV) and bovine viral diarrhea virus (BVDV) containing the HCV 5′UTR confirmed that the HCV IRES could functionally replace those of PV and BVDV (15, 26, 49, 50). A small Saccharomyces cerevisiae RNA inhibiting PV IRES-dependent translation was also shown to inhibit HCV IRES function (12, 13). Clearly, the HCV IRES is a cis-acting RNA element that directs HCV RNA genome expression.
The HCV 5′UTR also plays an important role in viral RNA replication (14). The first 125 nucleotides of the 5′UTR was recently found to be the minimal sequence required for HCV RNA replication (14). However, the sequence and structural requirements of cis-acting RNA elements of the 5′UTR for HCV RNA replication have not been determined. The highly conserved 5′UTR harbors two distinct RNA elements, a short 5′-proximal RNA element (nucleotides 1 to 43) and a longer IRES element consisting of nucleotides 44 to 341 in the 5′UTR (36). The 5′-proximal RNA element can form a stem-loop structure with nucleotides 5 to 20 (Fig. 1). Whether the 5′-proximal stem-loop structure is essential for HCV RNA replication remains to be determined. Recent success in HCV reverse genetics provides us an excellent opportunity to perform genetic analysis of the 5′-proximal stem-loop RNA element in HCV RNA replication (5, 25).
FIG. 1.
Comparison of the 5′-proximal RNA element sequences derived from different HCV isolates. The 5′-terminal 43 nucleotides of the 5′UTR derived from known sequences of HCV variants in GenBank were compared and aligned by a BLAST program. One representative from each group with the same nucleotide sequence at the 5′ terminus is shown. The accession number of each sequence is shown on the left. Nucleotides identical to those in the I377/NS3-3′ replicon (GenBank accession numbers AJ242652 and AJ242654) (25) are indicated by dots. The divergent nucleotides are indicated or underlined.
To reveal the conservation of the 5′-proximal RNA element, we performed a BLAST search for known HCV 5′UTR sequences of different HCV isolates in GenBank. Among 172 BLAST hits, the short RNA element sequences are highly conserved, with minimal sequence divergence mainly in the loop of the 5′-proximal stem-loop structure and downstream of the stem-loop (Fig. 1). The conservation of the 5′-proximal RNA element suggests an important role in HCV RNA replication. The conserved RNA sequences and/or structures located at the ends of viral RNA genomes normally contain cis-acting RNA elements required for viral RNA replication (3, 8, 18, 30, 33).
To determine the sequence and/or structural requirements of the 5′-proximal stem-loop RNA element for HCV RNA replication, we introduced a number of mutations into the 5′-proximal stem-loop RNA element by nucleotide deletions and substitutions (Fig. 2A). Two deletion mutants were constructed by removal of the first 4 and 20 nucleotides from the 5′ end, resulting in replicon RNAs designated 5′UTR/d4 and 5′UTR/d20, respectively (Fig. 2A, mutants II and III). To examine the structural requirements of the 5′-proximal stem-loop, the stem was disrupted by either substitution of a stretch of five cytidines (C) at nucleotides 6 to 10 with five uridines (U) (5′UTR/SLUG) (Fig. 2A, mutant IV) or mutation of guanosines (G) at positions 15 to 19 to adenosine (A) (5′UTR/SLCA, Fig. 2A, mutant V). To restore the 5′-proximal stem-loop structure with different sequence, the base-paired nucleotides of the stem were switched by compensatory mutations so that the stem-loop structure was still retained but with nucleotides in opposite order (5′UTR/SLswitch, Fig. 2A, mutant VI). To avoid potential base pairing of the G at position 11 with the switched nucleotide C, the sequence in the loop was also mutated from GAUU to UGAU (Fig. 2A, mutant VI). In addition, the base-paired C-G nucleotides of the stem were mutated to U-A base pairs, resulting in a mutant replicon designated 5′UTR/SLUA (Fig. 2A, mutant VII). To examine the effects of mutations in the loop region introduced to the replicon 5′UTR/SLswitch on HCV RNA replication, we made a separate replicon with mutations from GAUU to UGAU in the loop (5′UTR/M-loop) (Fig. 2A, mutant VIII). These mutations will reveal whether the sequence and/or structure of the 5′-proximal stem-loop is important for HCV RNA replication.
FIG. 2.
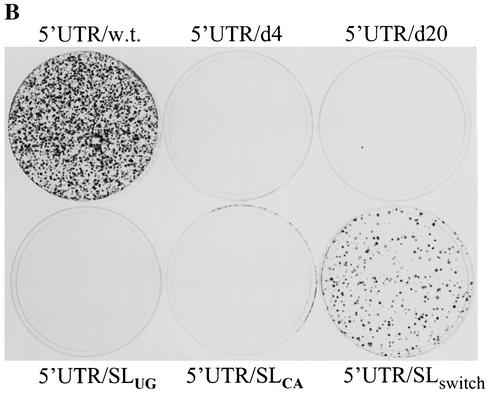
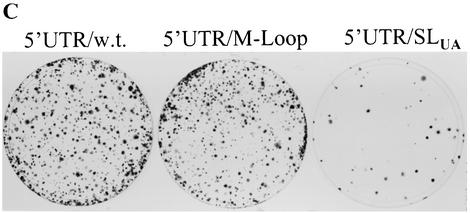
Mutagenesis analysis of the 5′-proximal stem-loop RNA element. (A) Sequences and structures of wild-type and mutant 5′-proximal stem-loop RNA element. Mutations of the 5′-proximal stem-loop RNA element were introduced by PCR using synthetic oligonucleotides (IDT, Coralville, Iowa). The PCR DNA fragment was cloned between ClaI and AscI sites of an HCV replicon cDNA vector pBR322/I377-NS3-3′/S117I (unpublished data). The wild-type 5′-proximal RNA element is shown in I. Deletion of the first 4 and 20 nucleotides resulted in two mutants designated 5′UTR/d4 (II) and 5′UTR/d20 (III), respectively. The stem of the stem-loop structure was disrupted by nucleotide substitution of a stretch of either cytidines at positions 6 to 10 with uridines (IV) or guanosines at positions 15 to 19 with adenosines (V). The stem-loop structure was restored by either switching the side of the nucleotides formed in the stem (VI) or substitution with A/U base pairs (VII). The loop sequence was mutated from GAUU to UGAU (VIII), which was found in HCV variants (Fig. 1). Nucleotide substitutions in the stem are highlighted by open boxes. The nucleotide position is numbered on the top of the sequence as the first 5′ end nucleotide is counted as 1. Underlined nucleotides indicate divergent sequences found in different HCV isolates. (B) Effects of mutations in the 5′-proximal stem-loop RNA element on cell colony formation. The in vitro-transcribed RNAs (2 μg each) were transfected into 8 × 106 Huh7 cells by electroporation. At 24 h posttransfection, cell culture medium was replaced with Dulbecco's modified Eagle medium containing 10% fetal bovine serum (FBS) and G418 sulfate (500 μg/ml). The medium was changed every 3 to 4 days. After a 4-week selection with G418 sulfate, cells were fixed and stained with a solution containing 0.01% crystal violet and 19% methanol. (C) Effects of the A/U base paired stem and mutations in the loop on cell colony formation. RNA transfection and selection of Huh7 cells resistant to G418 were the same as in panel B.
To determine the effects of mutations introduced into the 5′UTR on HCV RNA replication, we constructed a subgenomic HCV replicon based on the published sequence (GenBank accession number AJ242652 and AJ242654) (25). The subgenomic HCV RNA contains the 5′UTR, the neomycin phosphotransferase gene (neo) fused to sequences encoding the first 12 amino acids of the core protein, the IRES from encephalomyocarditis virus, nonstructural genes NS3 to NS5B of HCV genotype 1b, and the 3′UTR (5, 25). Since a highly adapted mutation from serine to isoleucine in NS5A at position 1179 (S1179I) was reported to dramatically enhance the level of HCV RNA replication (5), we introduced this mutation into the original replicon I377/NS3-3′, resulting in a replicon designated I377/NS3-3′/S1179I. This HCV replicon was used for introduction of the above-described mutations in the 5′-proximal stem-loop RNA element. Subgenomic HCV replicon RNAs were transcribed by a T7 RNA polymerase and transfected into Huh7 cells by electroporation. The replicon RNA-transfected Huh7 cells were then selected for resistance to antibiotics G418 sulfate. Results are shown in Fig. 2B and C. Deletion of the first 4 or first 20 nucleotides from the 5′ end failed to induce cell colony formation (Fig. 2B). Likewise, disruption of the stem of the 5′-proximal stem-loop by nucleotide substitutions resulted in elimination of cell colony formation (Fig. 2B, 5′UTR/SLUG and 5′UTR/SLCA). Restoration of the 5′-proximal stem-loop structure by compensatory mutations with different nucleotides rescued the ability of the RNA to replicate in Huh7 cells, as revealed by cell colony formation (Fig. 2B, 5′UTR/SLswitch). However, the efficiency of cell colony formation resulting from the replicon 5′UTR/SLswitch replication was reduced by approximately 10-fold, suggesting that the sequence of the stem also plays a role in efficient HCV RNA replication. Consistent with this finding, substitution of the C/G base-paired stem with A/U base pairs dramatically decreased the efficiency of cell colony formation (Fig. 2C, 5′UTR/SLUA). The findings indicate that the 5′-proximal stem-loop structure is essential for HCV RNA replication. When the loop sequence was mutated from GAUU to UGAU, however, the efficiency of cell colony formation was not affected (Fig. 2C, 5′UTR/M-loop). This finding further confirms that the loop of the 5′-proximal stem-loop can tolerate nucleotide mutations, which were found in natural HCV isolates (Fig. 1).
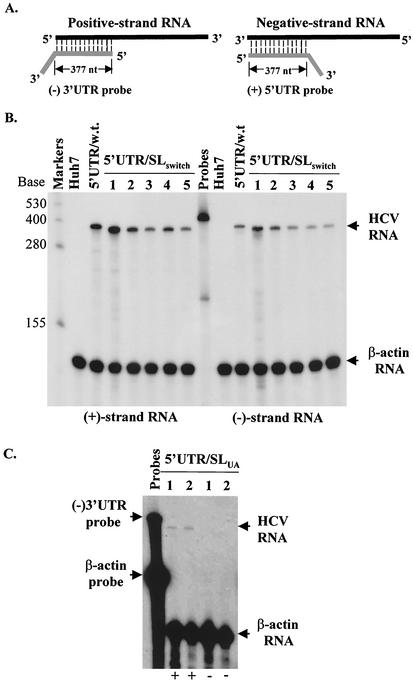
To further determine whether formation of cell colonies was the result of HCV RNA replication, we measured both positive- and negative-strand RNAs of the HCV replicons isolated from the replicon-bearing Huh7 cells using an RNase protection assay (RPA). As shown in Fig. 3B, both positive- and negative-strand RNAs of the replicon 5′UTR/SLswitch were detected at levels similar to those present in a wild-type replicon-harboring cell line (Fig. 3B). The RNA products of the replicon 5′UTR/SLswitch migrated slightly faster than those of the wild-type replicon RNA as the first 20 nucleotides of the mutant replicon 5′UTR/SLswitch RNA would not match the sequence of the wild-type RNA probes. However, the levels of positive-strand RNA of the replicon 5′UTR/SLUA were dramatically reduced (Fig. 3C). The negative-strand RNA of the replicon 5′UTR/SLUA present in Huh7 cells resistant to G418 was not detectable by RPA but was detected by strand-specific reverse transcription-PCR, suggesting that a more stable C/G base-paired stem-loop RNA element is required for efficient RNA replication (data not shown). Sequence analysis of the replicon RNAs isolated from G418-resistant Huh7 cells confirmed mutations introduced to the 5′-proximal stem-loop RNA element (data not shown). Taken together, these findings demonstrate that the 5′-proximal stem-loop structure plays an essential role in HCV RNA replication.
FIG. 3.
Determination of the positive- and negative-strand RNAs of subgenomic HCV replicons isolated from Huh7 cell lines resistant to G418 sulfate. (A) Schematic diagram of RPA. An RNA probe contains 377 nucleotides complementary to the positive-strand 5′UTR (for detection of positive-strand RNA) or the negative-strand 3′UTR (for detection of negative-strand RNA) and unpaired nucleotides from the vector (3′-end tail). After hybridization and digestion with RNase A/T1, the region (377 nucleotides) complementary to the (+)5′UTR or (−)3′UTR is protected from RNase digestion. (B) Quantitation of positive- and negative-strand RNAs of the replicon 5′UTR/SLswitch by RPA. Total cellular RNA was extracted from the replicon-containing Huh7 cells with Trizol reagent (Invitrogen). Fifteen micrograms of total RNA was used for hybridization with 5 × 104 cpm of [α-32P]UTP-labeled β-actin RNA probe (Ambion) and either 105 cpm of [α-32P]UTP-labeled (−)3′UTR (for detection of positive-strand RNA) or (+)5′UTR (for detection of negative-strand RNA) RNA probe. After digestion with RNase A/T1, RNA products were analyzed in a 6% polyacrylamide-7.7 M urea gel and visualized by autoradiography. The RNA level was quantitated with a PhosphorImager (Molecular Dynamics). The sizes of the RNA markers are indicated on the left, and arrows on the right highlight the RNA products. β-Actin RNA was used as an internal control to normalize the amount of total RNA used in RPA. Numbers on the top indicate different cell colonies (lines) resulting from replication of the HCV replicon 5′UTR/SLswitch. Negative control (Huh7), total RNA extracted from Huh7 cells; 5′UTR/w.t., total RNA from a wild-type replicon-containing cell line as a positive control. (C) Quantitation of positive- and negative-strand RNAs of the replicon 5′UTR/SLUA by RPA. Total RNA was from the replicon 5′UTR/SLUA-bearing cell lines; otherwise, the procedure was the same as that for B. Positive- and negative-strand RNAs are indicated by + and −, respectively, at the bottom.
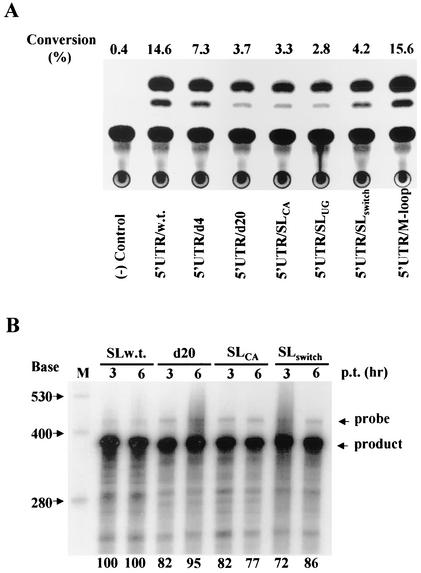
It was previously reported by others that the 5′-proximal RNA element formed by nucleotides 1 to 43 (Fig. 1) is dispensable for IRES function (20, 21, 34, 50). To determine the effects of mutations in the 5′-proximal stem-loop RNA element on translation, we replaced the neo gene with the chloramphenicol acetyltransferase (CAT) gene in the HCV replicon. Wild-type and mutant 5′UTR-CAT RNAs were transfected into Huh7 cells by lipofection. The levels of CAT expression were determined by a CAT assay (Fig. 4A). Deletions and nucleotide substitutions of the stem-loop structure resulted in reduction of CAT expression by two- to five-fold except that mutations of the loop sequences had no effect on RNA translation (Fig. 4A). Restoration of the stem-loop structure by compensatory mutations did not rescue translation of the impaired RNA (Fig. 4A, 5′UTR/SLswitch). To determine whether the transfected RNAs reached the same levels in the cell, RPA was used to quantitate the levels of 5′UTR-CAT RNAs extracted from Huh7 cells at 3 and 6 h posttransfection. As shown in Fig. 4B, there is no significant difference in the levels of wild-type and mutant 5′UTR-CAT RNAs at 3 and 6 h posttransfection. As the levels of CAT expression were also determined at 6 h posttransfection, reduction of CAT expression was indeed due to impairment of RNA translation by mutations introduced into the 5′-proximal stem-loop RNA element. These findings demonstrate that sequence of the 5′-proximal stem-loop RNA element is also important for optimal RNA translation. This conclusion was further supported by similar findings derived from a minigenome-like CAT RNA, which consists of the 5′UTR, a reporter gene CAT, and 3′UTR. In the presence of the 3′UTR, deletion of the first 20 nucleotides from the 5′end (5′UTR/d20), disruption of the stem-loop structure (5′UTR/SLCA and 5′UTR/SLUG), or restoration of the stem by compensatory mutations (5′UTR/ SLswitch) all resulted in reduction of CAT expression by 50%, as determined by immunoprecipitation (data not shown). These results confirmed that the sequence of the 5′-proximal stem-loop RNA element also modulates HCV RNA translation.
FIG. 4.
(A) Effects of mutations in the 5′-proximal stem-loop RNA element on translation. To facilitate the determination of the effects of mutations on translation, a selectable marker, the neo gene, in the HCV replicon RNA was replaced with a reporter gene, that encoding CAT. The 5′UTR-CAT RNAs were transcribed by a T7 RNA polymerase from PmeI-linearized HCV replicon cDNAs. The in vitro-transcribed and purified RNAs (2 μg each) were transfected into 35-mm-diameter dishes of Huh7 cells using TransMessenger reagent (Qiagen). At 6 h posttransfection, cells were harvested and lysed. The levels of CAT expression were determined by CAT assay, as described previously (28, 29). Conversion (percent) of the acetylated [14C]chloramphenicol is shown at top. The names of different RNAs are indicated at the bottom. (B) Quantitation of the RNA levels by RPA. RNA transfection was carried out as described for panel A. At 3 and 6 h posttransfection, total RNA was extracted from RNA-transfected cells by Trizol reagent. The levels of transfected RNAs in the cell were determined by RPA, as described in the legend to Fig. 3. Five micrograms of total RNA was used in RPA. The sizes of RNA markers are shown on the left, and arrows on the right highlight the RNA probe and products. The names of different RNAs are indicated on the top. The RNA levels were determined by quantitation with a PhosphorImager (Molecular Dynamics). The percentage of RNA levels relative to wild-type RNA (100%) is labeled at the bottom.
The question arose whether mutations in the 5′-proximal stem-loop RNA element affect the RNA stability. It was reported that some mutations introduced to the 5′UTRs of PV and BVDV RNAs destabilize the viral RNA and thereby affect its translatability and replication in the cell (3, 48). To address this concern, we determined the levels of wild-type and mutant HCV replicon RNAs by RPA at different time points after RNA transfection. Full-length HCV replicon RNAs were transfected into Huh7 cells, and total RNA was extracted at 3, 24, and 48 h, respectively, after RNA transfection. It was found that deletion of the first 20 nucleotides (5′UTR/d20), disruption of the stem-loop structure by nucleotide substitutions (5′UTR/SLCA), as well as the restored stem-loop mutant (5′UTR/SLswitch) did not significantly affect the levels of HCV RNAs compared to the level of a wild-type replicon (data not shown). Therefore, mutations of the 5′-proximal RNA element did not appear to alter the RNA stability.
Several lines of evidence derived from this study demonstrate that the 5′-proximal stem-loop structure is essential for HCV RNA replication and its sequence also modulates optimal HCV RNA translation. Cell colony formation resulting from HCV RNA replication was completely eliminated by either removal of the entire stem-loop (5′UTR/d20) or disruption of the stem-loop structure (5′UTR/SLCA and 5′UTR/SLUG) (Fig. 2). Restoration of the stem-loop structure by compensatory mutations with different nucleotides, however, was able to rescue HCV RNA replication (Fig. 2). The loop of the 5′-proximal RNA element can tolerate nucleotide variation, since mutations from GAUU to UGAU had no effect on the efficiency of cell colony formation (Fig. 2). In addition, the sequence of the 5′-proximal stem-loop structure also plays an important role in HCV RNA replication. The replicon RNA with an A/U base paired stem of the 5′-proximal stem-loop was replicated at an extremely low level (Fig. 3C). Moreover, the 5′-proximal stem-loop RNA element also modulates HCV RNA translation. Deletion and nucleotide substitutions of the 5′-proximal stem-loop RNA all decreased the HCV IRES-mediated translation by two- to fivefold (Fig. 4). These results are consistent with the findings reported by others that the 5′-proximal stem-loop RNA element was required for optimal HCV RNA translation (14, 16).
With many positive-strand RNA viruses, the 5′UTR has multiple important roles in the regulation of viral RNA translation and/or replication (3, 4, 10, 18, 19, 22, 45, 48). In the case of PV, a cloverleaf structure at the 5′ end of the viral RNA genome controls both viral RNA translation and replication through differential interactions with a cellular protein, poly(C) binding protein, and the viral protein 3CD (3, 17, 18, 31, 37, 41). A recent study also has shown that the cloverleaf structure is required for initiation of negative-strand RNA synthesis of PV RNA (3). For BVDV, a stem-loop structure at the immediate 5′ terminus of the BVDV genome is critical for not only viral RNA replication but also RNA translation (4, 48). The question of how the 5′-proximal stem-loop RNA element of HCV genome controls RNA replication and translation remains to be determined. Little is known about the molecular mechanisms of HCV RNA replication. In general, it is believed that HCV probably follows a replication strategy similar to that of other positive-stranded RNA viruses. The genomic RNA initially serves as an mRNA for translation of viral polyprotein. Following translation, RNA replication is initiated by synthesis of complementary negative-strand RNA that in turn acts as a template for synthesis of nascent positive-strand RNA genome (1, 33, 46). Therefore, the HCV 5′UTR, upon replication, becomes the complementary 3′UTR of the negative-strand RNA, which initiates positive-strand RNA replication. Thus, it is most likely that mutations of the 5′-proximal stem-loop RNA impaired the positive-strand RNA replication. It is also possible that the 5′-proximal stem-loop element might play a role in the negative-strand RNA synthesis like the cloverleaf RNA structure of PV (3). In this scenario, the 5′-proximal stem-loop element would play important roles in both positive- and negative-strand RNA replication. Another possible role of the 5′-proximal stem-loop RNA element might be to regulate HCV RNA translation and replication. Since HCV RNA acts as a template for both protein synthesis and negative-strand RNA replication, the question arises how these two different processes are regulated. In PV, the actively translating ribosomes were found to inhibit viral RNA replication (2, 18). A cloverleaf RNA structure in the 5′UTR, however, controls the switch of PV RNA translation to replication (18). As revealed by results derived from this study, the sequence of the 5′-proximal stem-loop RNA element is also required for optimal translation. Whether and how the 5′-proximal stem-loop RNA element regulates both HCV RNA translation and replication through interactions with viral and cellular proteins remain to be determined.
Acknowledgments
We thank Ralf Bartenschlager (University of Heidelberg, Heidelberg, Germany) for kindly providing the Huh7 cell line used in this study. We thank Amiya Banerjee (Cleveland Clinic), who suggested that we replace the C/G stem of the 5′-proximal stem-loop with an A/U stem. We also thank Bob Geraghty for critical reading of the manuscript.
This work was partially supported by NIH grant AI51592.
REFERENCES
- 1.Bartholomeusz, A., and P. Thompson. 1999. Flaviviridae polymerase and RNA replication. J. Viral Hepat. 6:261-270. [DOI] [PubMed] [Google Scholar]
- 2.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1999. Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J. Virol. 73:10104-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ Cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becher, P., M. Orlich, and H. J. Thiel. 2000. Mutations in the 5′ nontranslated region of bovine viral diarrhea virus result in altered growth characteristics. J. Virol. 74:7884-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1975. [DOI] [PubMed] [Google Scholar]
- 6.Brown, E. A., A. J. Zajac, and S. M. Lemon. 1994. In vitro characterization of an internal ribosomal entry site (IRES) present within the 5′ nontranslated region of hepatitis A virus RNA: comparison with the IRES of encephalomyocarditis virus. J. Virol. 68:1066-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, E. A., H. Zhang, L. H. Ping, and S. M. Lemon. 1992. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 20:5041-5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bukh, J., R. H. Purcell, and R. H. Miller. 1992. Sequence analysis of the 5′ noncoding region of hepatitis C virus. Proc. Natl. Acad. Sci. USA 89:4942-4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, J., A. Noueiry, and P. Ahlquist. 2001. Brome mosaic virus protein 1a recruits viral RNA2 to RNA replication through a 5′ proximal RNA2 signal. J. Virol. 75:3207-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 12.Das, S., M. Ott, A. Yamane, W. Tsai, M. Gromeier, F. Lahser, S. Gupta, and A. Dasgupta. 1998. A small yeast RNA blocks hepatitis C virus internal ribosome entry site (HCV IRES)-mediated translation and inhibits replication of a chimeric poliovirus under translational control of the HCV IRES element. J. Virol. 72:5638-5647. (Erratum, 72:9419.) [DOI] [PMC free article] [PubMed]
- 13.Das, S., M. Ott, A. Yamane, A. Venkatesan, S. Gupta, and A. Dasgupta. 1998. Inhibition of internal entry site (IRES)-mediated translation by a small yeast RNA: a novel strategy to block hepatitis C virus protein synthesis. Front. Biosci. 3:D1241-D1252. [DOI] [PubMed]
- 14.Friebe, P., V. Lohmann, N. Krieger, and R. Bartenschlager. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frolov, I., M. S. McBride, and C. M. Rice. 1998. cis-acting RNA elements required for replication of bovine viral diarrhea virus-hepatitis C virus 5′ nontranslated region chimeras. RNA 4:1418-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukushi, S., K. Katayama, C. Kurihara, N. Ishiyama, F. B. Hoshino, T. Ando, and A. Oya. 1994. Complete 5′ noncoding region is necessary for the efficient internal initiation of hepatitis C virus RNA. Biochem. Biophys. Res. Commun. 199:425-432. [DOI] [PubMed] [Google Scholar]
- 17.Gamarnik, A. V., and R. Andino. 2000. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 74:2219-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guan, H., C. D. Carpenter, and A. E. Simon. 2000. Requirement of a 5′-proximal linear sequence on minus strands for plus-strand synthesis of a satellite RNA associated with turnip crinkle virus. Virology 268:355-363. [DOI] [PubMed] [Google Scholar]
- 20.Honda, M., L. H. Ping, R. C. Rijnbrand, E. Amphlett, B. Clarke, D. Rowlands, and S. M. Lemon. 1996. Structural requirements for initiation of translation by internal ribosome entry within genome-length hepatitis C virus RNA. Virology 222:31-42. [DOI] [PubMed] [Google Scholar]
- 21.Kamoshita, N., K. Tsukiyama-Kohara, M. Kohara, and A. Nomoto. 1997. Genetic analysis of internal ribosomal entry site on hepatitis C virus RNA: implication for involvement of the highly ordered structure and cell type-specific transacting factors. Virology 233:9-18. [DOI] [PubMed] [Google Scholar]
- 22.Kim, K. H., and C. Hemenway. 1996. The 5′ nontranslated region of potato virus X RNA affects both genomic and subgenomic RNA synthesis. J. Virol. 70:5533-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolykhalov, A. A., S. M. Feinstone, and C. M. Rice. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 70:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kruger, M., C. Beger, Q. X. Li, P. J. Welch, R. Tritz, M. Leavitt, J. R. Barber, and F. Wong-Staal. 2000. Identification of eIF2Bγ and eIF2gamma as cofactors of hepatitis C virus internal ribosome entry site-mediated translation using a functional genomics approach. Proc. Natl. Acad. Sci. USA 97:8566-8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 26.Lu, H. H., and E. Wimmer. 1996. Poliovirus chimeras replicating under the translational control of genetic elements of hepatitis C virus reveal unusual properties of the internal ribosomal entry site of hepatitis C virus. Proc. Natl. Acad. Sci. USA 93:1412-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lukavsky, P. J., G. A. Otto, A. M. Lancaster, P. Sarnow, and J. D. Puglisi. 2000. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat. Struct. Biol. 7:1105-1110. [DOI] [PubMed] [Google Scholar]
- 28.Luo, G. X., W. Luytjes, M. Enami, and P. Palese. 1991. The polyadenylation signal of influenza virus RNA involves a stretch of uridines followed by the RNA duplex of the panhandle structure. J. Virol. 65:2861-2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luytjes, W., M. Krystal, M. Enami, J. D. Pavin, and P. Palese. 1989. Amplification, expression, and packaging of foreign gene by influenza virus. Cell 59:1107-1113. [DOI] [PubMed] [Google Scholar]
- 30.O'Neill, R. E., and P. Palese. 1994. Cis-acting signals and trans-acting factors involved in influenza virus RNA synthesis. Infect. Agents Dis. 3:77-84. [PubMed] [Google Scholar]
- 31.Parsley, T. B., J. S. Towner, L. B. Blyn, E. Ehrenfeld, and B. L. Semler. 1997. Poly(rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3:1124-1134. [PMC free article] [PubMed] [Google Scholar]
- 32.Pestova, T. V., I. N. Shatsky, S. P. Fletcher, R. J. Jackson, and C. U. Hellen. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12:67-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pogue, G. P., C. C. Huntley, and T. C. Hall. 1994. Common replication strategies emerging from the study of diverse groups of positive-strand RNA viruses. Arch. Virol. Suppl. 9:181-194. [DOI] [PubMed] [Google Scholar]
- 34.Reynolds, J. E., A. Kaminski, A. R. Carroll, B. E. Clarke, D. J. Rowlands, and R. J. Jackson. 1996. Internal initiation of translation of hepatitis C virus RNA: the ribosome entry site is at the authentic initiation codon. RNA 2:867-878. [PMC free article] [PubMed] [Google Scholar]
- 35.Rice, C. M. 1996. Flaviviridae: the viruses and their replication. Lippincott-Raven, Philadelphia, Pa.
- 36.Rijnbrand, R. C., and S. M. Lemon. 2000. Internal ribosome entry site-mediated translation in hepatitis C virus replication. Curr. Top. Microbiol. Immunol. 242:85-116. [DOI] [PubMed] [Google Scholar]
- 37.Simoes, E. A., and P. Sarnow. 1991. An RNA hairpin at the extreme 5′ end of the poliovirus RNA genome modulates viral translation in human cells. J. Virol. 65:913-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sizova, D. V., V. G. Kolupaeva, T. V. Pestova, I. N. Shatsky, and C. U. Hellen. 1998. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J. Virol. 72:4775-4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanaka, T., N. Kato, M. J. Cho, K. Sugiyama, and K. Shimotohno. 1996. Structure of the 3′ terminus of the hepatitis C virus genome. J. Virol. 70:3307-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsukiyama-Kohara, K., N. Iizuka, M. Kohara, and A. Nomoto. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walter, B. L., J. H. Nguyen, E. Ehrenfeld, and B. L. Semler. 1999. Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA 5:1570-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, C., S. Y. Le, N. Ali, and A. Siddiqui. 1995. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA 1:526-537. [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, C., P. Sarnow, and A. Siddiqui. 1994. A conserved helical element is essential for internal initiation of translation of hepatitis C virus RNA. J. Virol. 68:7301-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, C., P. Sarnow, and A. Siddiqui. 1993. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J. Virol. 67:3338-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu, B., W. B. Vanti, and K. A. White. 2001. An RNA domain within the 5′ untranslated region of the tomato bushy stunt virus genome modulates viral RNA replication. J. Mol. Biol. 305:741-756. [DOI] [PubMed] [Google Scholar]
- 46.Xiang, W., A. V. Paul, and E. Wimmer. 1997. RNA signals in entero- and rhinovirus genome replication. Semin. Virol. 8:256-273. [Google Scholar]
- 47.Yamada, N., K. Tanihara, A. Takada, T. Yorihuzi, M. Tsutsumi, H. Shimomura, T. Tsuji, and T. Date. 1996. Genetic organization and diversity of the 3′ noncoding region of the hepatitis C virus genome. Virology 223:255-261. [DOI] [PubMed] [Google Scholar]
- 48.Yu, H., O. Isken, C. W. Grassmann, and S. E. Behrens. 2000. A stem-loop motif formed by the immediate 5′ terminus of the bovine viral diarrhea virus genome modulates translation as well as replication of the viral RNA. J. Virol. 74:5825-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao, W. D., F. C. Lahser, and E. Wimmer. 2000. Genetic analysis of a poliovirus/hepatitis C virus (HCV) chimera: interaction between the poliovirus cloverleaf and a sequence in the HCV 5′ nontranslated region results in a replication phenotype. J. Virol. 74:6223-6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao, W. D., and E. Wimmer. 2001. Genetic analysis of a poliovirus/hepatitis C virus chimera: new structure for domain II of the internal ribosomal entry site of hepatitis C virus. J. Virol. 75:3719-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]